Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery
Abstract
1. Introduction
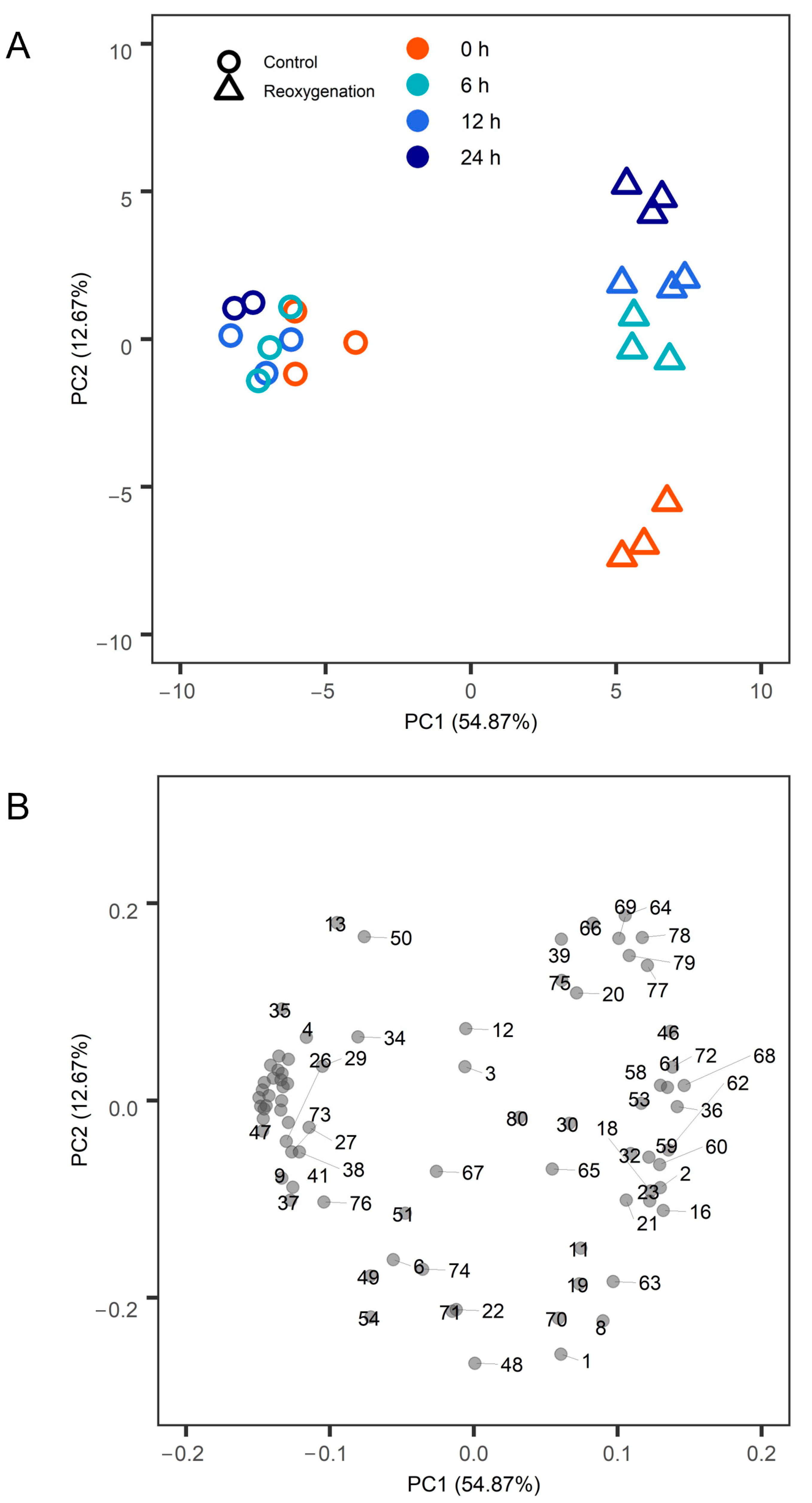
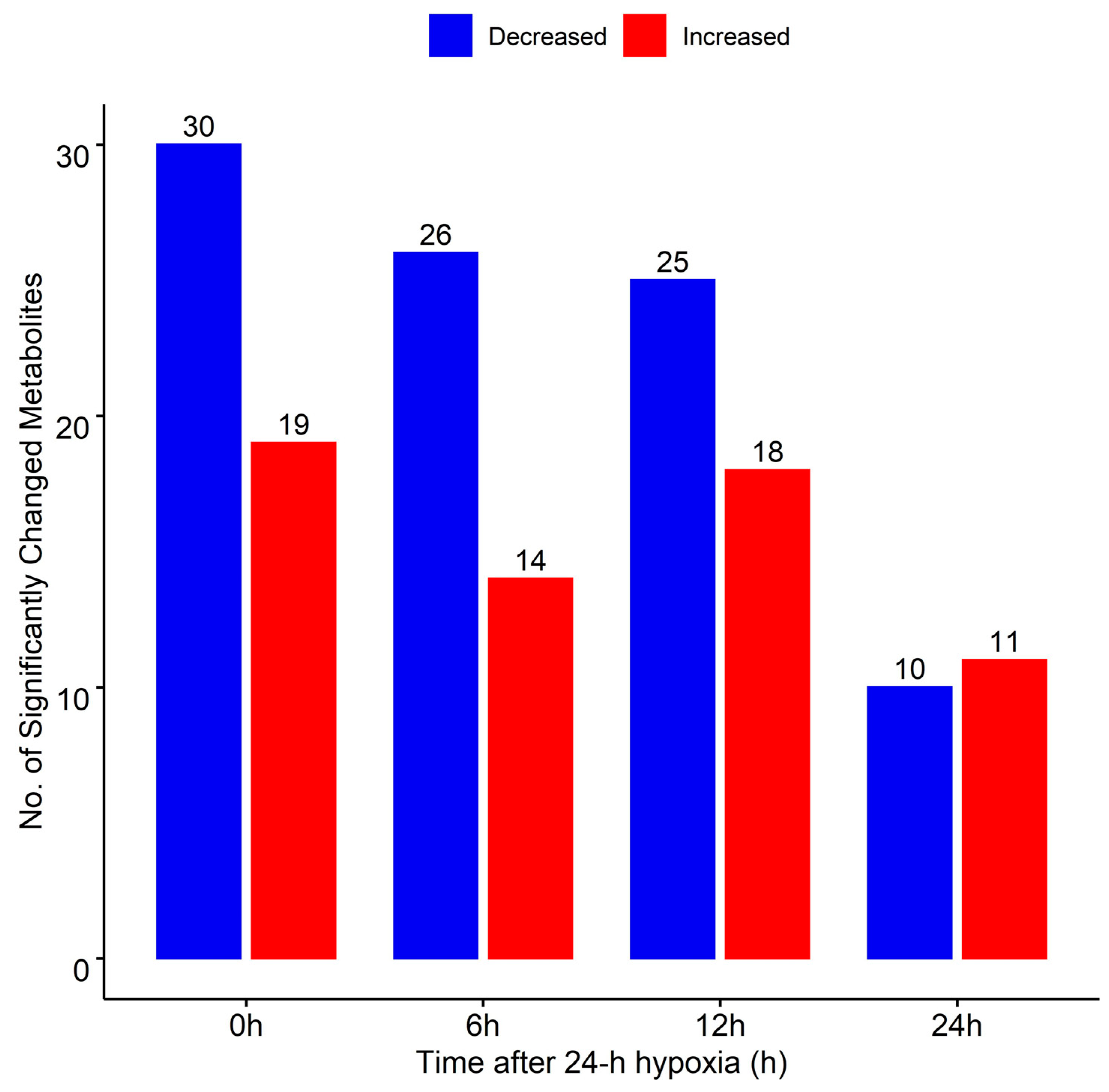
2. Results
3. Discussion
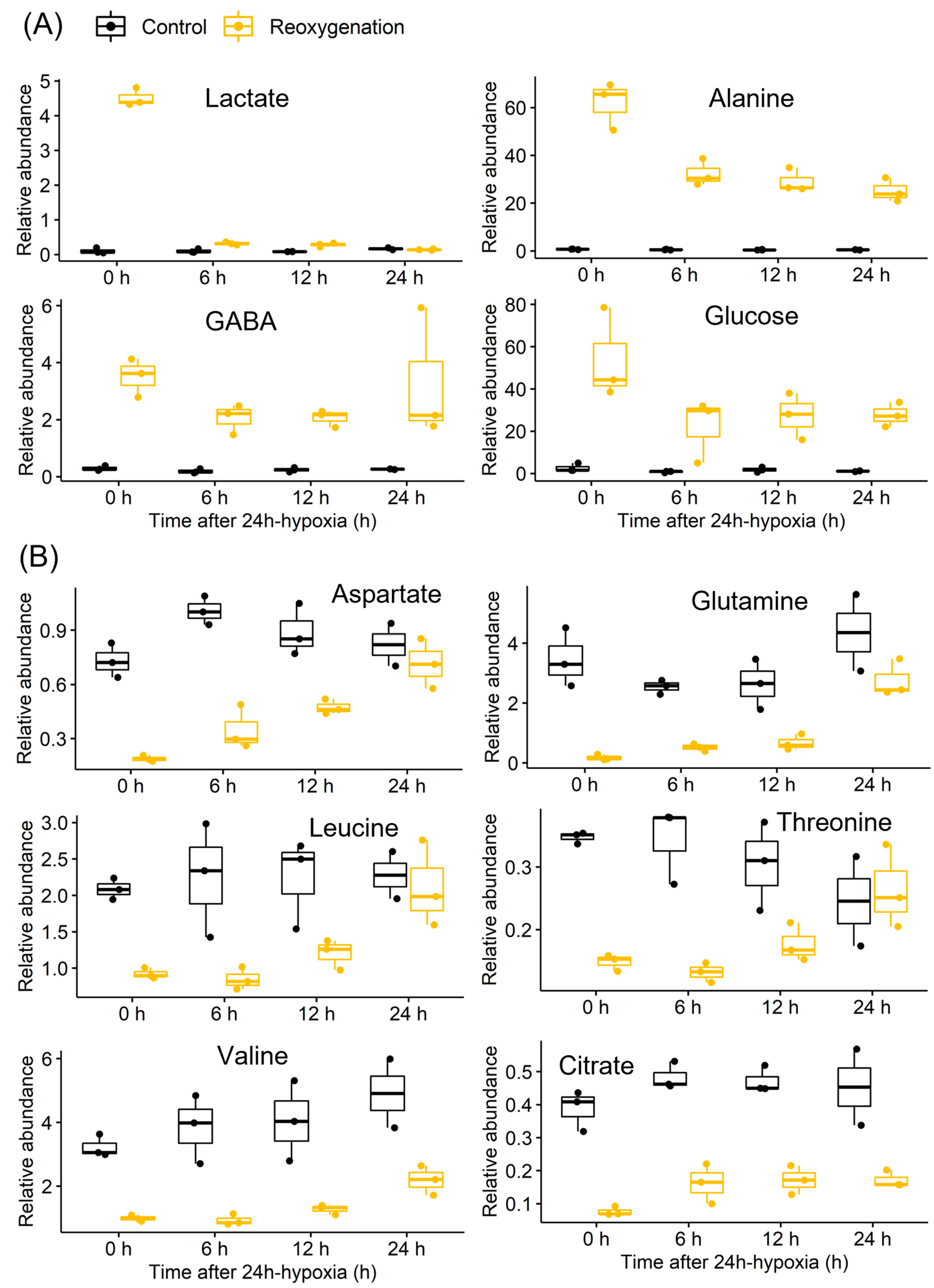
3.1. Ethanol and Lactate Fermentation
3.2. Nitrogen Metabolism
3.3. Mitochondrial Metabolism
3.4. Sugar Metabolism
3.5. Energy and Purine Metabolism
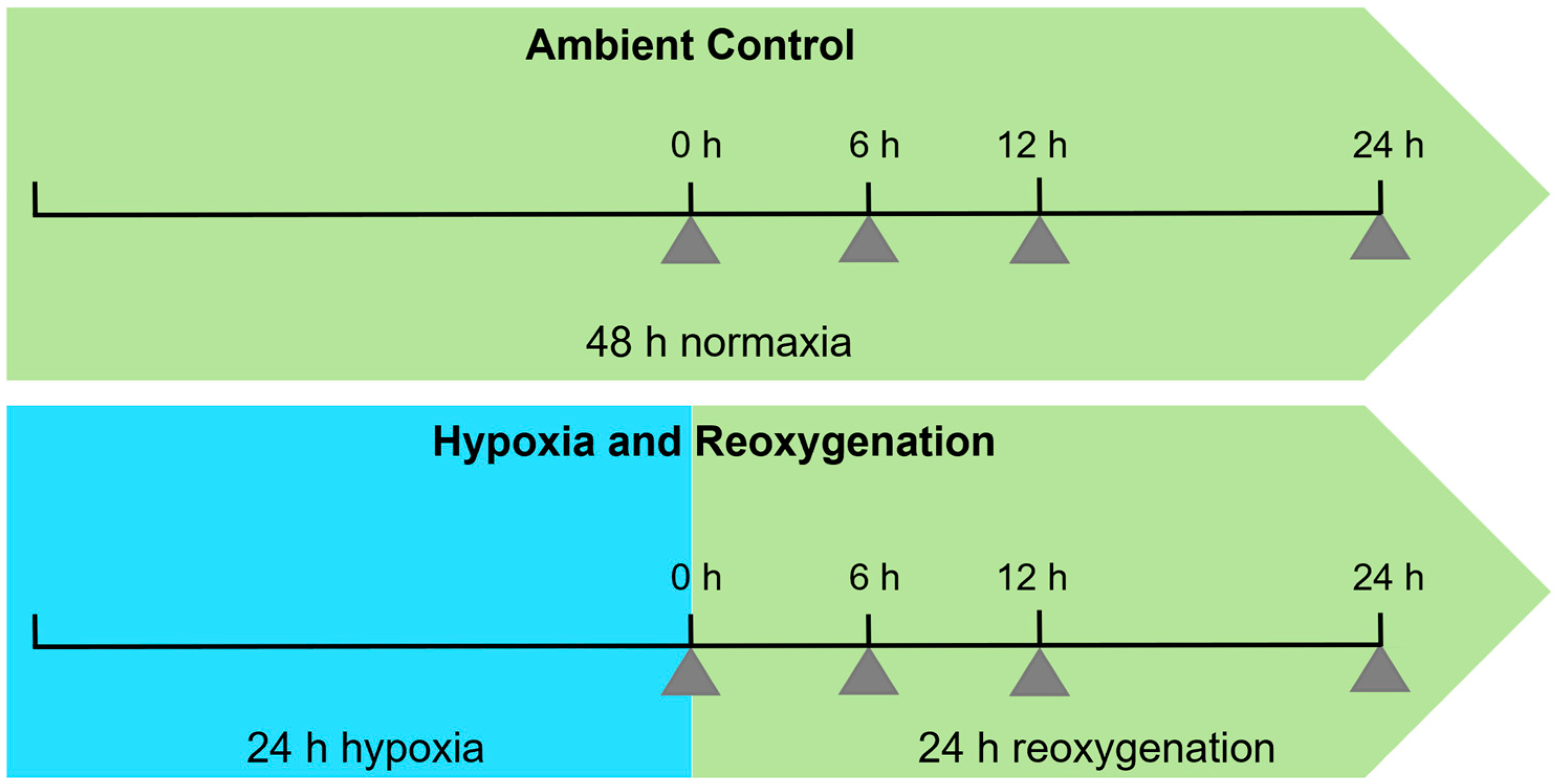
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Metabolite Extraction, Derivatization, and GC-MS Analysis
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, X.; Peng, S.; Yang, S.; Chen, Y.; Zhang, J.; Mo, W.; Zhu, J.; Ye, Y.; Huang, X. Effects of Flooding on Grafted Annona Plants of Different Scion/Rootstock Combinations. Agric. Sci. 2012, 3, 249–256. [Google Scholar] [CrossRef]
- Nakamura, M.; Noguchi, K. Tolerant Mechanisms to O2 Deficiency under Submergence Conditions in Plants. J. Plant Res. 2020, 133, 343–371. [Google Scholar] [CrossRef]
- Weits, D.A.; van Dongen, J.T.; Licausi, F. Molecular Oxygen as a Signaling Component in Plant Development. New Phytol. 2021, 229, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Walbot, V. Hypoxia Triggers Meiotic Fate Acquisition in Maize. Science 2012, 337, 345–348. [Google Scholar] [CrossRef]
- Deutsch, C.; Ferrel, A.; Seibel, B.; Pörtner, H.O.; Huey, R.B. Climate Change Tightens a Metabolic Constraint on Marine Habitats. Science 2015, 348, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.; Bailey-Serres, J.; Sasidharan, R. After The Deluge: Plant Revival Post-Flooding. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Gibbs, D.J.; MdIsa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-delaRosa, N.; VicenteConde, J.; SousaCorreia, C.; Pearce, S.P.; et al. Nitric Oxide Sensing in Plants Is Mediated by Proteolytic Control of Group VII ERF Transcription Factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Considine, M.J.; Diaz-Vivancos, P.; Kerchev, P.; Signorelli, S.; Agudelo-Romero, P.; Gibbs, D.J.; Foyer, C.H. Learning to Breathe: Developmental Phase Transitions in Oxygen Status. Trends Plant Sci. 2017, 22, 140–153. [Google Scholar] [CrossRef]
- Le Gac, A.L.; Laux, T. Hypoxia Is a Developmental Regulator in Plant Meristems. Mol. Plant 2019, 12, 1422–1424. [Google Scholar] [CrossRef]
- Abbas, M.; Berckhan, S.; Rooney, D.J.; Gibbs, D.J.; Vicente Conde, J.; Sousa Correia, C.; Bassel, G.W.; Marín-De La Rosa, N.; León, J.; Alabadí, D.; et al. Oxygen Sensing Coordinates Photomorphogenesis to Facilitate Seedling Survival. Curr. Biol. 2015, 25, 1483–1488. [Google Scholar] [CrossRef]
- Herzog, M.; Striker, G.G.; Colmer, T.D.; Pedersen, O. Mechanisms of Waterlogging Tolerance in Wheat—A Review of Root and Shoot Physiology. Plant Cell Environ. 2016, 39, 1068–1086. [Google Scholar] [CrossRef] [PubMed]
- Sjøgaard, K.S.; Valdemarsen, T.B.; Treusch, A.H. Responses of an Agricultural Soil Microbiome to Flooding with Seawater after Managed Coastal Realignment. Microorganisms 2018, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Zhong, J.; Ding, H.; Zheng, X.; Wang, Z.; Zhang, Y. Water-Level Alterations Modified Nitrogen Cycling across Sediment-Water Interface in the Three Gorges Reservoir. Environ. Sci. Pollut. Res. 2020, 27, 25886–25898. [Google Scholar] [CrossRef]
- Granger, D.N.; Kvietys, P.R. Reperfusion Injury and Reactive Oxygen Species: The Evolution of a Concept. Redox Biol. 2015, 6, 524–551. [Google Scholar] [CrossRef]
- Mallet, R.T.; Manukhina, E.B.; Ruelas, S.S.; Caffrey, J.L.; Downey, H.F. Cardioprotection by Intermittent Hypoxia Conditioning: Evidence, Mechanisms, and Therapeutic Potential. Am. J. Physiol.-Hear. Circ. Physiol. 2018, 315, H216–H232. [Google Scholar] [CrossRef]
- Tamang, B.G.; Fukao, T. Plant Adaptation to Multiple Stresses during Submergence and Following Desubmergence. Int. J. Mol. Sci. 2015, 16, 30164–30180. [Google Scholar] [CrossRef]
- Phukan, U.J.; Mishra, S.; Shukla, R.K. Waterlogging and Submergence Stress: Affects and Acclimation. Crit. Rev. Biotechnol. 2016, 36, 956–966. [Google Scholar] [CrossRef]
- Van Den Dries, N.; Gianní, S.; Czerednik, A.; Krens, F.A.; De Klerk, G.J.M. Flooding of the Apoplast Is a Key Factor in the Development of Hyperhydricity. J. Exp. Bot. 2013, 64, 5221–5230. [Google Scholar] [CrossRef]
- León, J.; Castillo, M.C.; Gayubas, B. The Hypoxia-Reoxygenation Stress in Plants. J. Exp. Bot. 2021, 72, 5841–5856. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kuroha, T.; Nagai, K.; Hattori, Y.; Kobayashi, M.; Nishizawa, T.; Kojima, M.; Utsumi, Y.; Oikawa, A.; Seki, M.; et al. Metabolite and Phytohormone Profiling Illustrates Metabolic Reprogramming as an Escape Strategy of Deepwater Rice during Partially Submerged Stress. Metabolites 2020, 10, 68. [Google Scholar] [CrossRef]
- Hwang, J.H.; Yu, S.I.; Lee, B.H.; Lee, D.H. Modulation of Energy Metabolism Is Important for Low-Oxygen Stress Adaptation in Brassicaceae Species. Int. J. Mol. Sci. 2020, 21, 1787. [Google Scholar] [CrossRef]
- Mignolli, F.; Todaro, J.S.; Vidoz, M.L. Internal Aeration and Respiration of Submerged Tomato Hypocotyls Are Enhanced by Ethylene-Mediated Aerenchyma Formation and Hypertrophy. Physiol. Plant. 2020, 169, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Rizi, V.; Herde, M.; Stöhr, C. RNA-Seq Reveals Novel Genes and Pathways Associated with Hypoxia Duration and Tolerance in Tomato Root. Sci. Rep. 2020, 10, 1692. [Google Scholar] [CrossRef] [PubMed]
- Shingaki-Wells, R.; Millar, A.H.; Whelan, J.; Narsai, R. What Happens to Plant Mitochondria under Low Oxygen? An Omics Review of the Responses to Low Oxygen and Reoxygenation. Plant Cell Environ. 2014, 37, 2260–2277. [Google Scholar] [CrossRef]
- Bui, L.T.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Licausi, F. Constitutively Expressed ERF-VII Transcription Factors Redundantly Activate the Core Anaerobic Response in Arabidopsis Thaliana. Plant Sci. 2015, 236, 37–43. [Google Scholar] [CrossRef]
- Papdi, C.; Pérez-Salamõ, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C.; Szabados, L. The Low Oxygen, Oxidative and Osmotic Stress Responses Synergistically Act through the Ethylene Response Factor VII Genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef]
- Licausi, F.; Van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, Two Hypoxia-Inducible Ethylene Response Factors, Affect Anaerobic Responses in Arabidopsis Thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Park, H.Y.; Seok, H.Y.; Woo, D.H.; Lee, S.Y.; Tarte, V.N.; Lee, E.H.; Lee, C.H.; Moon, Y.H. AtERF71/HRE2 Transcription Factor Mediates Osmotic Stress Response as Well as Hypoxia Response in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 414, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Hsu, F.C.; Li, J.P.; Wang, N.N.; Shih, M.C. The AP2/ERF Transcription Factor AtERF73/HRE1 Modulates Ethylene Responses during Hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Giuntoli, B.; Shukla, V.; Maggiorelli, F.; Giorgi, F.M.; Lombardi, L.; Perata, P.; Licausi, F. Age-Dependent Regulation of ERF-VII Transcription Factor Activity in Arabidopsis Thaliana. Plant Cell Environ. 2017, 40, 2333–2346. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, D. The Role of Long Noncoding Rnas in Plant Stress Tolerance. Methods Mol. Biol. 2017, 1631, 41–68. [Google Scholar] [PubMed]
- Moldovan, D.; Spriggs, A.; Yang, J.; Pogson, B.J.; Dennis, E.S.; Wilson, I.W. Hypoxia-Responsive MicroRNAs and Trans-Acting Small Interfering RNAs in Arabidopsis. J. Exp. Bot. 2010, 61, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Howell, K.A.; Carroll, A.; Ivanova, A.; Millar, H.; Whelan, J. Defining Core Metabolic and Transcriptomic Responses to Oxygen Availability in Rice Embryos and Young Seedlings. Plant Physiol. 2009, 151, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Secco, D.; Schultz, M.D.; Ecker, J.R.; Lister, R.; Whelan, J. Dynamic and Rapid Changes in the Transcriptome and Epigenome during Germination and in Developing Rice (Oryza Sativa) Coleoptiles under Anoxia and Re-Oxygenation. Plant J. 2017, 89, 805–824. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.H.; Larive, C.K.; Bailey-Serres, J. Selective MRNA Translation Coordinates Energetic and Metabolic Adjustments to Cellular Oxygen Deprivation and Reoxygenation in Arabidopsis Thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Rohr, K. Oxygen Is the High-Energy Molecule Powering Complex Multicellular Life: Fundamental Corrections to Traditional Bioenergetics. ACS Omega 2020, 5, 2221–2233. [Google Scholar] [CrossRef]
- Bui, L.T.; Novi, G.; Lombardi, L.; Iannuzzi, C.; Rossi, J.; Santaniello, A.; Mensuali, A.; Corbineau, F.; Giuntoli, B.; Perata, P.; et al. Conservation of Ethanol Fermentation and Its Regulation in Land Plants. J. Exp. Bot. 2019, 70, 1815–1827. [Google Scholar] [CrossRef]
- Cho, H.Y.; Loreti, E.; Shih, M.C.; Perata, P. Energy and Sugar Signaling during Hypoxia. New Phytol. 2021, 229, 57–63. [Google Scholar] [CrossRef]
- Mustroph, A.; Barding, G.A.; Kaiser, K.A.; Larive, C.K.; Bailey-Serres, J. Characterization of Distinct Root and Shoot Responses to Low-Oxygen Stress in Arabidopsis with a Focus on Primary C- and N-Metabolism. Plant Cell Environ. 2014, 37, 2366–2380. [Google Scholar] [CrossRef]
- Wany, A.; Gupta, A.K.; Kumari, A.; Mishra, S.; Singh, N.; Pandey, S.; Vanvari, R.; Igamberdiev, A.U.; Fernie, A.R.; Gupta, K.J. Nitrate Nutrition Influences Multiple Factors in Order to Increase Energy Efficiency under Hypoxia in Arabidopsis. Ann. Bot. 2019, 123, 691–705. [Google Scholar] [CrossRef]
- Gupta, P.; De, B. Metabolomics Analysis of Rice Responses to Salinity Stress Revealed Elevation of Serotonin, and Gentisic Acid Levels in Leaves of Tolerant Varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Mur, L.A.J.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The Role of Nitrite and Nitric Oxide under Low Oxygen Conditions in Plants. New Phytol. 2020, 225, 1143–1151. [Google Scholar] [CrossRef]
- Liu, B.; Rennenberg, H.; Kreuzwieser, J. Hypoxia Affects Nitrogen Uptake and Distribution in Young Poplar (Populus × Canescens) Trees. PLoS ONE 2015, 10, e0136579. [Google Scholar] [CrossRef] [PubMed]
- Horchani, F.; Aschi-Smiti, S. Prolonged Root Hypoxia Effects on Enzymes Involved in Nitrogen Assimilation Pathway in Tomato Plants. Plant Signal. Behav. 2010, 5, 1583–1589. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.C.; Freschi, L.; Sodek, L. Nitrogen Metabolism and Translocation in Soybean Plants Subjected to Root Oxygen Deficiency. Plant Physiol. Biochem. 2013, 66, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Mommer, L.; Visser, E.J.W. Underwater Photosynthesis in Flooded Terrestrial Plants: A Matter of Leaf Plasticity. Ann. Bot. 2005, 96, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for Plant Stress Response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y.; Walker, B.J. The Metabolic Origins of Non-Photorespiratory CO2 Release during Photosynthesis: A Metabolic Flux Analysis. Plant Physiol. 2021, 186, 297–314. [Google Scholar] [CrossRef]
- Xu, Y.; Wieloch, T.; Kaste, J.A.M.; Shachar-Hill, Y.; Sharkey, T.D. Reimport of Carbon from Cytosolic and Vacuolar Sugar Pools into the Calvin-Benson Cycle Explains Photosynthesis Labeling Anomalies. Proc. Natl. Acad. Sci. USA 2022, 119, e2121531119. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A Metabolomics View. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making Sense of Low Oxygen Sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Dupuis, I.; Kuhlemeier, C. Ethanolic Fermentation: New Functions for an Old Pathway. Trends Plant Sci. 1999, 4, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Meguro, N.; Suzuki, Y.; Tsutsumi, N.; Hirai, A.; Nakazono, M. Induction of Mitochondrial Aldehyde Dehydrogenase by Submergence Facilitates Oxidation of Acetaldehyde during Re-Aeration in Rice. FEBS Lett. 2003, 546, 369–373. [Google Scholar] [CrossRef]
- Fu, X.; Yang, H.; Pangestu, F.; Nikolau, B.J. Failure to Maintain Acetate Homeostasis by Acetate-Activating Enzymes Impacts Plant Development. Plant Physiol. 2020, 182, 1256–1271. [Google Scholar] [CrossRef]
- Davies, D.D.; Grego, S.; Kenworthy, P. The Control of the Production of Lactate and Ethanol by Higher Plants. Planta 1974, 118, 297–310. [Google Scholar] [CrossRef]
- Tsai, K.J.; Lin, C.Y.; Ting, C.Y.; Shih, M.C. Ethylene-Regulated Glutamate Dehydrogenase Fine-Tunes Metabolism during Anoxia-Reoxygenation. Plant Physiol. 2016, 172, 1548–1562. [Google Scholar] [CrossRef]
- Diab, H.; Limami, A.M. Reconfiguration of N Metabolism upon Hypoxia Stress and Recovery: Roles of Alanine Aminotransferase (AlaAt) and Glutamate Dehydrogenase (GDH). Plants 2016, 5, 25. [Google Scholar] [CrossRef]
- António, C.; Päpke, C.; Rocha, M.; Diab, H.; Limami, A.M.; Obata, T.; Fernie, A.R.; van Dongen, J.T. Regulation of Primary Metabolism in Response to Low Oxygen Availability as Revealed by Carbon and Nitrogen Isotope Redistribution1[OPEN]. Plant Physiol. 2016, 170, 43–56. [Google Scholar] [CrossRef]
- Walker, B.J.; Kramer, D.M.; Fisher, N.; Fu, X. Flexibility in the Energy Balancing Network of Photosynthesis Enables Safe Operation under Changing Environmental Conditions. Plants 2020, 9, 301. [Google Scholar] [CrossRef]
- Xu, Y.; Freund, D.M.; Hegeman, A.D.; Cohen, J.D. Metabolic Signatures of Arabidopsis Thaliana Abiotic Stress Responses Elucidate Patterns in Stress Priming, Acclimation, and Recovery. Stress Biol. 2022, 2, 1238812. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Huang, X.; Cui, J.; Shabala, L.; Zhou, M.; Yu, M.; Shabala, S. Hypoxia-Induced Increase in GABA Content Is Essential for Restoration of Membrane Potential and Preventing ROS-Induced Disturbance to Ion Homeostasis. Plant Commun. 2021, 2, 100188. [Google Scholar] [CrossRef] [PubMed]
- Reggiani, R.; Mattana, M.; Aurisano, N.; Bertani, A. Utilization of Stored Nitrate during the Anaerobic Germination of Rice Seeds. Plant Cell Physiol. 1993, 34, 379–383. [Google Scholar] [CrossRef]
- Reggiani, R.; Nebuloni, M.; Mattana, M.; Brambilla, I. Anaerobic Accumulation of Amino Acids in Rice Roots: Role of the Glutamine Synthetase/Glutamate Synthase Cycle. Amino Acids 2000, 18, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Howell, K.A.; Heazlewood, J.L.; Tan, T.Y.W.; Narsai, R.; Huang, S.; Whelan, J.; Millar, A.H. Analysis of the Rice Mitochondrial Carrier Family Reveals Anaerobic Accumulation of a Basic Amino Acid Carrier Involved in Arginine Metabolism during Seed Germination. Plant Physiol. 2010, 154, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Couée, I.; Defontaine, S.; Carde, J.P.; Pradet, A. Effects of Anoxia on Mitochondrial Biogenesis in Rice Shoots: Modification of in Organello Translation Characteristics. Plant Physiol. 1992, 98, 411–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Howell, K.A.; Cheng, K.; Murcha, M.W.; Jenkin, L.E.; Millar, A.H.; Whelan, J. Oxygen Initiation of Respiration and Mitochondrial Biogenesis in Rice. J. Biol. Chem. 2007, 282, 15619–15631. [Google Scholar] [CrossRef]
- Narsai, R.; Rocha, M.; Geigenberger, P.; Whelan, J.; Van Dongen, J.T. Comparative Analysis between Plant Species of Transcriptional and Metabolic Responses to Hypoxia. New Phytol. 2011, 190, 472–487. [Google Scholar] [CrossRef]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-Kingdom Comparison of Transcriptomic Adjustments to Low-Oxygen Stress Highlights Conserved and Plant-Specific Responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef]
- Millar, A.H.; Trend, A.E.; Heazlewood, J.L. Changes in the Mitochondrial Proteome during the Anoxia to Air Transition in Rice Focus around Cytochrome-Containing Respiratory Complexes. J. Biol. Chem. 2004, 279, 39471–39478. [Google Scholar] [CrossRef]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript Profiling of the Anoxic Rice Coleoptile. Plant Physiol. 2007, 144, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Shingaki-Wells, R.N.; Huang, S.; Taylor, N.L.; Carroll, A.J.; Zhou, W.; Harvey Miller, A. Differential Molecular Responses of Rice and Wheat Coleoptiles to Anoxia Reveal Novel Metabolic Adaptations in Amino Acid Metabolism for Tissue Tolerance. Plant Physiol. 2011, 156, 1706–1724. [Google Scholar] [CrossRef]
- Rocha, M.; Licausi, F.; Araújo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; van Dongen, J.T. Glycolysis and the Tricarboxylic Acid Cycle Are Linked by Alanine Aminotransferase during Hypoxia Induced by Waterlogging of Lotus Japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Menegus, F.; Cattaruzza, L.; Chersi, A.; Fronza, G. Differences in the Anaerobic Lactate-Succinate Production and in the Changes of Cell Sap PH for Plants with High and Low Resistance to Anoxia. Plant Physiol. 1989, 90, 29–32. [Google Scholar] [CrossRef]
- Fox, T.C.; Kennedy, R.A. Mitochondrial Enzymes in Aerobically and Anaerobically Germinated Seedlings of Echinochloa and Rice. Planta 1991, 184, 510–514. [Google Scholar] [CrossRef]
- Shelp, B.J.; Bown, A.W.; McLean, M.D. Metabolism and Functions of Gamma-Aminobutyric Acid. Trends Plant Sci. 1999, 4, 446–452. [Google Scholar] [CrossRef]
- Guan, X.; Nikolau, B.J. AAE13 Encodes a Dual-Localized Malonyl-CoA Synthetase That Is Crucial for Mitochondrial Fatty Acid Biosynthesis. Plant J. 2016, 85, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Garnczarska, M.; Bednarski, W.; Morkunas, I. Re-Aeration—Induced Oxidative Stress and Antioxidative Defenses in Hypoxically Pretreated Lupine Roots. J. Plant Physiol. 2004, 161, 415–422. [Google Scholar] [CrossRef]
- Fu, X.; Guan, X.; Garlock, R.; Nikolaua, B.J. Mitochondrial Fatty Acid Synthase Utilizes Multiple Acyl Carrier Protein Isoforms. Plant Physiol. 2020, 183, 547–557. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A Is an Ethylene-Response-Factor-like Gene That Confers Submergence Tolerance to Rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The Ethylene Response Factors SNORKEL1 and SNORKEL2 Allow Rice to Adapt to Deep Water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An Ethylene Response Transcription Factor That Is Important for Hypoxia Survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Vartapetian, B.B.; Andreeva, I.N.; Kozlova, G.I. The Resistance to Anoxia and the Mitochondrial Fine Structure of Rice Seedlings. Protoplasma 1976, 88, 215–224. [Google Scholar] [CrossRef]
- Bieniawska, Z.; Paul Barratt, D.H.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the Sucrose Synthase Gene Family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef] [PubMed]
- Santaniello, A.; Loreti, E.; Gonzali, S.; Novi, G.; Perata, P. A Reassessment of the Role of Sucrose Synthase in the Hypoxic Sucrose-Ethanol Transition in Arabidopsis. Plant Cell Environ. 2014, 37, 2294–2302. [Google Scholar] [CrossRef]
- Loreti, E.; Valeri, M.C.; Novi, G.; Perata, P. Gene Regulation and Survival under Hypoxia Requires Starch Availability and Metabolism. Plant Physiol. 2018, 176, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Ciccarelli, R.; Di Iorio, P.; Giuliani, P.; D’Alimonte, I.; Ballerini, P.; Caciagli, F.; Rathbone, M.P. Rat Cultured Astrocytes Release Guanine-Based Purines in Basal Conditions and after Hypoxia/Hypoglycemia. Glia 1999, 25, 93–98. [Google Scholar] [CrossRef]
- McVey, P.A.; Alexander, L.E.; Fu, X.; Xie, B.; Galayda, K.J.; Nikolau, B.J.; Houk, R.S. Light-Dependent Changes in the Spatial Localization of Metabolites in Solenostemon Scutellarioides (Coleus Henna) Visualized by Matrix-Free Atmospheric Pressure Electrospray Laser Desorption Ionization Mass Spectrometry Imaging. Front. Plant Sci. 2018, 9, 1348. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Xu, Y. Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery. Stresses 2023, 3, 86-101. https://doi.org/10.3390/stresses3010008
Fu X, Xu Y. Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery. Stresses. 2023; 3(1):86-101. https://doi.org/10.3390/stresses3010008
Chicago/Turabian StyleFu, Xinyu, and Yuan Xu. 2023. "Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery" Stresses 3, no. 1: 86-101. https://doi.org/10.3390/stresses3010008
APA StyleFu, X., & Xu, Y. (2023). Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery. Stresses, 3(1), 86-101. https://doi.org/10.3390/stresses3010008




