Selective and Mild Transcriptional Modulation of Lectin Genes in Soy Leaves Under Drought Stress
Abstract
1. Introduction
2. Results
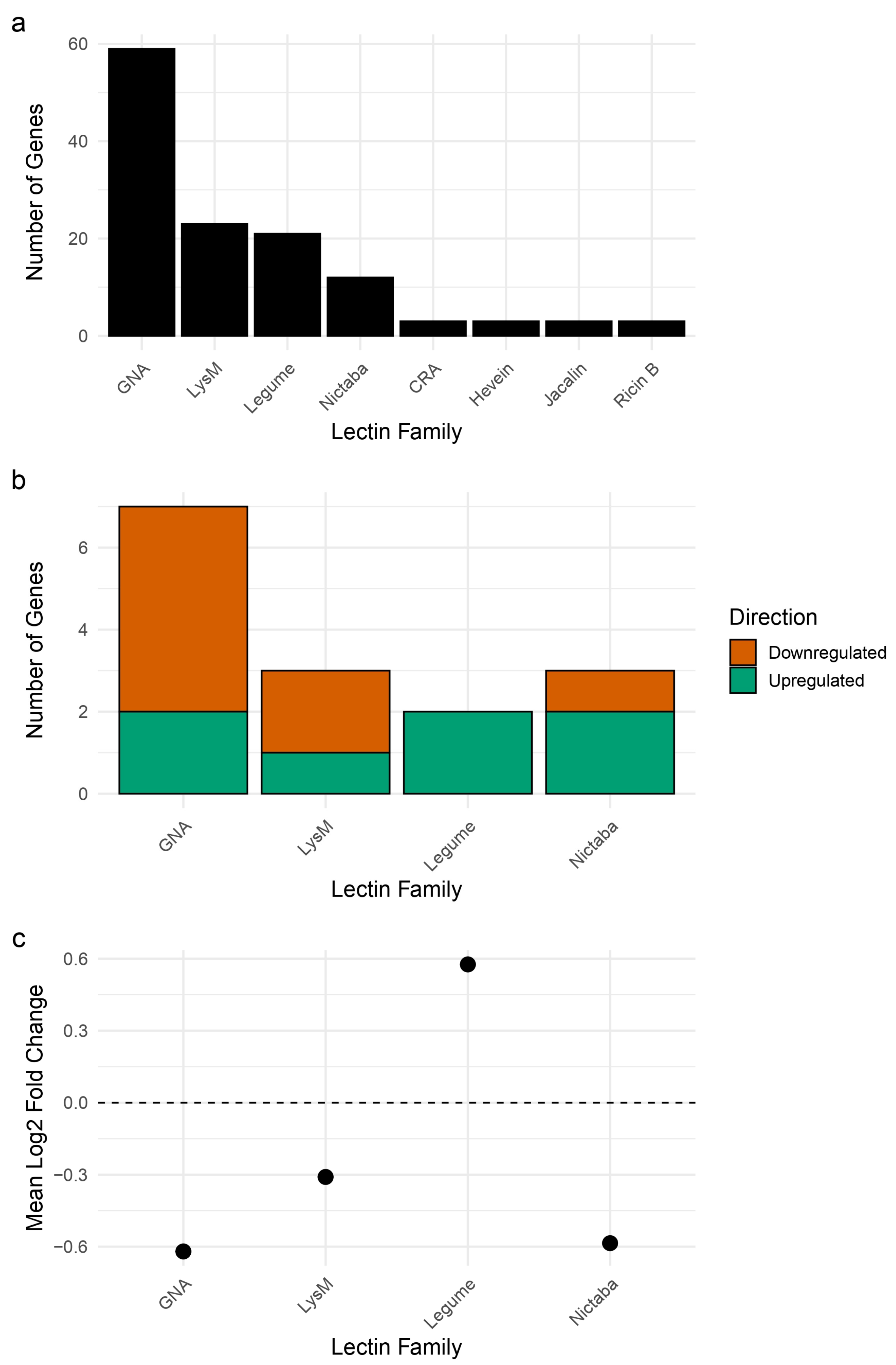
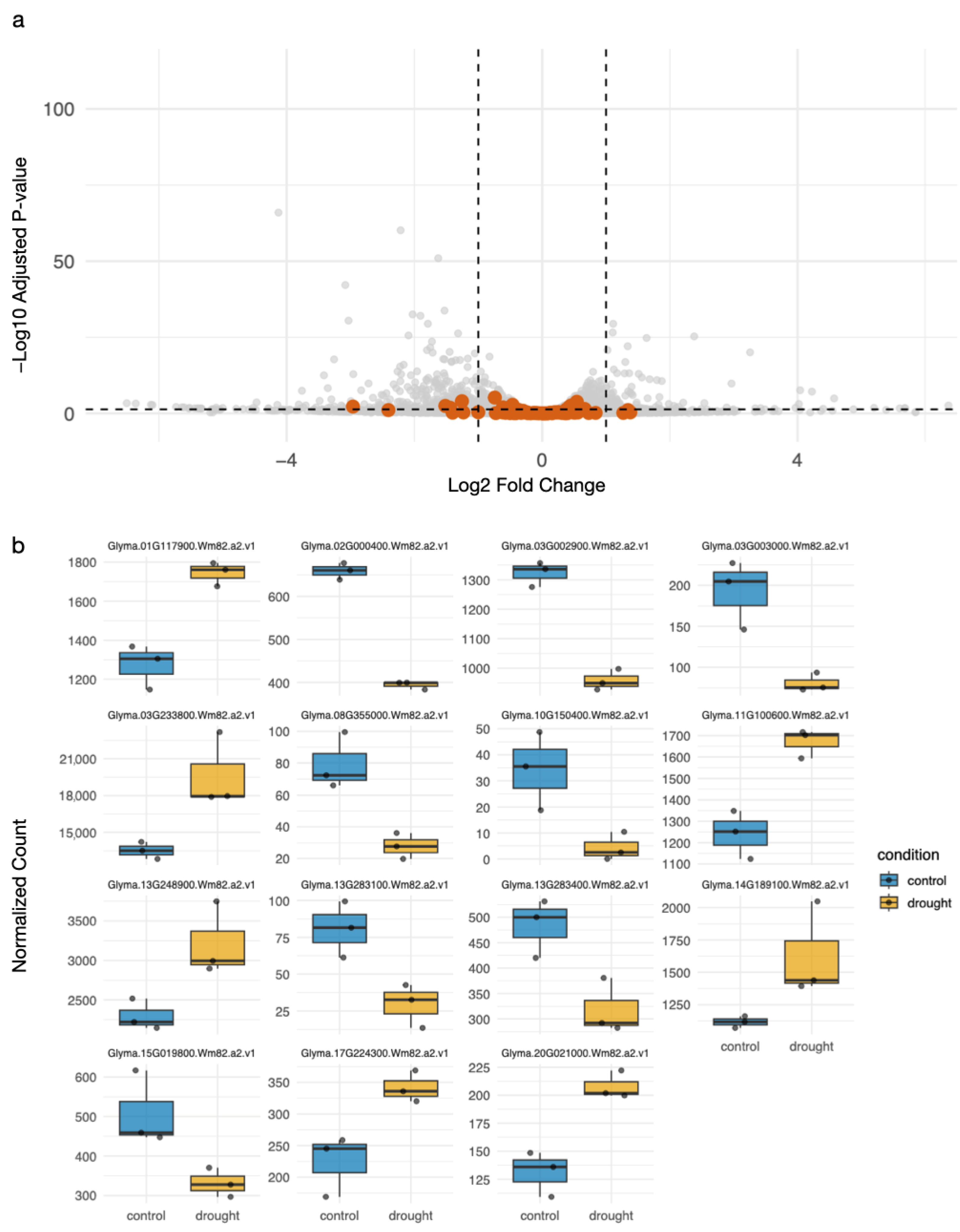
2.1. Differential Expression and Family Distribution of Lectin Genes Under Drought in Soy
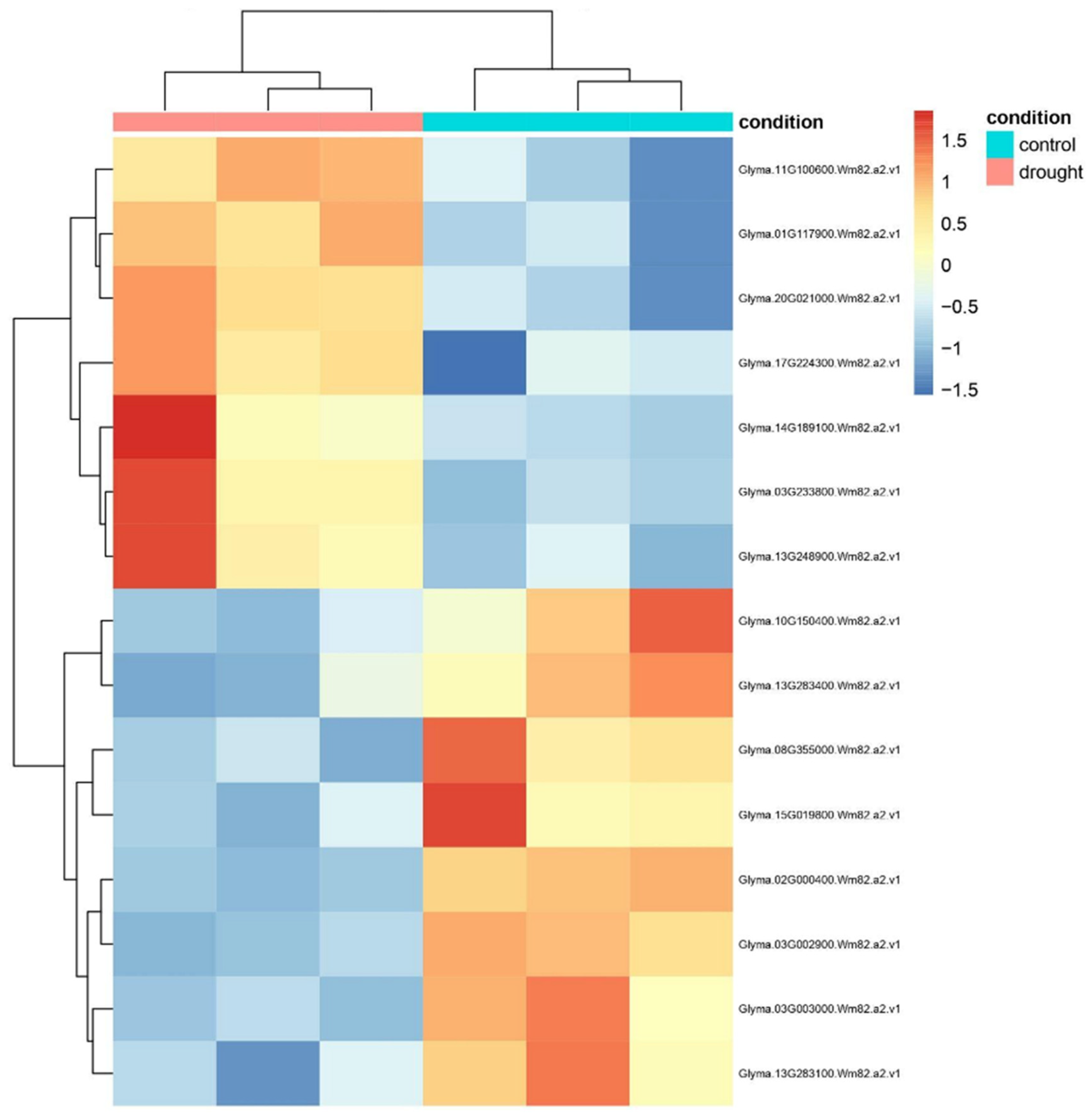
2.2. Transcriptional Clustering Reveals Co-Expression Modules Among DE Lectins
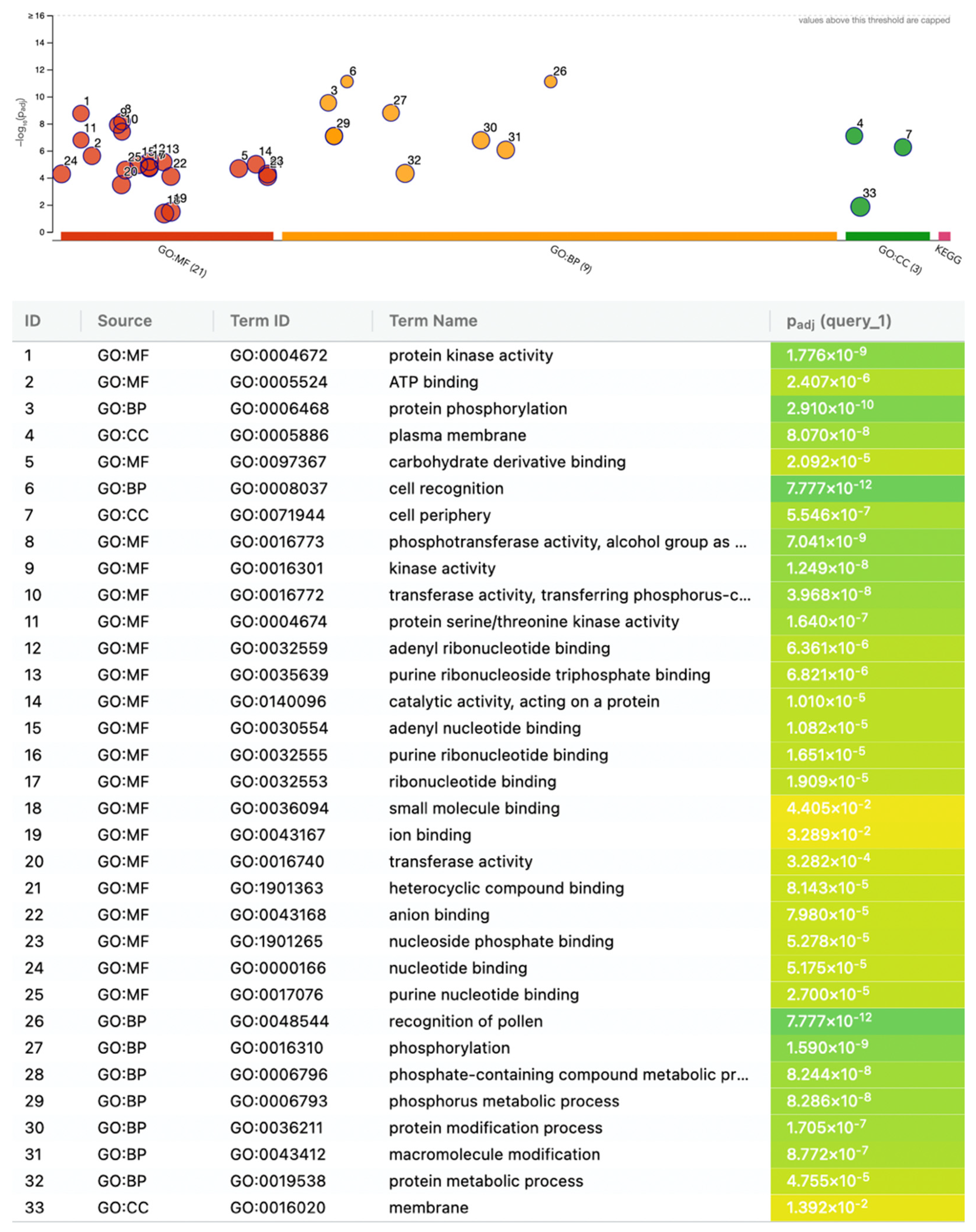
2.3. Functional Enrichment of Differentially Expressed Lectins Suggests Roles in Signaling and Kinase Activity
2.4. Domain Architecture, Localization, and Structural Modeling of Differentially Expressed Lectins
3. Discussion
4. Materials and Methods
4.1. RNA-Seq Data Acquisition and Preprocessing
4.2. Read Alignment and Quantification
4.3. Differential Gene Expression Analysis
4.4. Gene Ontology (GO) Enrichment Analysis
4.5. Domain Architecture and Functional Classification of DE Lectins
4.6. Subcellular Localization Prediction of DE Lectins
4.7. Structural Modeling and Comparison
4.8. Data Visualization
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kumari, S.; Dambale, A.S.; Samantara, R.; Jincy, M.; Bains, G. Introduction, history, geographical distribution, importance, and uses of soybean (Glycine max L.). In Soybean Production Technology; Springer Nature: Singapore, 2025; pp. 1–17. [Google Scholar]
- Hamza, M.; Basit, A.W.; Shehzadi, I.; Tufail, U.; Hassan, A.; Hussain, T.; Siddique, M.U.; Hayat, H.M. Global impact of soybean production: A review. Asian J. Biochem. Genet. Mol. Biol. 2024, 16, 12–20. [Google Scholar] [CrossRef]
- Wei, Y.; Jin, J.; Jiang, S.; Ning, S.; Liu, L. Quantitative response of soybean development and yield to drought stress during different growth stages in the Huaibei Plain, China. Agronomy 2018, 8, 97. [Google Scholar] [CrossRef]
- Ferreira, R.C.; Sibaldelli, R.N.R.; Crusiol, L.G.T.; Neumaier, N.; Farias, J.R.B. Soybean yield losses related to drought events in Brazil: Spatial–temporal trends over five decades and management strategies. Agriculture 2024, 14, 2144. [Google Scholar] [CrossRef]
- Mourtzinis, S.; Specht, J.E.; Lindsey, L.E.; Wiebold, W.J.; Ross, J.; Nafziger, E.D.; Kandel, H.J.; Mueller, N.; Devillez, P.L.; Arriaga, F.J.; et al. Climate-induced reduction in US-wide soybean yields underpinned by region- and in-season-specific responses. Nat. Plants 2015, 1, 14026. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought Tolerance in Plants: Physiological and Molecular Responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef]
- Peumans, W.J.; Van Damme, E.J. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, T.; Van Damme, E.J.M. Review: The multiple roles of plant lectins. Plant Sci. 2021, 313, 111096. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cordewener, J.H.G.; America, A.H.P.; Shan, W.; Bouwmeester, K.; Govers, F. Arabidopsis Lectin Receptor Kinases LecRK-IX.1 and LecRK-IX.2 Are Functional Analogs in Regulating Phytophthora Resistance and Plant Cell Death. Mol. Plant Microbe Interact. 2015, 28, 1032–1048. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 79. [Google Scholar] [CrossRef]
- Marothia, D.; Kaur, N.; Jhamat, C.; Sharma, I.; Pati, P.K. Plant lectins: Classical molecules with emerging roles in stress tolerance. Int. J. Biol. Macromol. 2023, 244, 125272. [Google Scholar] [CrossRef]
- Yang, X.; Chen, Z.; Lu, J.; Wei, X.; Yao, Y.; Lv, W.; Han, J.; Fei, J. Identification and Characterization of the Gene Family in Maize, and Its Role Under Biotic and Abiotic Stress. Biology 2024, 14, 20. [Google Scholar] [CrossRef]
- Bellande, K.; Bono, J.-J.; Savelli, B.; Jamet, E.; Canut, H. Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef] [PubMed]
- Van Holle, S.; Van Damme, E.J.M. Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 2015, 20, 2868–2891. [Google Scholar] [CrossRef]
- Li, X.; Qi, S.; Meng, L.; Su, P.; Sun, Y.; Li, N.; Wang, D.; Fan, Y.; Song, Y. Genome-wide identification of the wall-associated kinase gene family and their expression patterns under various abiotic stresses in soybean (Glycine max (L.) Merr). Front. Plant Sci. 2024, 15, 1511681. [Google Scholar] [CrossRef]
- Peláez-Vico, M.Á.; Sinha, R.; Induri, S.P.; Lyu, Z.; Venigalla, S.D.; Vasireddy, D.; Singh, P.; Immadi, M.S.; Pascual, L.S.; Shostak, B.; et al. The impact of multifactorial stress combination on reproductive tissues and grain yield of a crop plant. Plant J. 2024, 117, 1728–1745. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.E.M.; Osman, R.S.H.; Elmubarak, S.A.A.; Ibrahim, M.A.; Abakar, H.B.M.; Dirar, A.I.; Konozy, E.H.E. In silico analysis of L- and G-type lectin receptor kinases in tomato: Evolution, diversity, and abiotic responses. BMC Genom. 2024, 25, 1143. [Google Scholar] [CrossRef]
- Balagué, C.; Gouget, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 2017, 18, 937–948. [Google Scholar] [CrossRef]
- De Coninck, T.; Van Damme, E.J.M. Plant lectins: Handymen at the cell surface. Cell Surf. 2022, 8, 100091. [Google Scholar] [CrossRef]
- Schouppe, D.; Ghesquière, B.; Menschaert, G.; De Vos, W.H.; Bourque, S.; Trooskens, G.; Proost, P.; Gevaert, K.; Van Damme, E.J.M. Interaction of the tobacco lectin with histone proteins. Plant Physiol. 2011, 155, 1091–1102. [Google Scholar] [CrossRef]
- Eggermont, L.; Stefanowicz, K.; Van Damme, E.J.M. Nictaba Homologs from Arabidopsis thaliana Are Involved in Plant Stress Responses. Front. Plant Sci. 2017, 8, 2218. [Google Scholar] [CrossRef]
- Yao, X.; Humphries, J.; Johnson, K.L.; Chen, J.; Ma, Y. Function of WAKs in Regulating Cell Wall Development and Responses to Abiotic Stress. Plants 2025, 14, 343. [Google Scholar] [CrossRef]
- Pi, L.; Zhang, Y.; Wang, J.; Wang, N.; Yin, Z.; Dou, D. A G-type lectin receptor-like kinase in Nicotiana benthamiana enhances resistance to the fungal pathogen Sclerotinia sclerotiorum by complexing with CERK1/LYK4. Phytopathol. Res. 2023, 5, 27. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Wang, F.; Liu, Y.; Li, M.; Wang, H.; Zheng, Y.; Zhao, K.; Ji, Z. PWL1, a G-type lectin receptor-like kinase, positively regulates leaf senescence and heat tolerance but negatively regulates resistance to Xanthomonas oryzae in rice. Plant Biotechnol. J. 2023, 21, 2525–2545. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qi, Y.; Mu, Y.; Zhou, J.; Lu, W.; Tian, Z. Potato StLecRK-IV.1 negatively regulates late blight resistance by affecting the stability of a positive regulator StTET8. Hortic. Res. 2022, 9, uhac010. [Google Scholar] [CrossRef]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- FastQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 July 2025).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Heazlewood, J.L.; Verboom, R.E.; Tonti-Filippini, J.; Small, I.; Millar, A.H. SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 2007, 35, D213–D218. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Science & Business Media: New York, NY, USA, 2009. [Google Scholar]
- Kolde, R. pheatmap: Pretty Heatmaps, CRAN; The R Foundation: Vienna, Austria, 2010. [Google Scholar]
| Gene ID | Lectin Family | Domain(s) Detected | TM | Subcellular localization | Class |
|---|---|---|---|---|---|
| Glyma.03G003000 | GNA-related | GNA; S-locus glycoprotein; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.03G002900 | GNA-related | GNA; S-locus glycoprotein; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.01G117900 | GNA-related | GNA; S-locus glycoprotein; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.13G248900 | GNA-related | GNA; S-locus glycoprotein; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.08G355000 | GNA-related | GNA; EGF-like; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.13G283400 | GNA-related | GNA; S-locus glycoprotein; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.13G283100 | GNA-related | GNA; S-locus glycoprotein; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.14G189100 | Legume | Legume lectin; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.17G224300 | Legume | Legume lectin; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.02G000400 | LysM-related | LysM; kinase | Yes | Plasma membrane | LysM-RLK |
| Glyma.15G019800 | LysM-related | LysM; kinase | Yes | Plasma membrane | LysM-RLK |
| Glyma.11G100600 | LysM-related | GNA; S-locus glycoprotein; PAN/AP; kinase | Yes | Plasma membrane | LecRLK |
| Glyma.03G233800 | Nictaba-related | F-box; Nictaba | No | Cytosol; Endoplasmic reticulum | LecP |
| Glyma.10G150400 | Nictaba-related | F-box; Nictaba | No | Cytosol; Endoplasmic reticulum | LecP |
| Glyma.20G021000 | Nictaba-related | Nictaba | No | Plastid | LecP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osterne, V.J.S.; Leite, R.A.F.; Cavada, B.S.; Nascimento, K.S. Selective and Mild Transcriptional Modulation of Lectin Genes in Soy Leaves Under Drought Stress. Stresses 2025, 5, 54. https://doi.org/10.3390/stresses5030054
Osterne VJS, Leite RAF, Cavada BS, Nascimento KS. Selective and Mild Transcriptional Modulation of Lectin Genes in Soy Leaves Under Drought Stress. Stresses. 2025; 5(3):54. https://doi.org/10.3390/stresses5030054
Chicago/Turabian StyleOsterne, Vinicius J. S., Rafaela A. F. Leite, Benildo S. Cavada, and Kyria S. Nascimento. 2025. "Selective and Mild Transcriptional Modulation of Lectin Genes in Soy Leaves Under Drought Stress" Stresses 5, no. 3: 54. https://doi.org/10.3390/stresses5030054
APA StyleOsterne, V. J. S., Leite, R. A. F., Cavada, B. S., & Nascimento, K. S. (2025). Selective and Mild Transcriptional Modulation of Lectin Genes in Soy Leaves Under Drought Stress. Stresses, 5(3), 54. https://doi.org/10.3390/stresses5030054






