Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Proteins Preparation
2.2. Contact Analysis
2.3. Protein–Protein Docking
2.4. Align Complexes
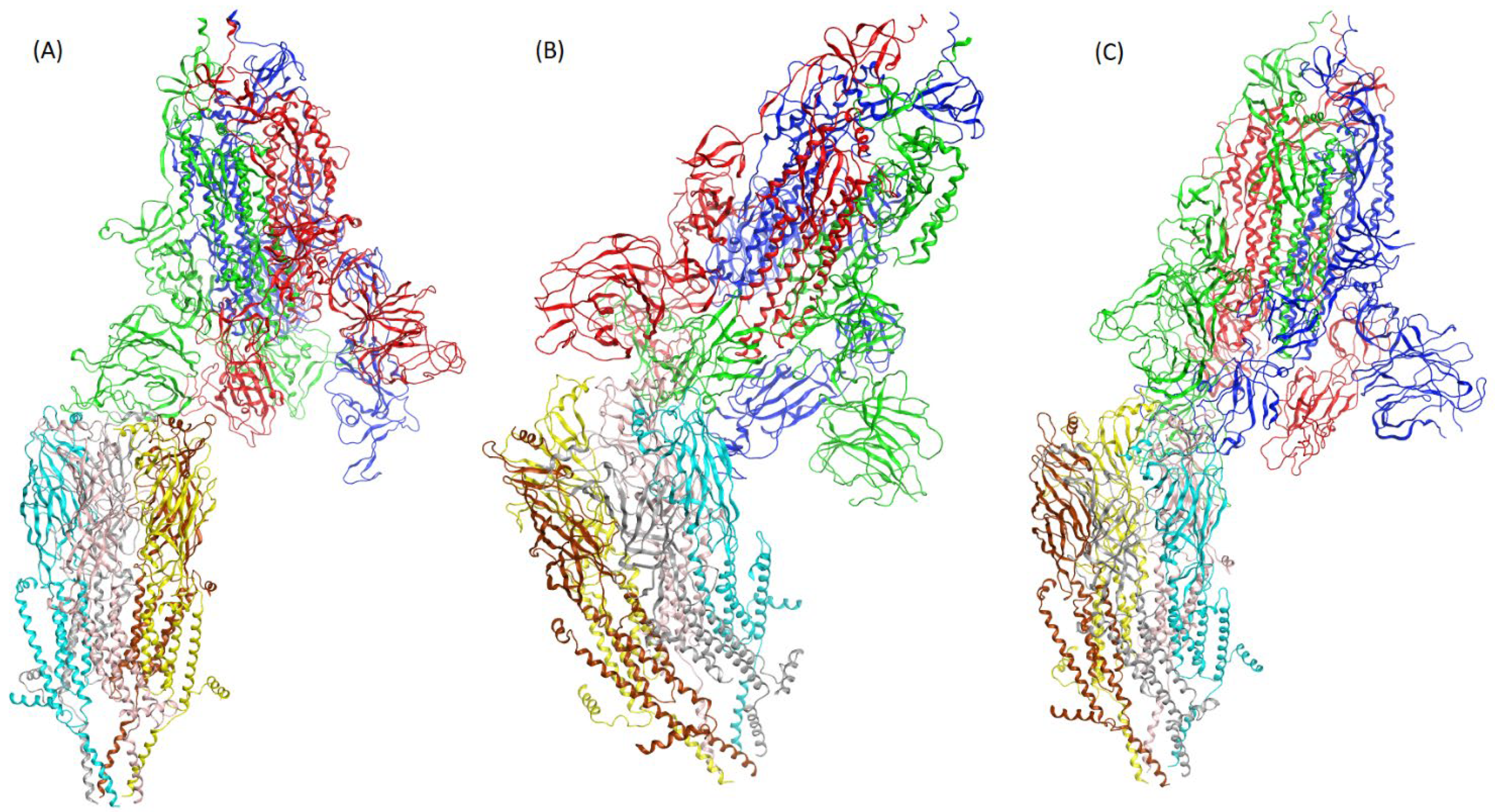
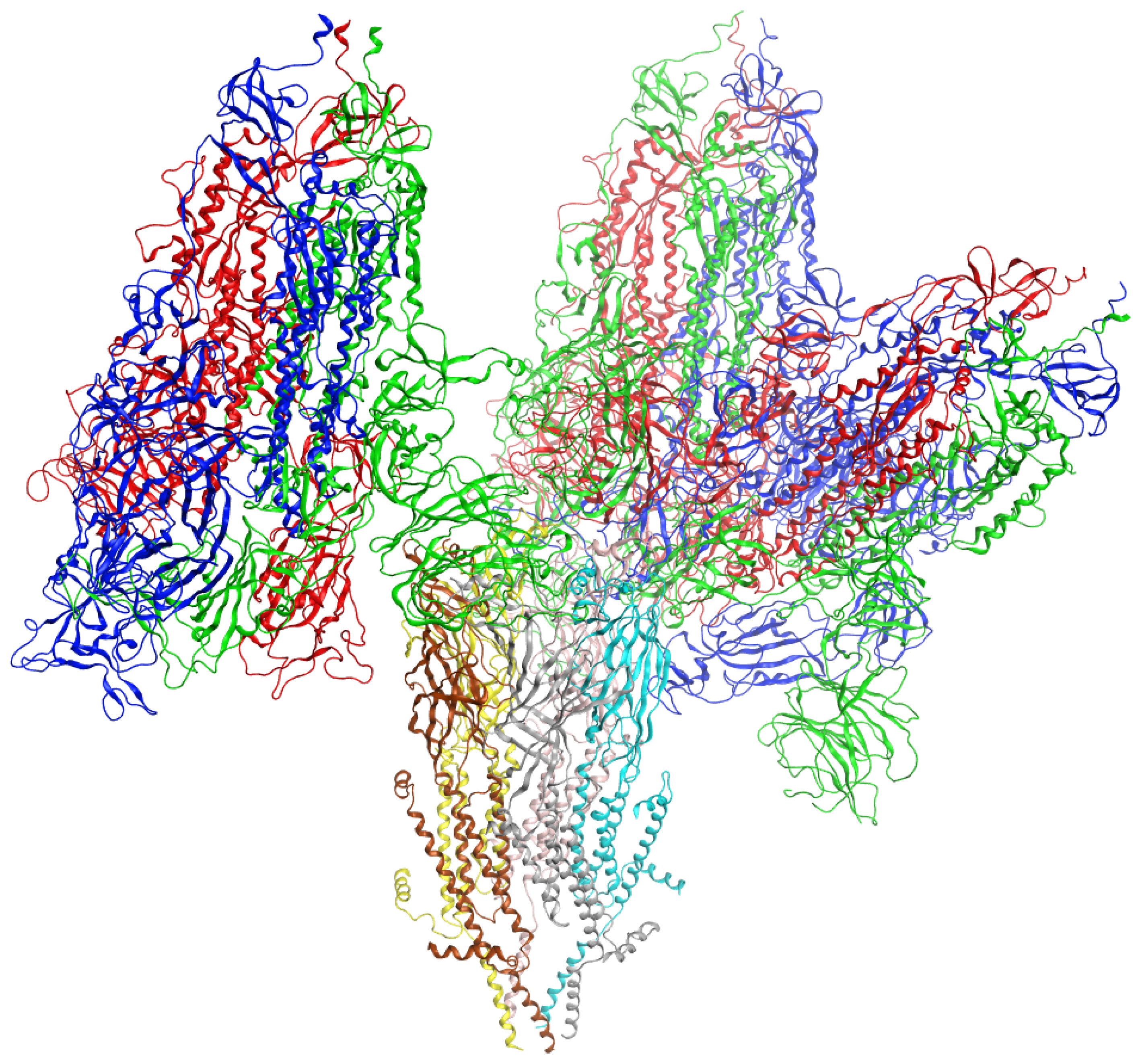
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 7nAChr | Alpha-7 nicotinic acetylcholine receptor |
| COVID-19 | Coronavirus disease 2019 |
| IL-1 | Interleukin 1 |
| IL-6 | Interleukin 6 |
| MOE | Molecular Operating Environment |
| NTD | N-terminal domain |
| PDB | Protein Data Bank |
| RBD | Receptor binding domain |
| RMSD | Root mean square deviation |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SPR | Surface plasmon resonance |
| TNF | Tumor necrosis factor |
References
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Maslo, C.; Toubkin, M. Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave-Reply. JAMA 2022, 327, 2148. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Kwatra, G.; Myers, J.E.; Jassat, W.; Dhar, N.; Mukendi, C.K.; Nana, A.J.; Blumberg, L.; Welch, R.; Ngorima-Mabhena, N.; et al. Population Immunity and Covid-19 Severity with Omicron Variant in South Africa. N. Engl. J. Med. 2022, 386, 1314–1326. [Google Scholar] [CrossRef]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Bentley, E.G.; Kirby, A.; Sharma, P.; Kipar, A.; Mega, D.F.; Bramwell, C.; Penrice-Randal, R.; Prince, T.; Brown, J.C.; Zhou, J.; et al. SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.; Hu, B.; Chai, Y.; Yuen, T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Zhang, X.; Lemmens, V.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Weynand, B.; Dallmeier, K.; et al. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. Antivir. Res. 2022, 198, 105253. [Google Scholar] [CrossRef] [PubMed]
- Aminpour, M.; Cannariato, M.; Safaeeardebili, M.E.; Preto, J.; Moracchiato, A.; Doria, D.; Donato, F.; Zizzi, E.A.; Deriu, M.A.; Scheim, D.E.; et al. In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds. Computation 2022, 10, 51. [Google Scholar] [CrossRef]
- Changeux, J.P.; Amoura, Z.; Rey, F.A.; Miyara, M. A nicotinic hypothesis for Covid-19 with preventive and therapeutic implications. Comptes Rendus Biol. 2020, 343, 33–39. [Google Scholar]
- Lagoumintzis, G.; Chasapis, C.T.; Alexandris, N.; Kouretas, D.; Tzartos, S.; Eliopoulos, E.; Farsalinos, K.; Poulas, K. Nicotinic cholinergic system and COVID-19: In silico identification of interactions between alpha7 nicotinic acetylcholine receptor and the cryptic epitopes of SARS-Co-V and SARS-CoV-2 Spike glycoproteins. Food Chem. Toxicol. 2021, 149, 112009. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, M.; Ochani, M.; Amella, C.A.; Tanovic, M.; Susarla, S.; Li, J.H.; Wang, H.; Yang, H.; Ulloa, L.; et al. Nicotinic acetylcholine receptor α7 subunit is an essential regulator of inflammation. Nature 2003, 421, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.S.F.; Ibarra, A.A.; Bermudez, I.; Casalino, L.; Gaieb, Z.; Shoemark, D.K.; Gallagher, T.; Sessions, R.B.; Amaro, R.E.; Mulholland, A.J. A potential interaction between the SARS-CoV-2 spike protein and nicotinic acetylcholine receptors. Biophys. J. 2021, 120, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281-292.e6. [Google Scholar] [CrossRef]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; Shoemark, D.; Carroll, M.W.; Heesom, K.J.; Zambon, M.; Ellis, J.; Lewis, P.A.; Hiscox, J.A.; et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020, 12, 68. [Google Scholar] [CrossRef]
- Ren, C.; Tong, Y.L.; Li, J.C.; Lu, Z.Q.; Yao, Y.M. The Protective Effect of Alpha 7 Nicotinic Acetylcholine Receptor Activation on Critical Illness and Its Mechanism. Int. J. Biol. Sci. 2017, 13, 46–56. [Google Scholar] [CrossRef]
- Gahring, L.C.; Myers, E.J.; Dunn, D.M.; Weiss, R.B.; Rogers, S.W. Nicotinic alpha 7 receptor expression and modulation of the lung epithelial response to lipopolysaccharide. PLoS ONE 2017, 12, e0175367. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Lv, Z.; Meng, L.; Xu, J.; Yuan, S.; Fu, Z. Activation of Alpha-7 Nicotinic Acetylcholine Receptors (α7nAchR) Promotes the Protective Autophagy in LPS-Induced Acute Lung Injury (ALI) In Vitro and In Vivo. Inflammation 2019, 42, 2236–2245. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Lentz, T.L.; Burrage, T.G.; Smith, A.L.; Crick, J.; Tignor, G.H. Is the acetylcholine receptor a rabies virus receptor? Science 1982, 215, 182–184. [Google Scholar] [CrossRef]
- Courties, A.; Boussier, J.; Hadjadj, J.; Yatim, N.; Barnabei, L.; Péré, H.; Veyer, D.; Kernéis, S.; Carlier, N.; Pène, F.; et al. Regulation of the acetylcholine/α7nAChR anti-inflammatory pathway in COVID-19 patients. Sci. Rep. 2021, 11, 11886. [Google Scholar] [CrossRef]
- Noviello, C.M.; Gharpure, A.; Mukhtasimova, N.; Cabuco, R.; Baxter, L.; Borek, D.; Sine, S.M.; Hibbs, R.E. Structure and gating mechanism of the α7 nicotinic acetylcholine receptor. Cell 2021, 184, 2121-2134.e13. [Google Scholar] [CrossRef]
- Ni, D.; Lau, K.; Turelli, P.; Raclot, C.; Beckert, B.; Nazarov, S.; Pojer, F.; Myasnikov, A.; Stahlberg, H.; Trono, D. Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Zhang, C.; Wang, Y.; Hong, Q.; Xu, S.; Li, Z.; Yang, Y.; Huang, Z.; Cong, Y. Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun. 2022, 13, 871. [Google Scholar] [CrossRef]
- Duhovny, D.; Nussinov, R.; Wolfson, H.J. Efficient Unbound Docking of Rigid Molecules; Springer: Berlin/Heidelberg, Germany, 2002; pp. 185–200. [Google Scholar]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Krause, R.M.; Buisson, B.; Bertrand, S.; Corringer, P.J.; Galzi, J.L.; Changeux, J.P.; Bertrand, D. Ivermectin: A positive allosteric effector of the alpha7 neuronal nicotinic acetylcholine receptor. Mol. Pharmacol. 1998, 53, 283–294. [Google Scholar] [CrossRef]
- Zemkova, H.; Tvrdonova, V.; Bhattacharya, A.; Jindrichova, M. Allosteric modulation of ligand gated ion channels by ivermectin. Physiol. Res. 2014, 63 (Suppl. 1), S215–S224. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Larrous, F.; Feige, L.; Kornobis, E.; Levallois, S.; Marchio, A.; Kergoat, L.; Hardy, D.; Cokelaer, T.; et al. Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin. EMBO Mol. Med. 2021, 13, e14122. [Google Scholar]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef]
- Liu, H.; Wu, N.C.; Yuan, M.; Bangaru, S.; Torres, J.L.; Caniels, T.G.; van Schooten, J.; Zhu, X.; Lee, C.D.; Brouwer, P.J.M.; et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity 2020, 53, 1272-1280.e5. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, R.D.; Vijayaraghavan, S. Paying attention to smell: Cholinergic signaling in the olfactory bulb. Front. Synaptic Neurosci. 2014, 6, 21. [Google Scholar] [PubMed]
- Coelho, D.H.; Reiter, E.R.; French, E.; Costanzo, R.M. Decreasing Incidence of Chemosensory Changes by COVID-19 Variant. Otolaryngol. Head Neck Surg. 2022, 1945998221097656. [Google Scholar] [CrossRef] [PubMed]
- Debroy, S. Severe Immune Effect Seen in Delta Absent in Omicron Cases in Mumbai: City Hospital Data. Times of India, 17 January 2022. Available online: https://timesofindia.indiatimes.com/city/mumbai/severe-immune-effect-seen-in-delta-absent-in-omicron-city-hosp-data/articleshow/88939499.cms (accessed on 5 September 2022).
- Piovesana, R.; Salazar Intriago, M.S.; Dini, L.; Tata, A.M. Cholinergic Modulation of Neuroinflammation: Focus on alpha7 Nicotinic Receptor. Int. J. Mol. Sci. 2021, 22, 4912. [Google Scholar] [CrossRef]
- Davies, M.A.; Kassanjee, R.; Rousseau, P.; Morden, E.; Johnson, L.; Solomon, W.; Hsiao, N.Y.; Hussey, H.; Meintjes, G.; Paleker, M.; et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection in the Omicron-driven fourth wave compared with previous waves in the Western Cape Province, South Africa. Trop. Med. Int. Health 2022, 27, 564–573. [Google Scholar] [CrossRef]
| B.1.1.7 | B.1.351 | P.1 | B.1.617.2 |
|---|---|---|---|
| (United Kingdom, Alpha) | (South Africa, Beta) | (Brazil, Gamma) | (India, Delta) |
| H69-V70 deletion (NTD) | L18F (NTD) | L18F (NTD) | T19R (NTD) |
| Y144 deletion (NTD) | D80A (NTD) | T20N (NTD) | 157–158 Deletion (NTD) |
| N501Y (RBD) | D215G (NTD) | P26S (NTD) | L452R (RBD) |
| 242–244 deletion (NTD) | D138Y (NTD) | T478K (RBD) | |
| R2461 (NTD) | R190S (NTD) | ||
| K417N (RBD) | K417T (RBD) | ||
| E484K (RBD) | E484K (RBD) | ||
| N501Y (RBD) | N501Y (RBD) | ||
| B.1.1.529 | |||
| (Omicron) | |||
| A67V (NTD) | |||
| H69-V70 deletion (NTD) | |||
| T95I (NTD) | |||
| G142D (NTD) | |||
| V143-Y144-Y145 deletion (NTD) | |||
| N211I (NTD) | |||
| L212V (NTD) | |||
| E214-P215-E216 addition (NTD) | |||
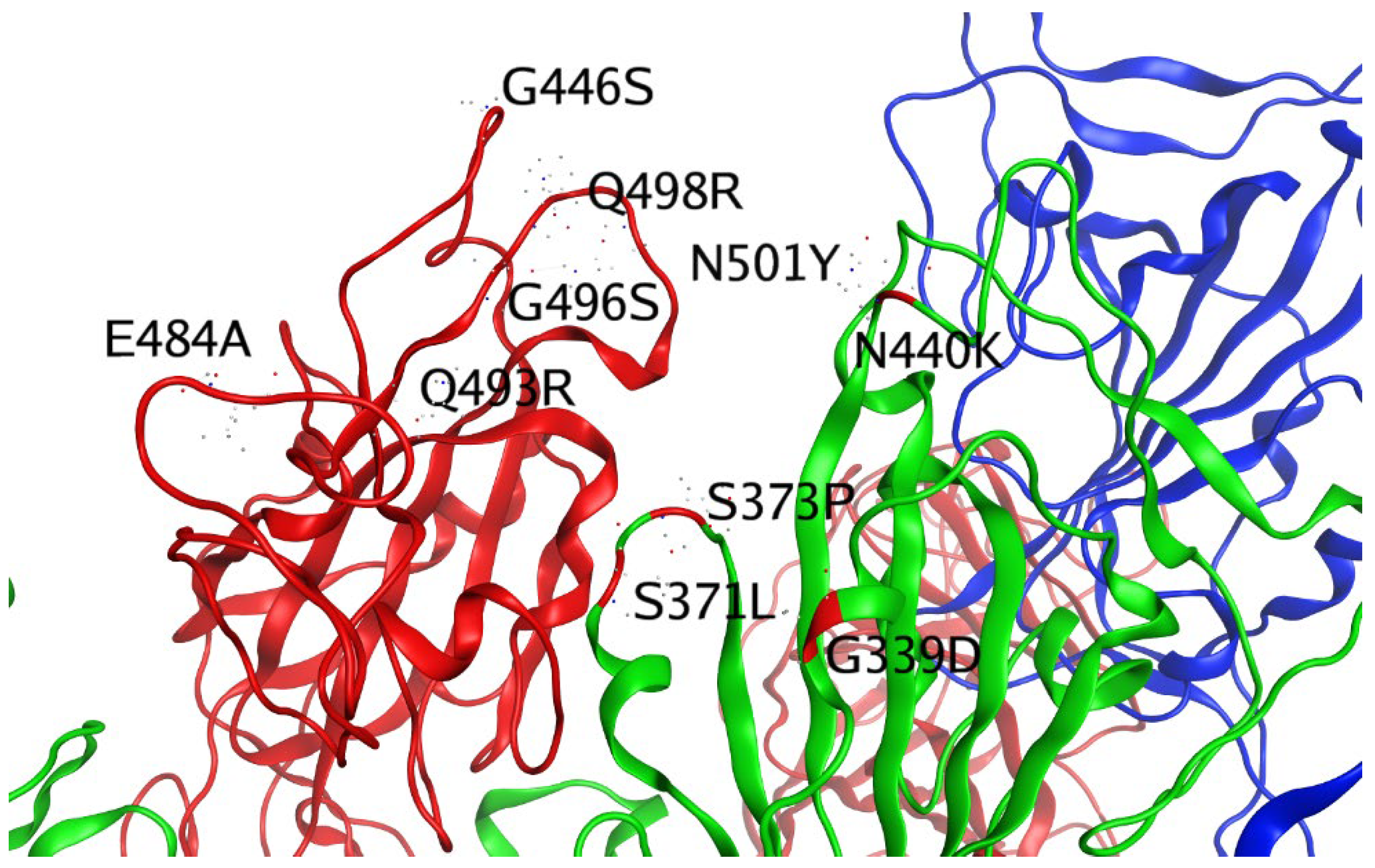
| G339D (RBD) | |||
| S371L (RBD) | |||
| S373P (RBD) | |||
| S375F (RBD) | |||
| K417N (RBD) | |||
| N440K (RBD) | |||
| G446S (RBD) | |||
| S447N (RBD) | |||
| T478K (RBD) | |||
| E484A (RBD) | |||
| Q493R (RBD) | |||
| G496S (RBD) | |||
| Q498R (RBD) | |||
| N501Y (RBD) | |||
| Glob | Global Energy, the Binding Energy of the Solution |
|---|---|
| aVdW | attractive van der Waals energy |
| rVdW | repulsive van der Waals energy |
| ACE | atomic contact energy (ACE) |
| Bond Type | Set A | Set B | Bond Type | Set A | Set B | Bond Type | Set A | Set B |
|---|---|---|---|---|---|---|---|---|
| DH | Asn23 | Arg403 | D | Ser69 | Ala372 | D | Val68 | Ser373 |
| DH | Asp156 | Thr500 | D | Ser69 | Trp436 | D | Glu9 | Phe486 |
| D | Val68 | Trp436 | D | His62 | Gln493 | D | Pro72 | Ser371 |
| D | Phe2 | Phe456 | D | Glu1 | Tyr489 | D | Lys191 | Lys444 |
| I | Ser25 | Arg403 | D | Pro27 | Asn501 | D | Asn13 | Gly485 |
| D | Phe186 | Thr500 | D | Asn67 | Gly339 | D | Leu28 | Gln498 |
| D | Trp153 | Pro499 | D | Val68 | Phe342 | D | Phe2 | Leu455 |
| D | Arg4 | Phe486 | D | Lys191 | Pro499 | D | Thr29 | Gly447 |
| D | Phe186 | Val445 | D | Lys12 | Glu484 | D | Ser25 | Tyr505 |
| DH | Ala22 | Gln493 | D | His62 | Leu492 | D | Tyr71 | Phe486 |
| D | His62 | Leu452 | D | Gln26 | Gln498 | D | Ser69 | Phe374 |
| D | Phe2 | Ala372 | D | Asp156 | Gln498 | D | Tyr31 | Gly446 |
| D | Leu28 | Gly446 | D | Asn23 | Tyr453 | D | Leu28 | Tyr449 |
| D | Ser69 | Phe342 | D | Pro27 | Tyr449 | D | Ser112 | Thr345 |
| D | Asn67 | Phe342 | D | Val30 | Val445 | D | His62 | Ser494 |
| D | Trp153 | Asn439 | D | Thr60 | Lys444 | D | Pro27 | Gln498 |
| D | Leu6 | Phe486 | D | Tyr63 | Gln493 | DH | Thr29 | Gln498 |
| D | Lys8 | Gly485 | D | Val68 | Ser371 | D | Gln158 | Val445 |
| D | Glu70 | Val367 | D | Lys191 | Ser443 | D | Thr60 | Tyr449 |
| D | Lys75 | Ser438 | D | Tyr71 | Ser371 | D | Pro72 | Ser373 |
| D | Asn23 | Ser494 | D | Thr60 | Gly446 | D | Pro72 | Ala372 |
| D | Asn110 | Asn440 | D | Gln65 | Tyr449 | D | Lys12 | Val483 |
| D | Lys75 | Asn439 | D | Asp156 | Val445 | D | Glu1 | Gln493 |
| D | Lys5 | Tyr489 | D | Asn23 | Tyr495 | D | Lys8 | Phe486 |
| D | Lys75 | Asn440 | D | Thr29 | Tyr449 | DH | Thr29 | Gly446 |
| D | Asn23 | Tyr505 | D | Ser69 | Asn343 | D | Ser69 | Ser373 |
| D | Gln26 | Gly496 | D | Val30 | Gly446 | D | Tyr31 | Val445 |
| D | His62 | Tyr449 | D | Lys191 | Val445 | D | Ala22 | Ser494 |
| D | Gln158 | Thr500 | D | Val21 | Tyr449 | D | Asn13 | Glu484 |
| D | Phe2 | Ser371 | D | Val68 | Asn343 | D | Lys5 | Asn487 |
| D | Trp66 | Asn343 | D | Glu70 | Ser373 | D | Gln26 | Tyr505 |
| D | Lys75 | Asn437 | D | Tyr31 | Lys444 | D | Glu1 | Phe456 |
| D | Ser112 | Leu441 | D | Asn13 | Val483 | D | Lys5 | Phe486 |
| D | Thr29 | Lys444 | D | Asn23 | Gly496 | D | Glu1 | Leu455 |
| D | Phe2 | Phe486 | D | Ala22 | Tyr449 | D | Thr29 | Val445 |
| D | Thr60 | Gly447 | D | Gln26 | Asn501 | D | Ser69 | Ser371 |
| D | Ser111 | Leu441 | D | Glu70 | Ser371 | D | Asn67 | Asn343 |
| D | Pro20 | Tyr449 | D | Ala22 | Tyr495 |
| Set A | Set B | Set B-Mutated | Set A | Set B | Set B-Mutated |
|---|---|---|---|---|---|
| Ala22 | Gln493 | Q493R | Glu70 | Ser373 | S373P |
| Leu28 | Gly446 | G446S | Asn23 | Gly496 | Q493R |
| Asn110 | Asn440 | N440K | Gln26 | Asn501 | N501Y |
| Lys75 | Asn440 | N440K | Glu70 | Ser371 | S371L |
| Gln26 | Gly496 | G496S | Val68 | Ser373 | S373P |
| Phe2 | Ser371 | S371L | Pro72 | Ser371 | S371L |
| His62 | Gln493 | Q493R | Leu28 | Gln498 | Q498R |
| Pro27 | Asn501 | N501Y | Tyr31 | Gly446 | G446S |
| Asn67 | Gly339 | G339D | Pro27 | Gln498 | Q498R |
| Lys12 | Glu484 | E484A | Thr29 | Gln498 | Q498R |
| Gln26 | Gln498 | Q498R | Pro72 | Ser373 | S373P |
| Asp156 | Gln498 | Q498R | Glu1 | Gln493 | Q493R |
| Tyr63 | Gln493 | Q493R | Thr29 | Gly446 | G446S |
| Val68 | Ser371 | S371L | Ser69 | Ser373 | S373P |
| Tyr71 | Ser371 | S371L | Asn13 | Glu484 | E484A |
| Thr60 | Gly446 | G446S | Ser69 | Ser371 | S371L |
| Val30 | Gly446 | G446S |
| Complex | Glob (Kcal/mol) | aVdW (Kcal/mol) | rVdW (Kcal/mol) | ACE (Kcal/mol) |
|---|---|---|---|---|
| α7-main spike | −2.45 | −31.08 | 18.5 | 13.58 |
| α7-Delta’s spike | −13.05 | −20.71 | 6.39 | 11.04 |
| α7-Omicron’s spike | 4.84 | −23.29 | 8.39 | 14.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doria, D.; Santin, A.D.; Tuszynski, J.A.; Scheim, D.E.; Aminpour, M. Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis. BioMedInformatics 2022, 2, 553-564. https://doi.org/10.3390/biomedinformatics2040035
Doria D, Santin AD, Tuszynski JA, Scheim DE, Aminpour M. Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis. BioMedInformatics. 2022; 2(4):553-564. https://doi.org/10.3390/biomedinformatics2040035
Chicago/Turabian StyleDoria, Domiziano, Alessandro D. Santin, Jack Adam Tuszynski, David E. Scheim, and Maral Aminpour. 2022. "Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis" BioMedInformatics 2, no. 4: 553-564. https://doi.org/10.3390/biomedinformatics2040035
APA StyleDoria, D., Santin, A. D., Tuszynski, J. A., Scheim, D. E., & Aminpour, M. (2022). Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis. BioMedInformatics, 2(4), 553-564. https://doi.org/10.3390/biomedinformatics2040035








