A Biomedical Case Study Showing That Tuning Random Forests Can Fundamentally Change the Interpretation of Supervised Data Structure Exploration Aimed at Knowledge Discovery
Abstract
1. Introduction
2. Materials
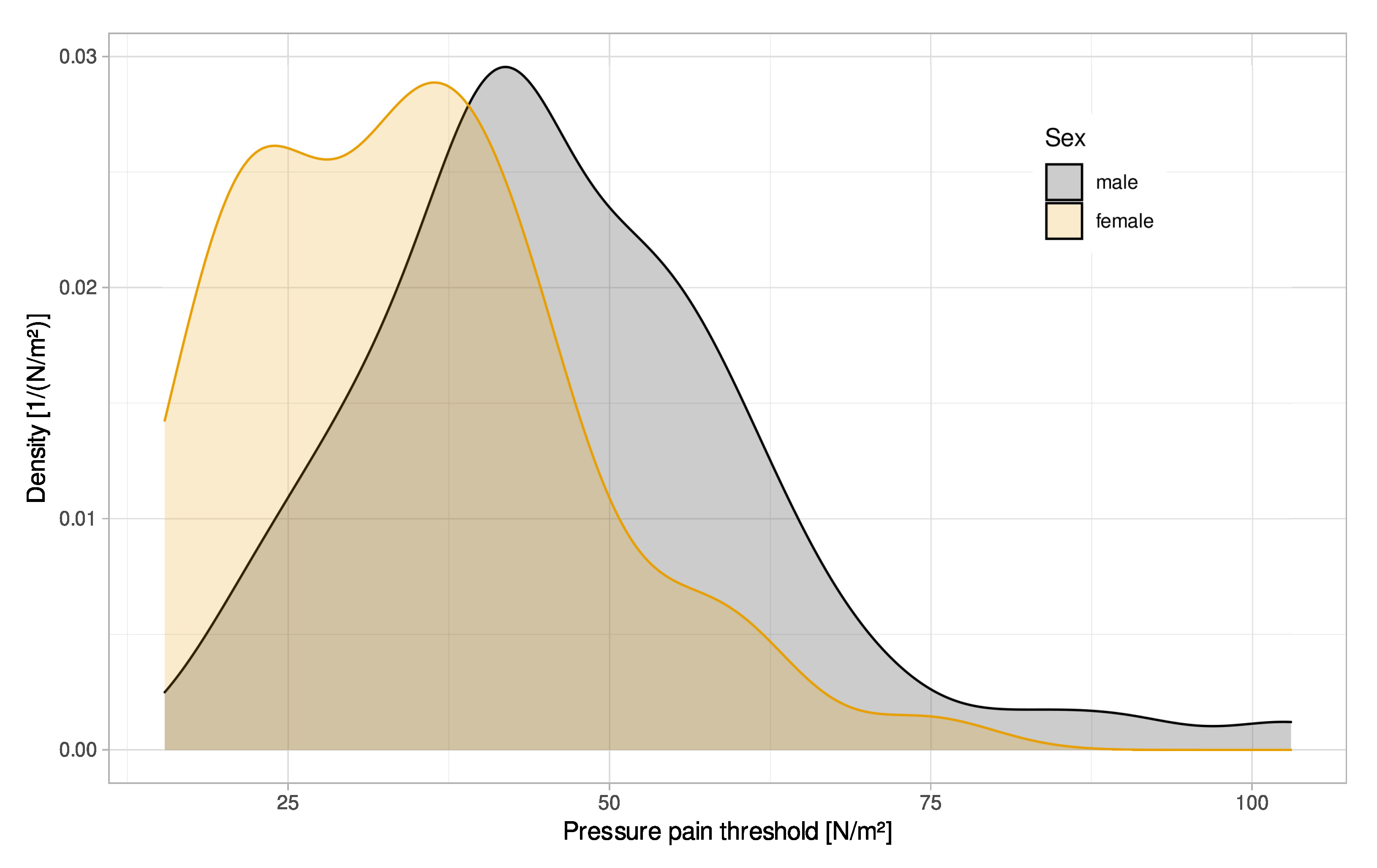
2.1. Data Set
2.2. Experimentation
2.2.1. Programming Environment
2.2.2. Design of the Experiments
3. Results
3.1. Random Forests Classification with Default Hyperparameters
| Listing 1: R code for classification with random forests using the default settings of the library “randomForest” (https://cran.r-project.org/package=randomForest (accessed on 9 October 2022) [23]). |
 |
3.2. Investigation of the Classification Failure
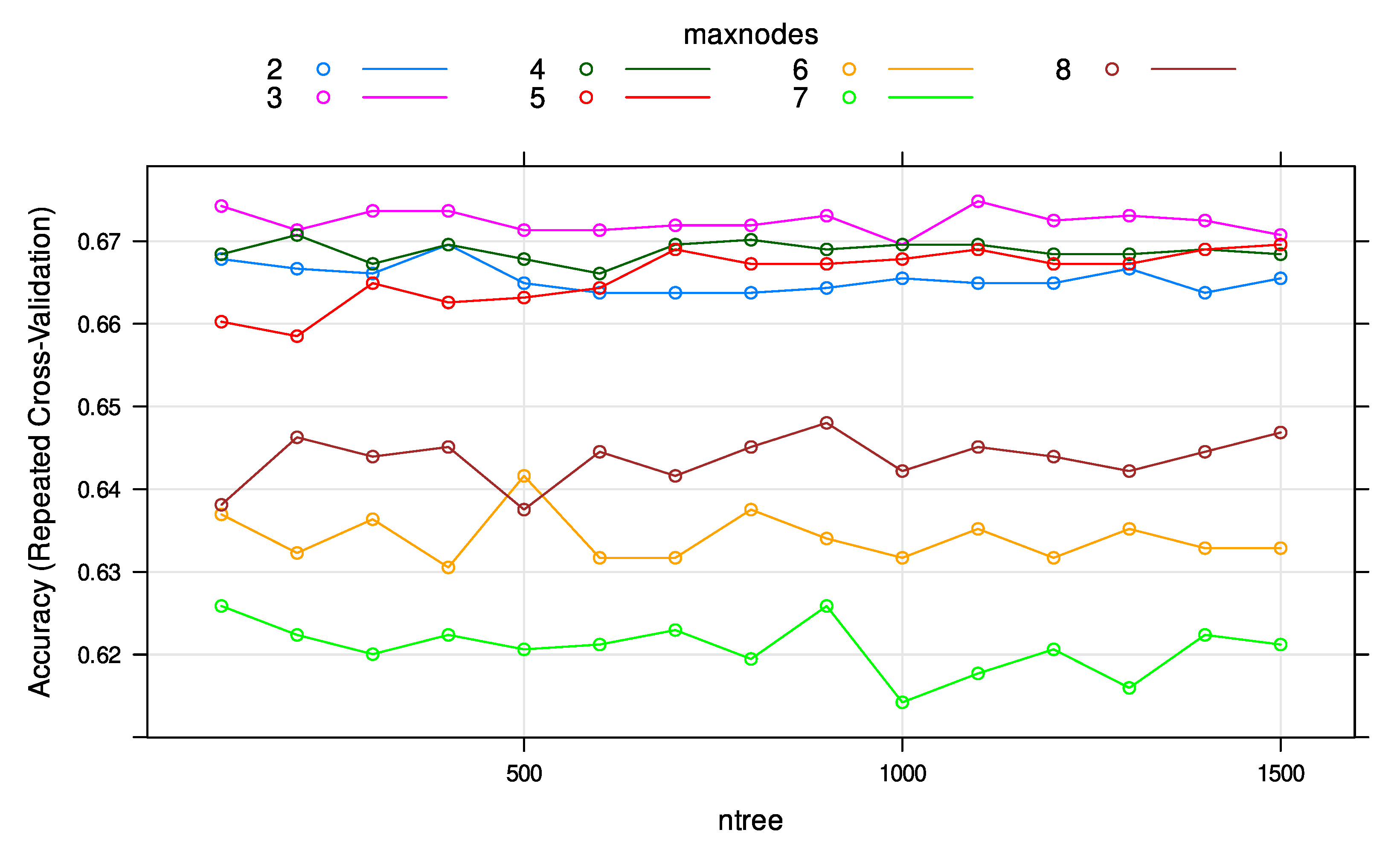
3.3. Random Forests Classification with Tuned Hyperparameters
| Listing 2: Python code for hyperparameter tuning and classification with random forests using the “RandomForestClassifier” method imported from the “scikit-learn” package (https://scikit-learn.org/stable/ (accessed on 9 October 2022) [30]). |
 |
4. Discussion
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spiliopoulou, M.; Schmidt-Thieme, L.; Janning, R. Data Analysis, Machine Learning and Knowledge Discovery; Studies in Classification, Data Analysis, and Knowledge Organization; Springer International Publishing: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, ICDAR ’95, Montreal, QC, Canada, 14–16 August 1995; Volume 1, p. 278. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Svetnik, V.; Wang, T.; Tong, C.; Liaw, A.; Sheridan, R.P.; Song, Q. Boosting: An Ensemble Learning Tool for Compound Classification and QSAR Modeling. J. Chem. Inf. Model. 2005, 45, 786–799. [Google Scholar] [CrossRef]
- Xu, H.; Kinfu, K.A.; LeVine, W.; Panda, S.; Dey, J.; Ainsworth, M.; Peng, Y.C.; Kusmanov, M.; Engert, F.; White, C.M.; et al. When are Deep Networks really better than Decision Forests at small sample sizes, and how? arXiv 2021, arXiv:2108.13637. [Google Scholar]
- Couronné, R.; Probst, P.; Boulesteix, A.L. Random forest versus logistic regression: A large-scale benchmark experiment. BMC Bioinform. 2018, 19, 270. [Google Scholar] [CrossRef] [PubMed]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random Forest: A Classification and Regression Tool for Compound Classification and QSAR Modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- Huang, B.F.; Boutros, P.C. The parameter sensitivity of random forests. BMC Bioinform. 2016, 17, 331. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Johnson, K. Feature Engineering and Selection: A Practical Approach for Predictive Models; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theor. 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Bryant, V. Metric Spaces: Iteration and Application; Cambridge University Press: Cambridge, UK, 1985. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Bennett, K.P.; Campbell, C. Support vector machines: Hype or hallelujah? ACM SIGKDD Explor. Newsl. 2000, 2, 1–13. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L., Jr. Python Tutorial; Centrum voor Wiskunde en Informatica: Amsterdam, The Netherlands, 1995; Volume 620. [Google Scholar]
- Doehring, A.; Küsener, N.; Flühr, K.; Neddermeyer, T.J.; Schneider, G.; Lötsch, J. Effect sizes in experimental pain produced by gender, genetic variants and sensitization procedures. PLoS ONE 2011, 6, e17724. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Routledge: New York, NY, USA, 1988. [Google Scholar] [CrossRef]
- Student. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Mogil, J.S. Sex differences in pain and pain inhibition: Multiple explanations of a controversial phenomenon. Nat. Rev. Neurosci. 2012, 13, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training; Astrophysics Source Code Library: Houghton, HI, USA, 2018. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference (SciPy 2010), Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- The Pandas Development Team Pandas-dev/pandas: Pandas. Zenodo 2020. [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Good, P.I. Resampling Methods: A Practical Guide to Data Analysis; Birkhäuser: Boston, MA, USA, 2006. [Google Scholar]
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The Balanced Accuracy and Its Posterior Distribution. In Proceedings of the Pattern Recognition (ICPR), 2010 20th International Conference on, Istanbul, Turkey, 23–26 August 2010; pp. 3121–3124. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In AI 2006: Advances in Artificial Intelligence, Lecture Notes in Computer Science; Sattar, A., Kang, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. [Google Scholar] [CrossRef]
- Bayes, M.; Price, M. An essay towards solving a problem in the doctrine of chances. By the late Rev. Mr. Bayes, FRS communicated by Mr. Price, in a letter to John Canton, AMFR S. Philos. Trans. 1763, 53, 370–418. [Google Scholar] [CrossRef]
- Ultsch, A.; Thrun, M.C.; Hansen-Goos, O.; Lötsch, J. Identification of Molecular Fingerprints in Human Heat Pain Thresholds by Use of an Interactive Mixture Model R Toolbox (AdaptGauss). Int. J. Mol. Sci. 2015, 16, 25897–25911. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D. Lattice: Multivariate Data Visualization with R; Springer: New York, NY, USA, 2008. [Google Scholar]
- Lotsch, J.; Ultsch, A. Random Forests Followed by Computed ABC Analysis as a Feature Selection Method for Machine Learning in Biomedical Data. In Advanced Studies in Classification and Data Science; Imaizumi, T., Okada, A., Miyamoto, S., Sakaori, F., Yoshiro, Y., Vichi, M., Eds.; Springer: Singapore, 2020; pp. 57–69. [Google Scholar]
- Ivakhnenko, A.G. Polynomial Theory of Complex Systems. IEEE Trans. Syst. Man Cybern. 1971, 4, 364–378. [Google Scholar] [CrossRef]
- Tkachenko, R.; Duriagina, Z.; Lemishka, I.; Izonin, I.; Trostianchyn, A. Development of machine learning method of titanium alloy properties identification in additive technologies. East.-Eur. J. Enterp. Technol. 2018, 3, 23–31. [Google Scholar] [CrossRef]
- Tripoliti, E.E.; Fotiadis, D.I.; Manis, G. Modifications of the construction and voting mechanisms of the Random Forests Algorithm. Data Knowl. Eng. 2013, 87, 41–65. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
| Classifier | Tuning | Performance Measure | ||
|---|---|---|---|---|
| Package | RandomForest (R) | RandomForest Classifier (Python) | ||
| RF | non-tuned | BA | 0.56 (0.42–0.68) | 0.55 (0.43–0.67) |
| auc-roc | 0.59 (0.45–0.72) | 0.58 (0.45–0.7) | ||
| tuned | BA | 0.65 (0.53–0.77) | 0.67 (0.55–0.78) | |
| auc-roc | 0.73 (0.59–0.85) | 0.72 (0.58–0.85) | ||
| Split | ad hoc | BA | 0.65 (0.54–0.76) | - |
| Bayes decision | ad hoc | BA | 0.67 (0.55–0.79) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lötsch, J.; Mayer, B. A Biomedical Case Study Showing That Tuning Random Forests Can Fundamentally Change the Interpretation of Supervised Data Structure Exploration Aimed at Knowledge Discovery. BioMedInformatics 2022, 2, 544-552. https://doi.org/10.3390/biomedinformatics2040034
Lötsch J, Mayer B. A Biomedical Case Study Showing That Tuning Random Forests Can Fundamentally Change the Interpretation of Supervised Data Structure Exploration Aimed at Knowledge Discovery. BioMedInformatics. 2022; 2(4):544-552. https://doi.org/10.3390/biomedinformatics2040034
Chicago/Turabian StyleLötsch, Jörn, and Benjamin Mayer. 2022. "A Biomedical Case Study Showing That Tuning Random Forests Can Fundamentally Change the Interpretation of Supervised Data Structure Exploration Aimed at Knowledge Discovery" BioMedInformatics 2, no. 4: 544-552. https://doi.org/10.3390/biomedinformatics2040034
APA StyleLötsch, J., & Mayer, B. (2022). A Biomedical Case Study Showing That Tuning Random Forests Can Fundamentally Change the Interpretation of Supervised Data Structure Exploration Aimed at Knowledge Discovery. BioMedInformatics, 2(4), 544-552. https://doi.org/10.3390/biomedinformatics2040034







