1. Introduction and Background
One of the biggest challenges to work with electronic health record (EHR) data is that there are many missing values. This issue incorporates uncertainty in the predictive model if the missing instances are imputed. Common imputation methods usually do not consider the temporal information, which is crucial for time series analysis. Moreover, most time series analysis methods ignore the time gap between measurements or assume that the time gaps are equal. In this study, we investigated time series imputation with irregular time gaps and propose a method based on neural ordinary differential equations (ODEs), recurrent neural networks (RNNs), and Bayesian estimation. This method offers a robust imputation of sporadically sampled multivariate time series measurements obtained from different patients.
Many data imputation techniques have been developed over the years. In real-life problems, it is very common to have multiple missing attributes for a particular dataset. In the literature, most datasets have 30% to 50% missing values, and they have been imputed using various techniques [
1]. There are mostly two techniques widely used for data imputation. These are statistical techniques and machine-learning-based techniques. Among the statistical techniques, expectation minimization (EM), the Gaussian mixture model (GMM), Markov chain Monte Carlo (MCMC), naive Bayes (NB), principal component analysis (PCA), etc., have been used frequently [
1]. Among the machine-learning-based techniques, the Gaussian process for machine learning (GPML) (see [
2]), support vector machines (SVMs) [
3,
4], k-nearest neighbors (k-NNs) [
5], decision trees (DTs) [
6], and artificial neural networks (ANNs) [
7] have been heavily used in the literature.
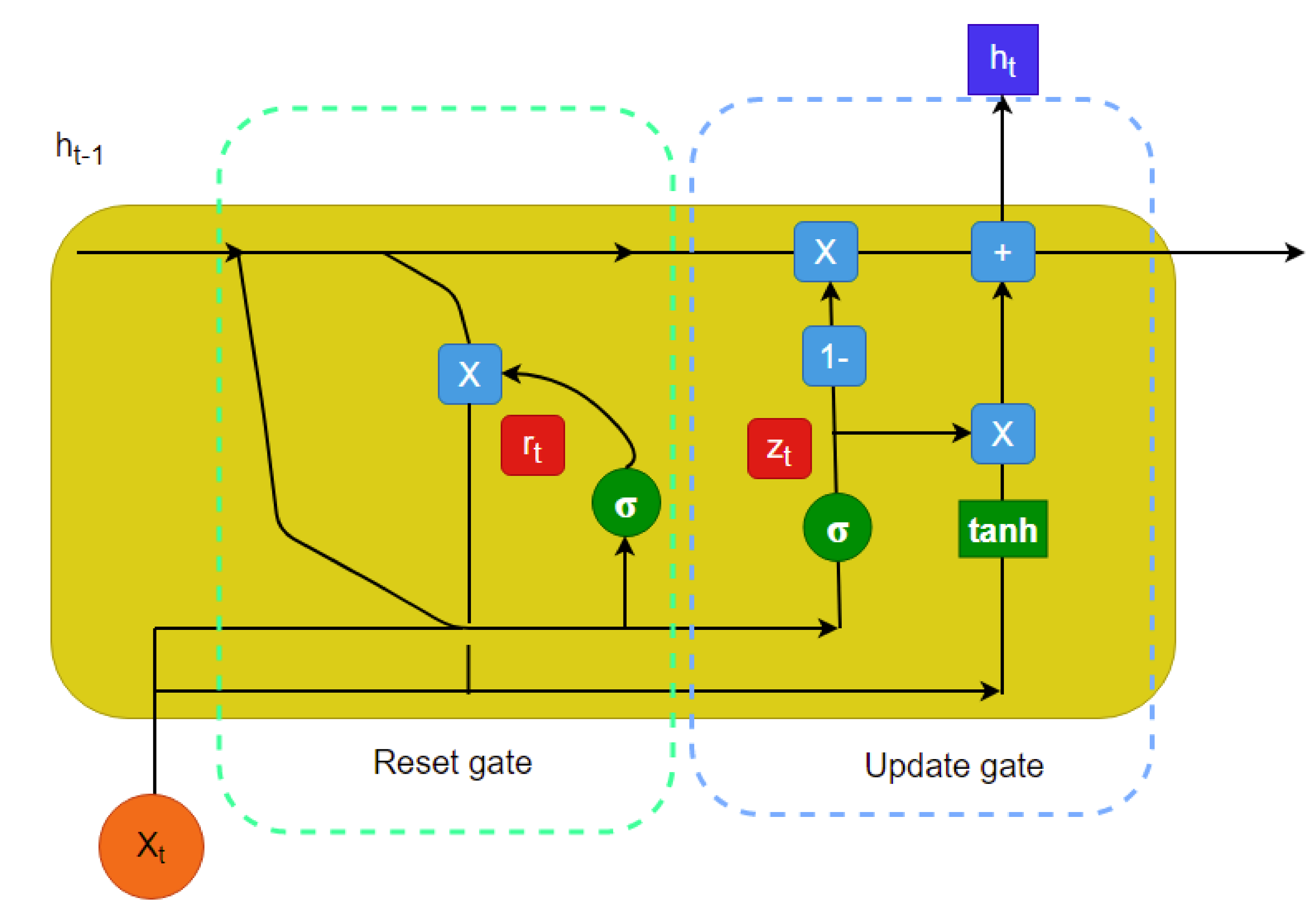
In recent years, longitudinal data imputation has been necessary specially in EHRs. However, many imputation methods only consider the data without the very important element–temporal information. However, there are many time series imputation methods that only consider equal time steps. Our research focuses on a time series imputation method that can deal with sporadically observed time series measurements obtained from EHRs. The major key components to implement this imputation method are neural ODEs [
8], which parameterize the derivative of a neural network’s hidden state. As compared to the popular residual neural networks, neural ODEs have superior memory and parameter efficiency. Neural ODEs can easily deal with continuous time series, unlike recurrent neural networks, which require discretization. In a follow-up study, latent ODEs were proposed for irregularly sampled (e.g., sporadic) time series [
9]. This method (called ODE-RNN) is presented as an alternative to autoregressive models. However, neural ODEs [
8] use RNNs as the recognition network to estimate the posterior probabilities. However, that approach is more appropriate for continuous time series modeling with regularly sampled data. Therefore, the ODE-RNN [
9] has been introduced as the recognition network to deal with irregularly sampled continuous time series analysis. Combined with neural ODEs and the GRU, a Bayesian update [
10] is proposed that uses a predictive (with observation masking) method (called the GRU-ODE-Bayes method) to include only the available observations to update the predicted values along the multivariate time series. However, this method does not impute the missing values; rather, it performs zero- or mean-value padding. The GRU-ODE-Bayes method assumes that the observations are sampled from a multidimensional stochastic process whose dynamics can be explained by a Weiner process. The examples include the stochastic Brusselator process [
11], the double-Ornstein–Uhlenbeck (OU) stochastic differential equations [
12], etc. The authors showed that their method achieved better results than the GRU-D [
13,
14], minimal gated unit or minimal GRU [
15], and other popular methods. In another recent study [
16], a bidirectional recurrent imputation for time series (BRITS) was proposed. This algorithm uses both a forward and backward feeding of inputs to the RNN and simultaneously imputes the missing values. However, the BRITS does not allow the stochastic imputation of time series data. The BRITS is composed of a recurrent component and a regression component for imputation. The authors also presented a unidirectional approach called the RITS and claimed that the process was slower compared to the BRITS. Among other RNN-based models, the multidirectional RNN (M-RNN) provides good imputation results [
17]. This study aims to develop a robust multivariate stochastic imputation technique for irregular time series that will fill this research gap.
2. Data Processing
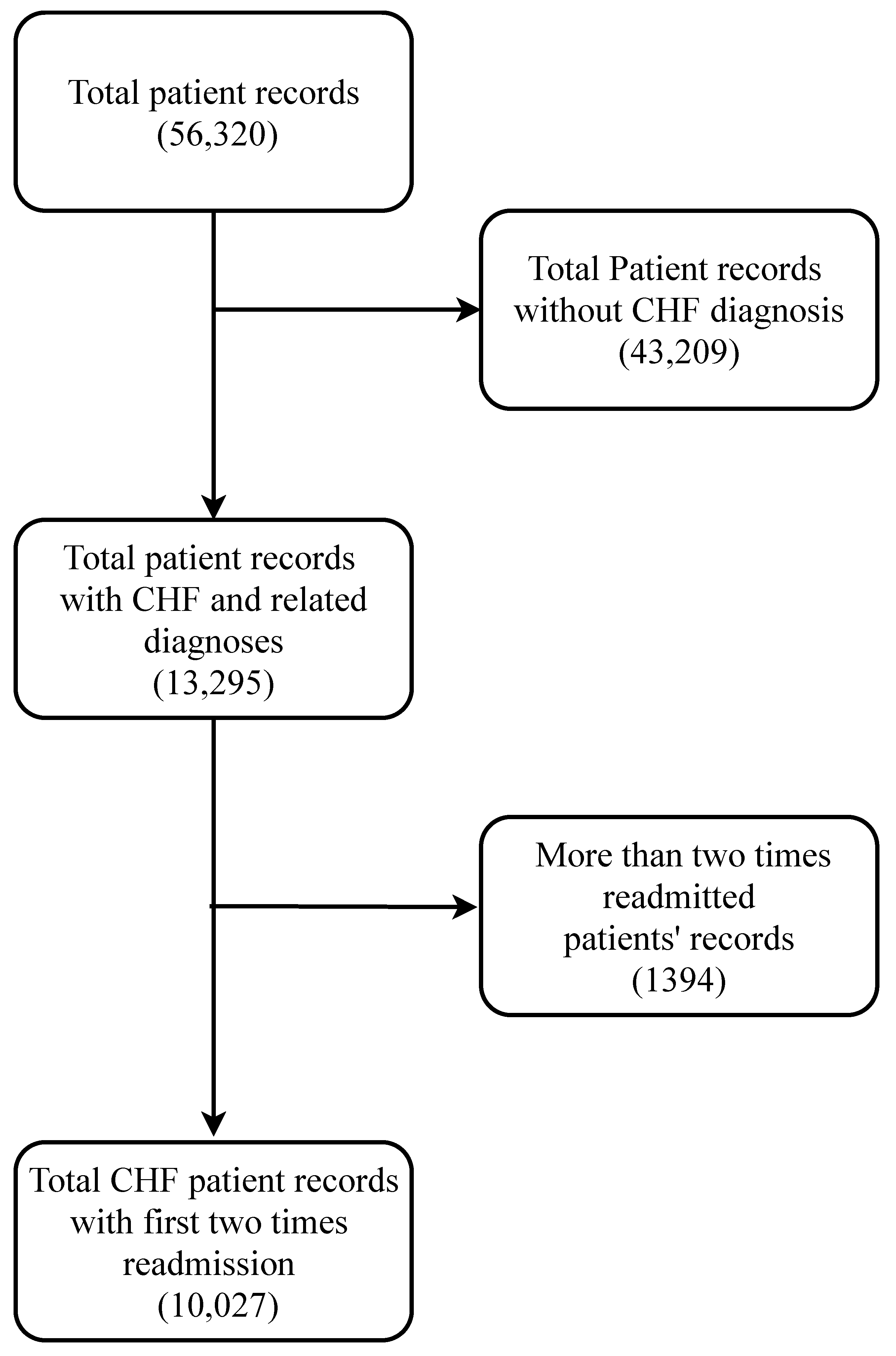
To develop a good predictive model, a reliable database is very crucial. The data need to be authentic and mostly accurate. Any big-data-driven research highly depends on the quality of the data being used. In this study, a very popular dataset [
13,
18,
19,
20] named the Medical Information Mart for Intensive Care (MIMIC) clinical database was explored and analyzed. This dataset contains intensive care unit (ICU) admission records of patients admitted to Beth Israel Deaconess Medical Center in Boston, Massachusetts, from 2001 to 2012. This database has several versions. In this study, MIMIC III Version 1.4 was used, which is the latest. It contains de-identified electronic medical records, demographic information, and billing information for ICU-admitted patients. Many of these records contain timestamps of clinical events, nurse-verified physiological measurements, routine vital signs, check-up information, etc. However, this database is only available after completing a required course and acknowledging a data use agreement.
After accessing the electronic records from the MIMIC III clinical database, a clean dataset needs to be formed that can be useful for analysis. As most other databases, MIMIC III provides its users with different types of structured and unstructured data records that must be cleaned prior to performing any data-driven analysis. Furthermore, the electronic records sometimes have different artifacts associated with them. Data cleaning tends to be more difficult in the case of large databases such as MIMIC III. The search algorithms for the desired attributes need to designed in a way that they can extract the necessary information efficiently. There are hardware and software limitations due to which it becomes very difficult to carry out large matrix operations in traditional computers. Since the analysis highly depends on the data quality, a proper cleaning process should be selected and performed carefully. There might be anomalies in the chosen dataset that should be properly dealt with before any analysis.
2.1. Data Description
The MIMIC-III database is an information storehouse for critical care patients. Therefore, it needs to deal with proper care and privacy. To access the data, one needs to request access formally through the Physionet website (
https://www.physionet.org) (last accessed on 15 November 2021). Two important steps need to be followed to access the data. The first one is to take a recognized course and comply with the Health Insurance Portability and Accountability Act (HIPAA) requirements. The second step is to sign a data use agreement that includes the appropriate data usage policy, security standards, and preventing identification efforts. Once the request is submitted, the approval comes within a week. Then, the data can be accessed from the server or can be stored in local storage. More information can be obtained by visiting the official MIMIC website (
https://physionet.org/content/mimiciii/1.4/) (last accessed on 15 November 2021).
There are 26 tables in total in the MIMIC-III database (see
Appendix A,
Table A1). In this subsection, we mainly discuss the tables that were directly used for the analysis. Since our goal was to extract as many CHF-related variables as possible, we needed to explore different tables and match records with mostly unique patient IDs and sometimes with admission IDs. Different tables are linked together to compile the dataset that we needed. The tables are described in the following paragraphs.
The “Admissions” table provides unique hospital admission information for each patient. It reports the admission ID, ICUstay ID, date of admission, admission type, discharge location, diagnosis, insurance, language, religion, marital status, age, ethnicity, etc. This can be linked with other tables via the admission ID and patient ID.
The “Patients” table provides demographic information for 46,520 unique patients. It contains the patient ID, admission ID, date of birth, date of death, gender, hospital expire flag, etc.
The “Chartevents” table is the largest table in the entire MIMIC III database. This has about 330,712,483 records. This table provides the patient ID, admission ID, item ID, and the corresponding routine physiological measurements of each patient from time to time.
The “D_ICD_diagnosis” table contains unique patient IDs, unique hospital admission IDs, and International Coding Definitions Version 9 (ICD-9) for each of the 14,567 diagnosis categories. The code for CHF diagnosis is 4280.
The “D_items” table has 12,487 records of items used to treat different patients. Routine vital signs such as blood pressure, heart rate, white blood cell count (WBC), respiratory rate, and other numerical variables are listed here with distinct item IDs.
6. Conclusions
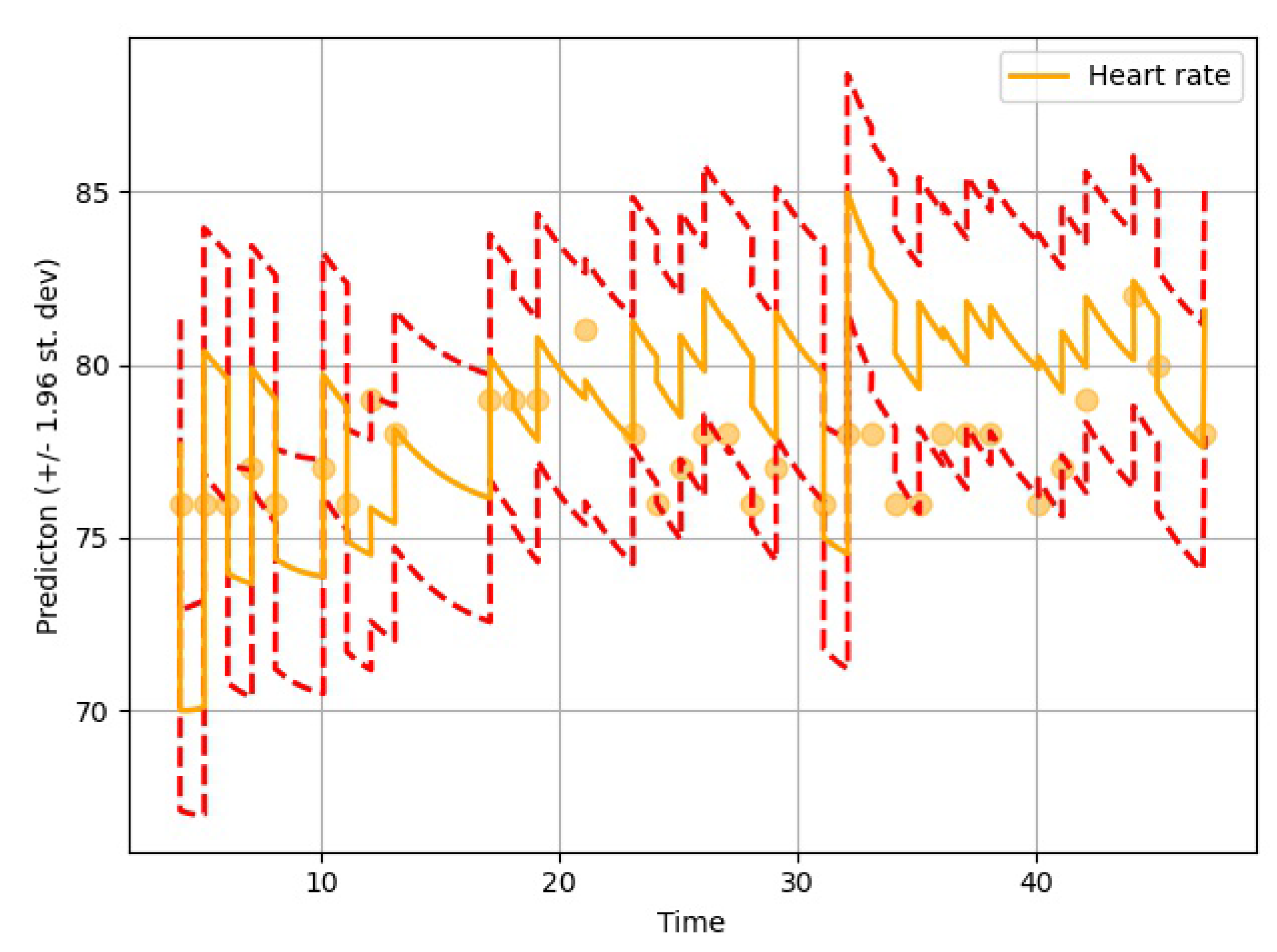
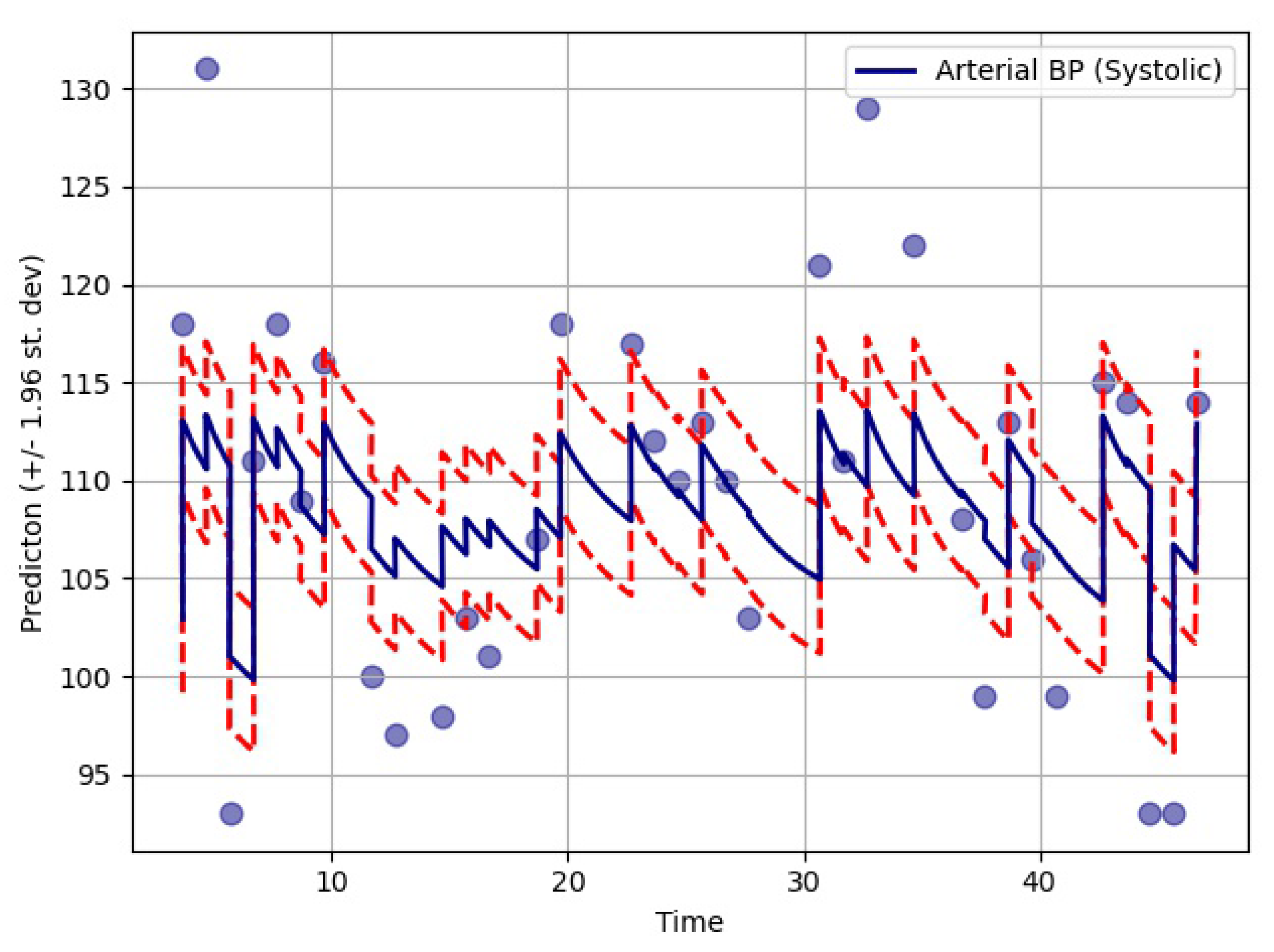
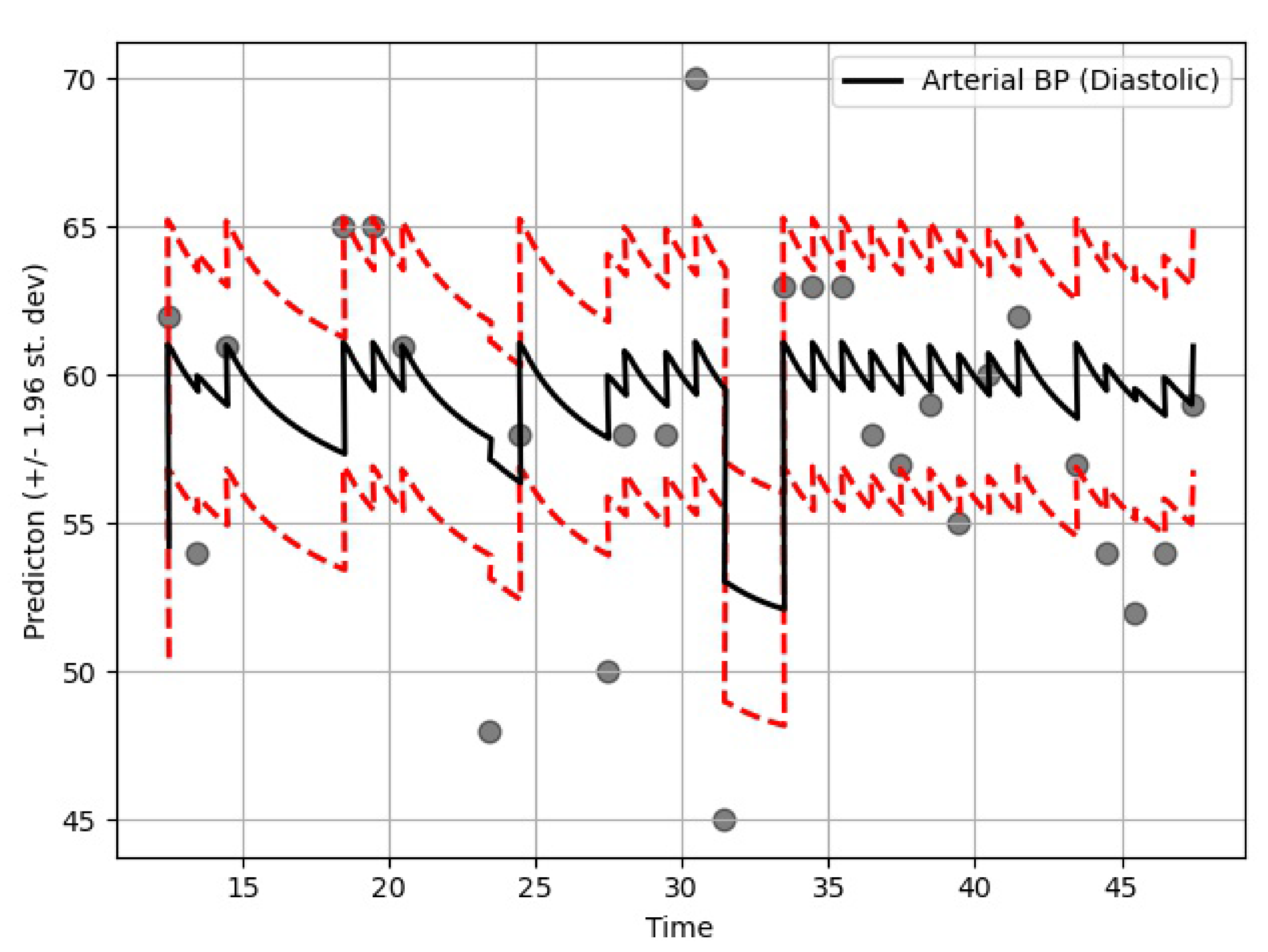
The proposed model was based on neural ODEs, RNN units, and Bayesian estimation and is suitable for imputation tasks involving temporal information. In most real-life scenarios, temporal information is very sensitive and determines many aspects of our day to day lives. Therefore, it is not always right to ignore the time-sensitive information found in EHRs or other similar databases. Furthermore, it makes more sense to have a probabilistic imputation rather than a deterministic one since there is always some level of uncertainty associated with the imputed data.
The novel contribution of this model is that it is tailored specifically for multivariate irregularly sampled time series imputation. As mentioned earlier, most other imputation methods deal with regular time series and provide deterministic imputation. Both of these issues are addressed by the GOBI method in this study.
The performance of the GOBI method was satisfactory, as shown in the comparative analysis section. Many state-of-the-art methods have been developed for classification tasks, but RNN-inspired stochastic imputation is still a growing area of data scientific research. In this study, only EHR data in the MIMIC databases were used for imputation. However, the GOBI method works well for many other datasets, as well, and provides fairly good estimation [
10]. As seen in
Table 6, the GOBI model has greater accuracy and lower variance.
GOBI method has several advantages, as well. It takes less time to train since GRU cells are simpler to compute and have fewer parameters. Even with very large datasets, it works relatively faster than most RNN-based techniques [
15,
26]. Besides, the GOBI method is quite accurate as compared to many algorithms currently available. Although the performance might vary from dataset to dataset, still it should be fairly competitive overall.
The GOBI method has high potential in data science sectors. It should be very useful for analyzing large datasets with a high amount of missing values. It might have broad impacts in healthcare, manufacturing industries, process improvement, etc., because most of these sectors typically deal with missing data or have physical constraints for time-sensitive data collection. The GOBI method can deal with these types of problems, providing a good solution with an acceptable error margin.
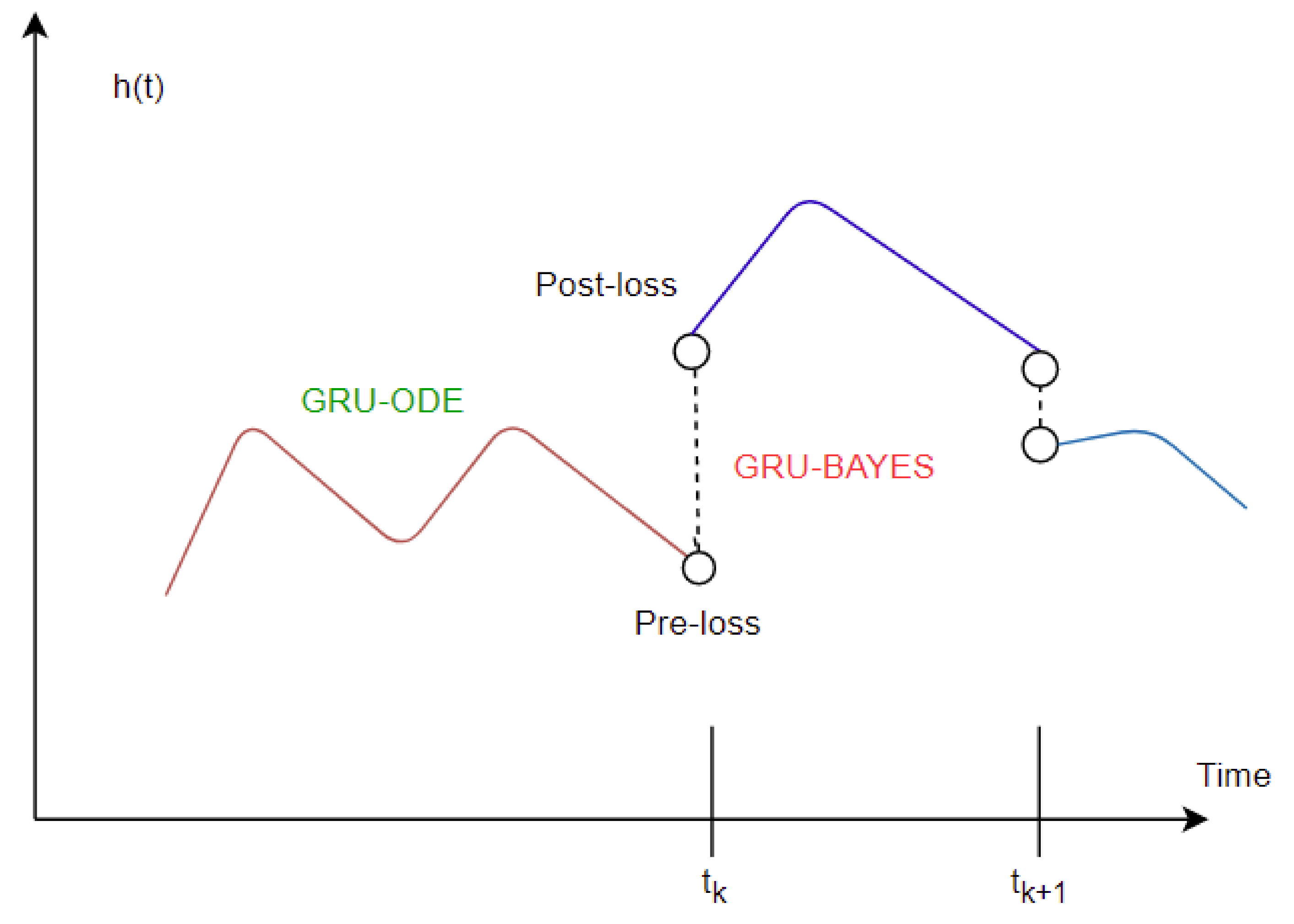
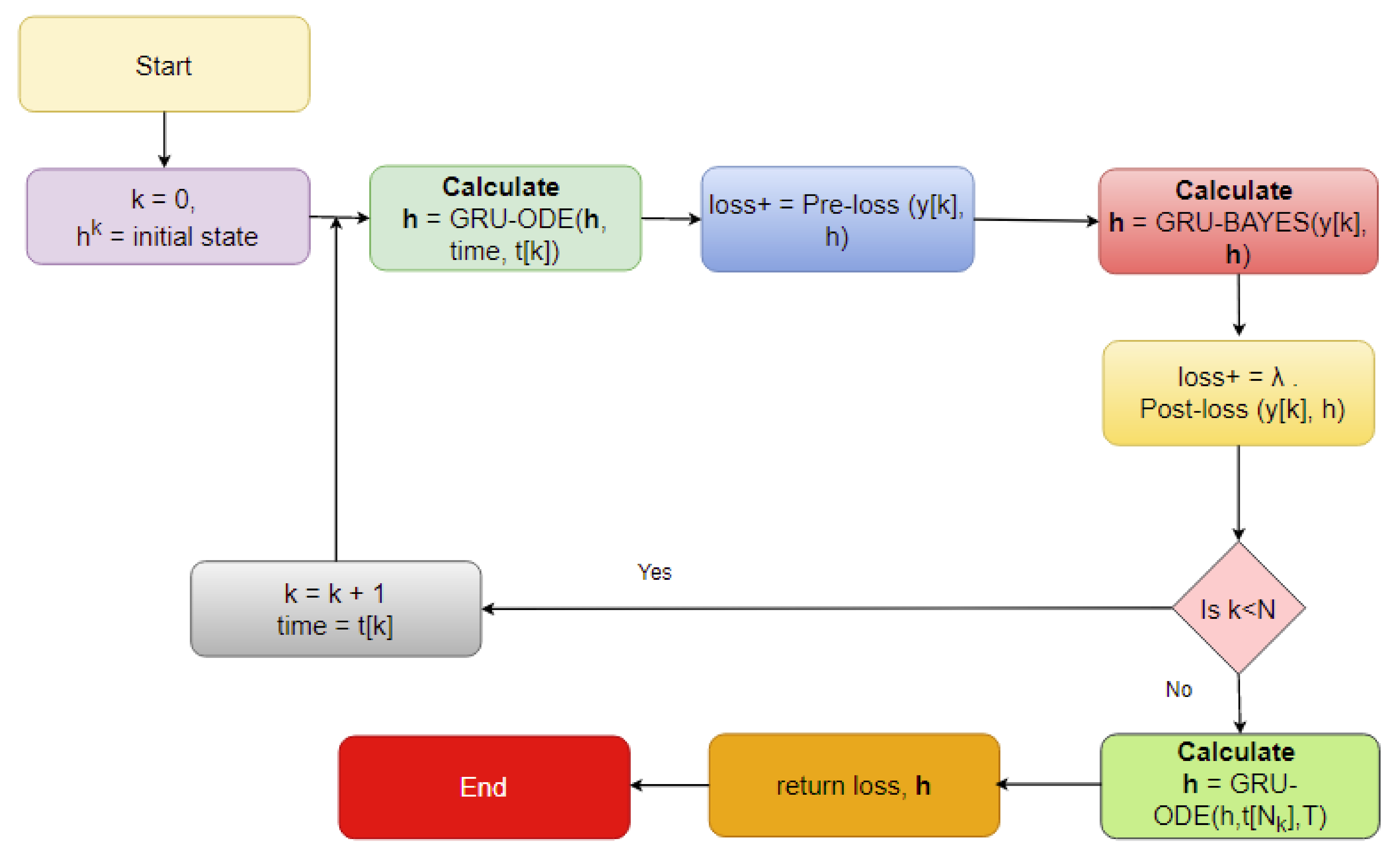
The imputation algorithm presented here is of great importance due to the increasing number of missing values in EHRs. It is very common to have more than 50–60% missing values per channel in EHR time series data. Each patient has some set of demographics, which vary greatly from one patient to the other. Therefore, it is difficult to impute records of one patient based on another. There is hardly any way around this since some patients might not have any measurement for a particular channel or item. As mentioned earlier, the EHR time series data might have some underlying dynamics that might not be approximated well by the stochastic Weiner process. This opens up a great opportunity for further research. As for standard oscillating behaviors, there are many well-established equations to represent the internal dynamics. As shown in many recent articles, the simulated data for standard oscillations can be accurately estimated by the Weiner process. However, this might not be the case for most real-life EHR time series data. The proposed method not only imputes the time series data, but also provides an estimation of the level of uncertainty that the imputed values represent. In the proposed model, the Bayesian estimation is coupled with an LSTM cell. This allows for the update to happen only when there are available values. The missing values are then inferred from the resulting imputed time series. The target is to predict the mean imputed values as close as possible to the actual values. The log variance needs to be smaller as well, since higher levels of uncertainty are much more difficult to propagate and can easily jeopardize the entire predictive model. In most common imputation methods, the mean-squared error is minimized. The proposed method uses two losses (pre and post) before and after Bayesian estimation, which are useful to decide whether the new observational update reduces the error. The two loss functions (negative log likelihood and KL divergence) are well established and widely used in ML techniques. Considering these issues, further research can be concentrated on developing an imputation method that could efficiently learn the dynamics of the underlying process instead of assuming a Weiner process in every case. This would significantly boost performance in complex EHR data.