Evaluation of Papaya Plants Tolerant to PRSV Obtained Through Conventional Genetic Improvement
Abstract
1. Introduction
2. Results
2.1. Symptomatology Evaluation
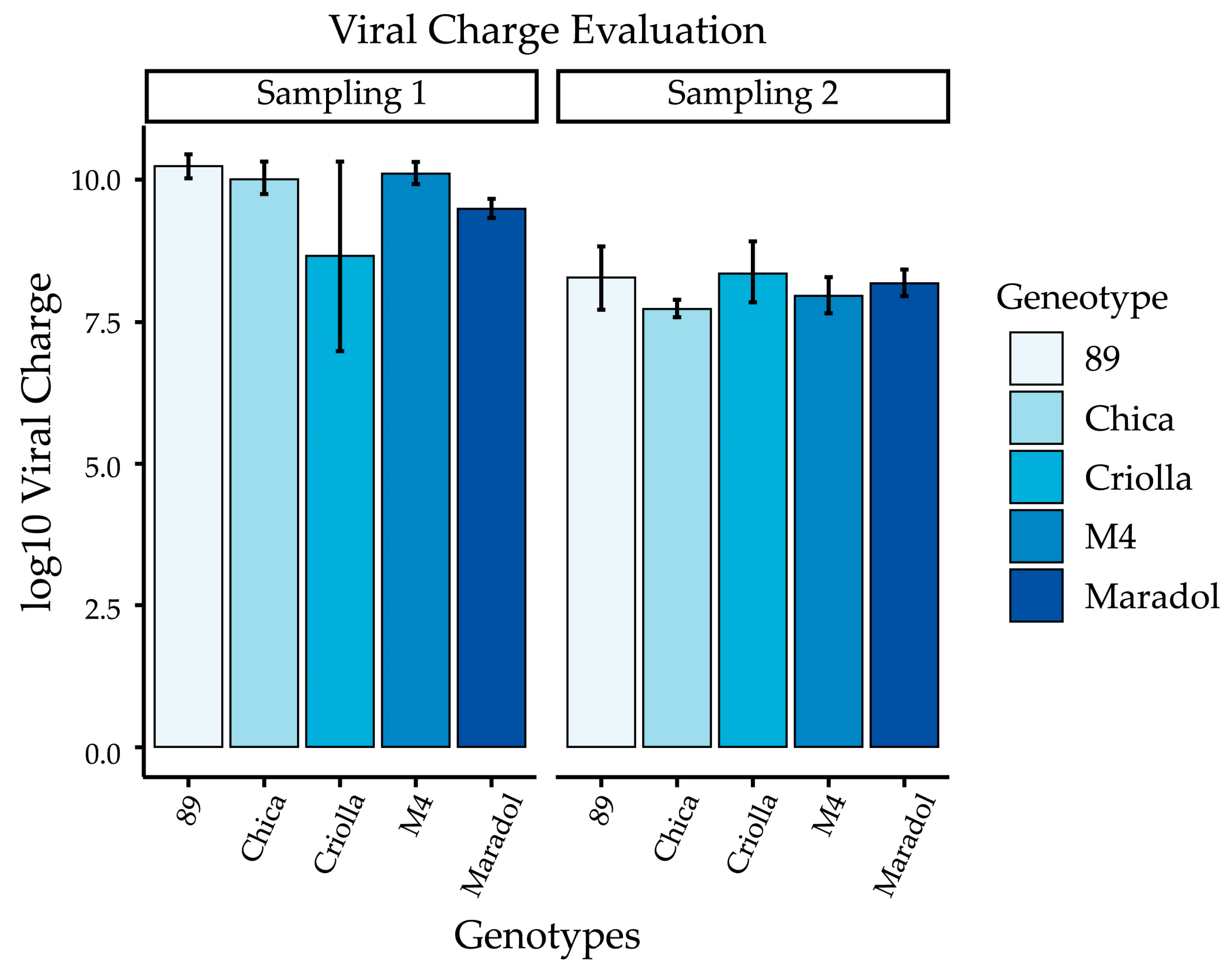
2.2. Evaluation of Viral Load
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sampling Collection
4.2. Quantification of PRSV Viral Load
4.3. Evaluation of Plant Symptoms and Calculation of the Disease Index
4.4. Statistical Analyisis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koul, B.; Pudhuvai, B.; Sharma, C.; Kumar, A.; Sharma, V.; Yadav, D.; Jin, J.-O. Carica papaya L.: A Tropical Fruit with Benefits beyond the Tropics. Diversity 2022, 14, 683. [Google Scholar] [CrossRef]
- Miranda-Ramírez, J.M.; Aguilar-García, O.; Miranda-Medina, D. Comparación de la productividad agrícola-económica sustentable y convencional de papaya, en Michoacán, México. Agron. Mesoam. 2020, 31, 384–403. [Google Scholar] [CrossRef]
- Arango, L.; Román, C. Aspectos botánicos. In Manual de Asistencia Técnica No. 4. Cultivo de la Papaya en los Llanos Orientales de Colombia; Corporación Colombiana de Investigación Agropecuaria; Asociación de Horticultores y Fruticultores de Colombia; Servicio Nacional de Aprendizaje: Villavicencio, Colombia, 1999; pp. 13–21. Available online: https://repository.agrosavia.co/bitstream/handle/20.500.12324/12830/42363_46132.pdf?sequence=1&isAllowed=y (accessed on 19 March 2025).
- Manshardt, R. Papaya. In Biotechnology of Perennial Fruit Crops; Hammerschlag, F.A., Litz, R.E., Eds.; CABI: Wallingford, UK, 1992; pp. 489–511. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19931635613 (accessed on 6 March 2025).
- Premchand, U.; Mesta, R.K.; Devappa, V.; Basavarajappa, M.P.; Venkataravanappa, V.; Narasimha Reddy, L.R.C.; Shankarappa, K.S. Survey, Detection, Characterization of Papaya Ringspot Virus from Southern India and Management of Papaya Ringspot Disease. Pathogens 2023, 12, 824. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cartón, I. La biotecnología al servicio del cultivo de la papaya. Vida Rural. 2002, 160, 35–37. Available online: https://www.mapa.gob.es/ministerio/pags/Biblioteca/Revistas/pdf_vrural%2FVrural_2002_160_35_37.pdf (accessed on 10 February 2025).
- Muhammad, U.; Mustansar, M.Y.I.; Haider, A.; Muhammad, Z.; Rafia, A.; Mazhar, A.; Malik, A.R.; Ernesto, A.M.E.; Yuejun, H. Papaya Ring Spot Virus: An Understanding of a Severe Positive-Sense Single. Stranded RNA Viral Disease and Its Management. Phyton Int. J. Exp. Bot. 2022, 91, 2099–2110. [Google Scholar] [CrossRef]
- Alcántara, J.J.A.; Hernández, C.E.; Ayvar, S.S.; Damián, N.A.; Brito, G.T. Características fenotípicas y agronómicas de seis genotipos de papaya (Carica papaya L.) de Tuxpan, Guerrero, México. Rev. Venez. Cienc. Tecnol. Aliment. 2010, 1, 035–046. Available online: https://oaji.net/articles/2017/4924-1495199597.pdf (accessed on 19 February 2025).
- Siar, S.V.; Beligan, G.A.; Sajise, A.J.C.; Villegas, V.N.; Drew, R.A. Papaya ringspot virus resistance in Carica papaya via introgression from Vasconcellea quercifolia. Euphytica 2011, 181, 159–168. [Google Scholar] [CrossRef]
- Razean, H.M.R.; Drew, R.A. Isolation and Characterization of PRSV-P Resistance Genes in Carica and Vasconcellea. Int. J. Genom. 2014, 2014, 145403. [Google Scholar] [CrossRef]
- Vasugi, C.; Ravishankar, K.V.; Kumar, A.; Poornima, K. Genetic Enhancement of Nutraceuticals in Papaya (Carica papaya L.). In Compendium of Crop Genome Designing for Nutraceuticals; Springer Nature: Singapore, 2023; pp. 1001–1031. [Google Scholar]
- Bruening, G. Resistance to Infection. In Natural Resistance Mechanism of Plant to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 211–240. [Google Scholar]
- Alviar, A.; Cruz, F.S.; Hautea, D. Assessing the responses of tolerant papaya (Carica papaya L.) varieties to Papaya RingSpot Virus (PRSV) infection and establishment of symptom severity rating scale for resistance screening. Crop Sci. Soc. Phillippines 2012, 37, 20–28. Available online: https://www.cabi.org/gara/mobile/FullTextPDF/2012/20123307085.pdf (accessed on 14 February 2025).
- Rubio, L.; Herrero, J.R.; Sarrió, J.; Moreno, P.; Guerri, J. A new approach to evaluate relative resistance and tolerance of tomato cultivars to begomoviruses causing the tomato yellow leaf curl disease in Spain. Plant Pathol. 2003, 52, 763–769. [Google Scholar] [CrossRef]
- Pagán, I.; Alonso-Blanco, C.; García-Arenal, F. Differential Tolerance to Direct and Indirect Density-Dependent Costs of Viral Infection in Arabidopsis thaliana. PLoS Pathog. 2009, 5, e1000531. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Tripathi, S. Horticultural characterization and papaya ringspot virus reaction of papaya Pune Selections. Indian J. Hortic. 2019, 76, 32–37. [Google Scholar] [CrossRef]
- Bengyella, L.; Waikhom, S.D.; Allie, F.; Rey, C. Virus tolerance and recovery from viral-induced symptoms in plants are associated with transcriptome reprogramming. Plant Mol. Biol. 2015, 89, 243–252. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, R. RNA silencing in productive virus infections. Annu. Rev. Phytopathol. 2005, 43, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Tripathi, S. Differential expression of microRNAs in response to apaya ringspot virus infection in differentially responding genotypes of papaya (Carica papaya L.) and its wild relative. Front. Plant Sci. 2024, 15, 1398437. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, X.; Zhang, P.; Mar, T.; Liu, Y.; Zhang, Z.; Han, C.; Wang, X. A one-step real-time RT-PCR assay for the quantification of Wheat yellow mosaic virus (WYMV). Virol. J. 2013, 10, 173. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Castro, S.; Silva-Rosales, L. Use of RT-PCR for papaya ringspotyvirus detection in papaya (Carica papaya) plants from Veracruz, Tabasco and Chiapas. Rev. Mex. Fitopatol 1997, 15, 83–90. [Google Scholar]
- Wang, X.; Seed, B. High-throughput primer and probe design. In Real-Time PCR; Dorak, M., Ed.; Taylor and Francis Group: Abingdon, UK, 2006; pp. 93–105. Available online: https://www.taylorfrancis.com/chapters/edit/10.4324/9780203967317-14/high-throughput-primer-probe-design-xiaowei-wang-brian-seed (accessed on 12 February 2025).
- Janthasri, R.; Chaiyaboon, W. Yellow Krang—A new cultivar of papaya for green consumption with tolerance to papaya ringspot virus. J. Hortic. Res. 2015, 23, 39–48. [Google Scholar] [CrossRef]
- Cabrera, D.; García, D.; Portal, O. Virus de la mancha anular de la papaya (PRSV-p): Biología, epifitología y diversidad genética como base para el manejo mediante técnicas biotecnológicas. Biotecnol. Veg. 2010, 10, 67–77. [Google Scholar]

| Sampling 1 (97 DAP) | Sampling 2 (532 DAP) | ||||
|---|---|---|---|---|---|
| Genetic Line | Number of Plants Evaluated | Incidence Mean | Severity Mean | Incidence Mean | Severity Mean |
| Maradol | 10 | 9 | 5 | 8 | 6 |
| Criolla | 10 | 9 | 3 | 3 | 3 |
| M4 | 11 | 8 | 4 | 7 | 2 |
| Chica-2 | 12 | 9 | 3 | 7 | 4 |
| 54 | 1 | 9 | 4 | 9 | 4 |
| 89 | 4 | 9 | 3 | 6 | 3 |
| 90 | 1 | 9 | 3 | 9 | 2 |
| Genetic Line | No. Plants Tested | Disease Index | Comment | |
|---|---|---|---|---|
| 97 Days | 532 Days | |||
| Maradol | 10 | 38.89 | 65.56 | Moderately susceptible |
| Criolla | 10 | 33.33 | 34.57 | Moderately tolerant |
| M4 | 11 | 41.41 | 28.28 | Moderately tolerant |
| Chica-2 | 12 | 35.19 | 50.93 | Moderately susceptible |
| 54 | 1 | 44.44 | 44.44 | Moderately tolerant |
| 89 | 4 | 36.11 | 22.22 | Highly tolerant |
| 90 | 1 | 33.33 | 22.22 | Highly tolerant |
| Sample | Cq (Quantification Cycle) | Copies/μg of Total RNA | Log10 of Copies/μg of Total RNA |
|---|---|---|---|
| Maradol | 17.799 | 3.31 × 109 | 9.493 ± 0.174 |
| Criolla | 20.845 | 5.59 × 109 | 8.660 ± 1.670 |
| Chica-2 | 20.168 | 1.25 × 1010 | 10.116 ± 0.195 |
| M4 | 23.545 | 1.41 × 1010 | 10.021 ± 0.290 |
| 89 | 22.9 | 1.81 × 1010 | 10.223 ± 0.211 |
| Sample | Cq (Quantification Cycle) | Copies/μg of Total RNA | Log10 of Copies/μg of Total RNA |
|---|---|---|---|
| Maradol | 19.92 | 1.67 × 108 | 8.186 ± 0.231 |
| Criolla | 18.94 | 3.79 × 108 | 8.373 ± 0.536 |
| Chica-2 | 21.75 | 1.84 × 107 | 7.965 ± 0.318 |
| M4 | 20.86 | 1.11 × 108 | 7.746 ± 0.150 |
| 89 | 19.85 | 3.34 × 108 | 8.282 ± 0.560 |
| Range Scale | Severity Percentage (%) | Description of Symptoms | Comments |
|---|---|---|---|
| 0 | 0 | No symptoms | Resistant |
| 1 | 0–0.9 | Very light speckled no bruises or stripes on petioles and stem. | Highly tolerant |
| 2 | 1–5 | Very light speckled yellow areas cover 1–5% of the leaf area, with some rings on the leaves but no noticeable symptoms on the fruit no bruises or streaks on petioles and stems. | |
| 3 | 6–25 | Very light speckled yellow areas cover 6–25% of the leaf area, with some rings on the leaves but no noticeable symptoms on the fruit no bruises or stripes on petioles and stem. | Moderately tolerant |
| 5 | 26–50 | Very light speckled yellow areas cover 26–50% of the leaf area soft fruit rings no bruises or stripes on petioles or stems. | |
| 7 | 51–75 | Speckled yellow areas cover 51–75% of the leaf area, with clear evidence of rings throughout the fruit bruises or streaks on petioles and stems. | Moderately susceptible |
| 9 | 75–100 | Severe speckled yellow areas cover 75–100% of the leaf area deformed and fragile leaves with severe distortion with visible ribbing clearly visible rings throughout the fruit crusty spots deformed fruit rough and bitter outer shell and granulated flesh. | Highly susceptible |
| Disease Index (%) | Plant Characteristics |
|---|---|
| 0–25 | Highly tolerant |
| 26–50 | Moderately tolerant |
| 51–75 | Moderately susceptible |
| 75–100 | Highly susceptible |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Noriega, L.M.; Kirchmayr, M.R.; Rodríguez-Garay, B.; Navarro-López, D.E.; Gutiérrez-Mora, A. Evaluation of Papaya Plants Tolerant to PRSV Obtained Through Conventional Genetic Improvement. Stresses 2025, 5, 51. https://doi.org/10.3390/stresses5030051
Sánchez-Noriega LM, Kirchmayr MR, Rodríguez-Garay B, Navarro-López DE, Gutiérrez-Mora A. Evaluation of Papaya Plants Tolerant to PRSV Obtained Through Conventional Genetic Improvement. Stresses. 2025; 5(3):51. https://doi.org/10.3390/stresses5030051
Chicago/Turabian StyleSánchez-Noriega, Luz María, Manuel R. Kirchmayr, Benjamín Rodríguez-Garay, Diego E. Navarro-López, and Antonia Gutiérrez-Mora. 2025. "Evaluation of Papaya Plants Tolerant to PRSV Obtained Through Conventional Genetic Improvement" Stresses 5, no. 3: 51. https://doi.org/10.3390/stresses5030051
APA StyleSánchez-Noriega, L. M., Kirchmayr, M. R., Rodríguez-Garay, B., Navarro-López, D. E., & Gutiérrez-Mora, A. (2025). Evaluation of Papaya Plants Tolerant to PRSV Obtained Through Conventional Genetic Improvement. Stresses, 5(3), 51. https://doi.org/10.3390/stresses5030051







