Quinaldehyde o-Nitrobenzoylhydrazone: Structure and Sensitization of HepG2 Cells to Anti-Cancer Drugs
Abstract
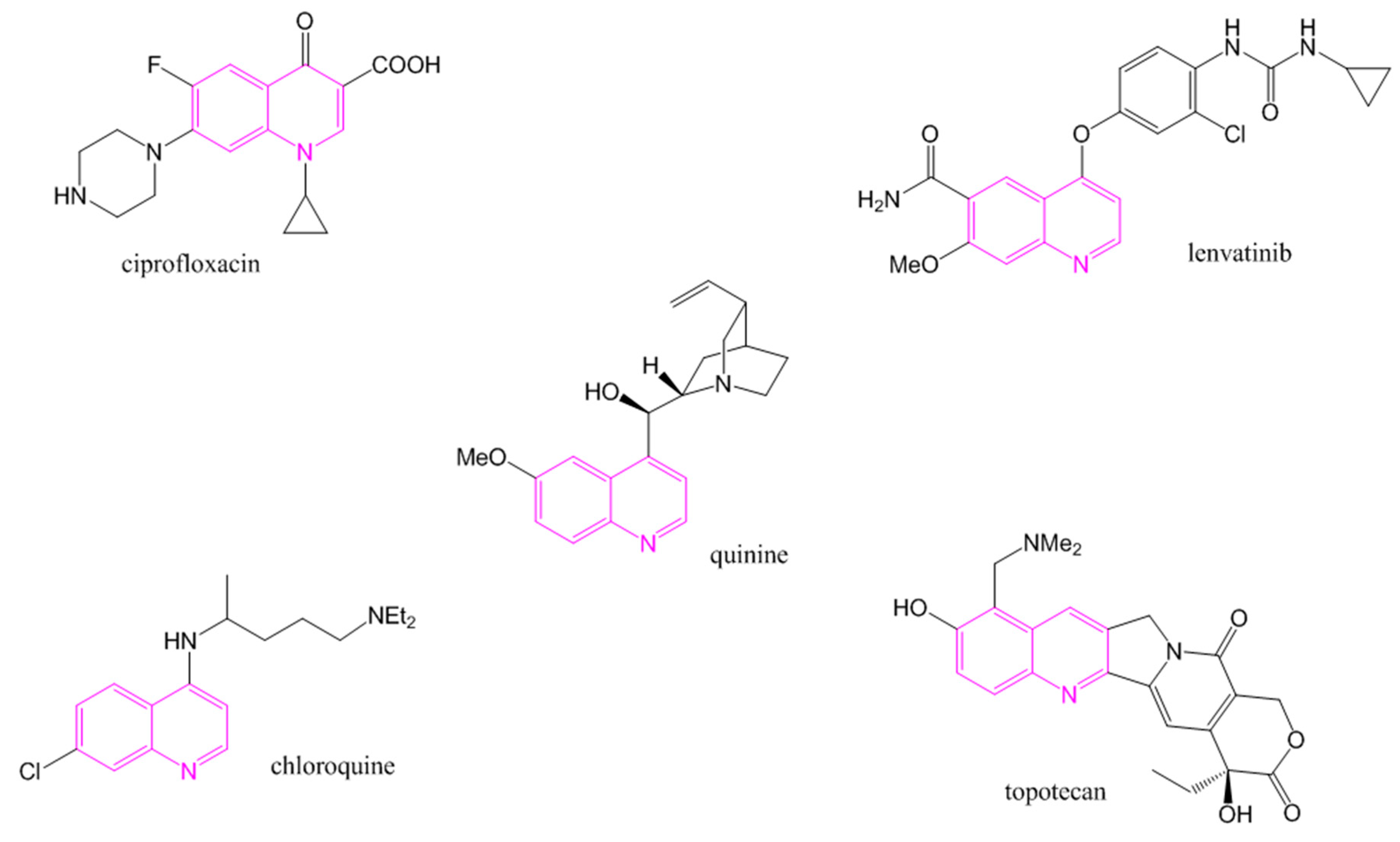
1. Introduction
2. Materials and Methods
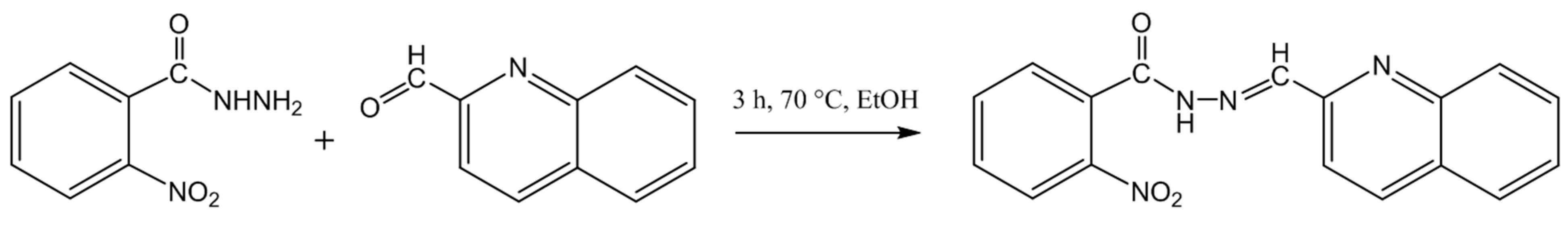
2.1. Synthesis and Crystallization of 2-Nitro-N’-(Quinolin-2-ylmethylene)benzohydrazide
2.2. Mass-Spectrometric and NMR Characterization
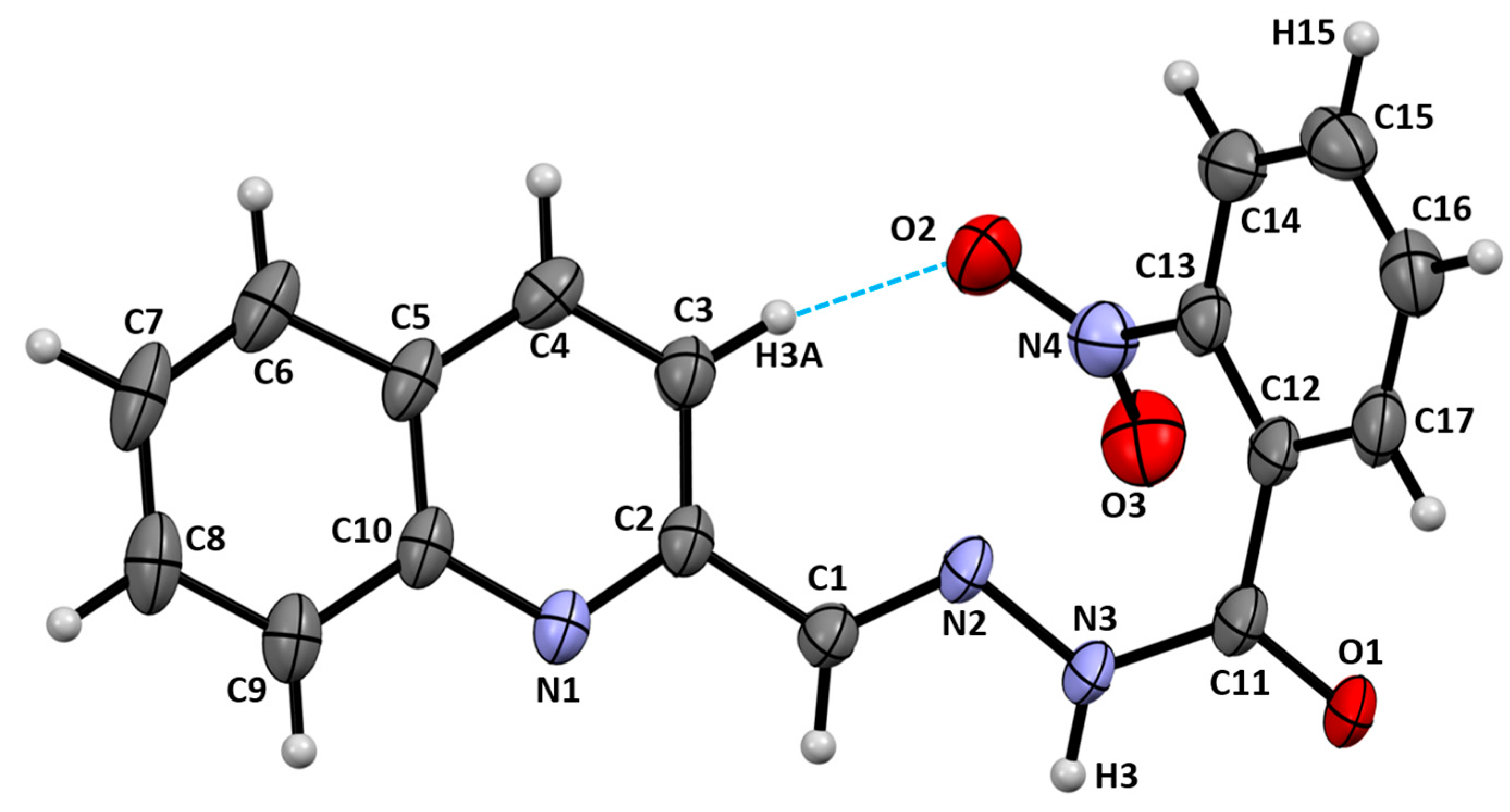
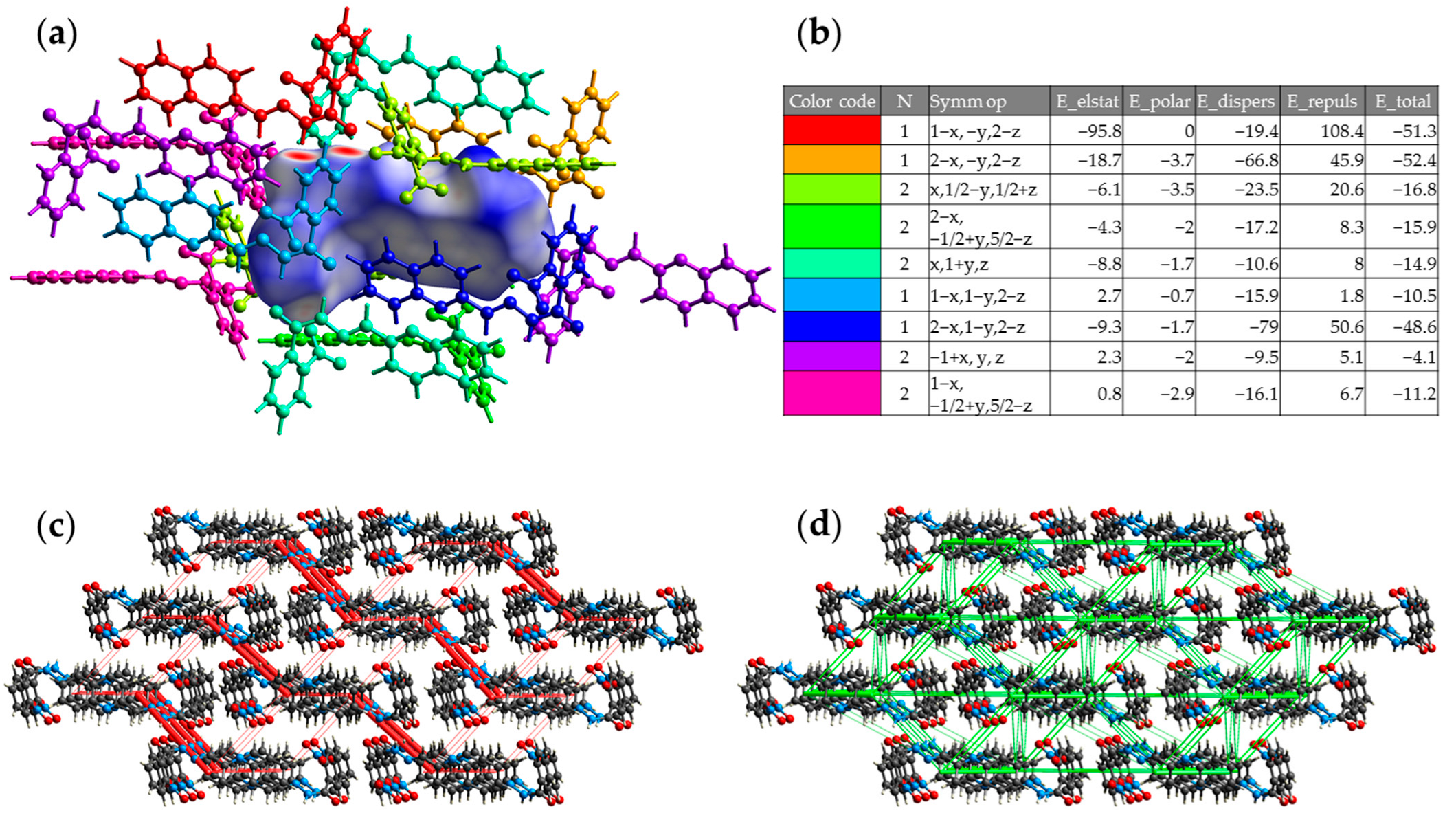
2.3. X-Ray Diffraction Studies
2.4. Cellular Proliferation Assay
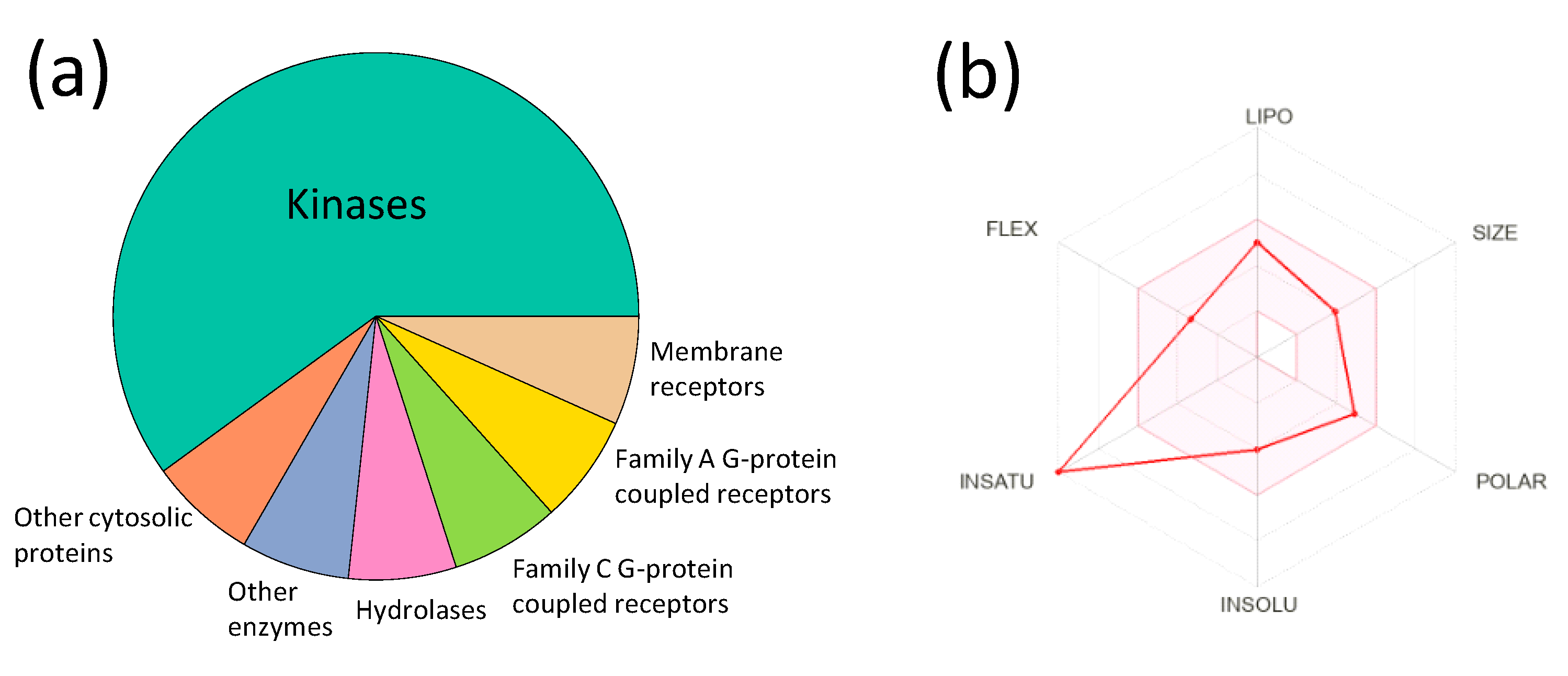
2.5. Molecular Modeling, Dose-Effect Analysis, ADME Predictions, and Statistical Analysis
3. Results
Interactions of QN and Cytotoxic Chemotherapeutic Agents in HepG2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.Y.; Danpanichkul, P.; Yong, J.N.; Yu, Z.; Tan, D.J.; Lim, W.H.; Koh, B.; Lim, R.Y.; Tham, E.K.; Mitra, K.; et al. Liver cancer in 2021: Global burden of disease study. J. Hepatol. 2025, 82, 851–860. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, H.; Zhang, L.; Zhu, A.X.; Bernards, R.; Qin, W.; Wang, C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 203–222. [Google Scholar] [CrossRef]
- PDQ Pediatric Treatment Editorial Board. Childhood Liver Cancer Treatment (PDQ®): Health Professional Version. In PDQ Cancer Information Summaries [Internet]; National Cancer Institute (US): Bethesda, MD, USA, 2005. Available online: https://www.ncbi.nlm.nih.gov/books/NBK65790/ (accessed on 7 June 2025).
- Avendaño, C.; Menéndez, J.C. Medicinal Chemistry of Anticancer Drugs, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 655–700. [Google Scholar]
- Al-Lazikani, B.; Banerji, U.; Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012, 30, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Vinod, B.S.; Maliekal, T.T.; Anto, R.J. Phytochemicals as chemosensitizers: From molecular mechanism to clinical significance. Antioxid. Redox Signal. 2013, 18, 1307–1348. [Google Scholar] [CrossRef] [PubMed]
- Verbaanderd, C.; Pantziarka, P.; Bouche, G.; Maes, H.; Schaaf Marco, B.; Agostinis, P.; Sukhatme Vikas, P.; Sukhatme, V. Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. eCancer 2017, 11, 781. [Google Scholar] [CrossRef]
- Ferreira, P.M.P.; Ferreira, J.R.d.O.; de Sousa, R.W.R.; Bezerra, D.P.; Militão, G.C.G. Aminoquinolines as translational models for drug repurposing: Anticancer adjuvant properties and toxicokinetic-related features. J. Oncol. 2021, 2021, 3569349. [Google Scholar] [CrossRef]
- Zuma, N.H.; Aucamp, J.; N’Da, D.D. An update on derivatisation and repurposing of clinical nitrofuran drugs. Eur. J. Pharm. Sci. 2019, 140, 105092. [Google Scholar] [CrossRef]
- Lopes, M.S.; Sena, C.F.d.A.; Silva, B.L.; de Souza, C.M.; Ramos, J.P.; Cassali, G.D.; de Souza-Fagundes, E.M.; Alves, R.J.; de Oliveira, M.C.; de Oliveira, R.B. Synthesis of nitroaromatic compounds as potential anticancer agents. Anti-Cancer Agents Med. Chem. 2015, 15, 206–216. [Google Scholar] [CrossRef]
- Song, P.; Yao, X.; Zhong, T.; Zhang, S.; Guo, Y.; Ren, W.; Huang, D.; Duan, X.-C.; Yin, Y.-F.; Zhang, S.-S.; et al. The anti-tumor efficacy of 3-(2-nitrophenyl) propionic acid-paclitaxel (NPPA-PTX): A novel paclitaxel bioreductive prodrug. Oncotarget 2016, 7, 48467–48480. [Google Scholar] [CrossRef]
- Mossine, V.V.; Chopra, P.; Mawhinney, T.P. Interaction of tomato lycopene and ketosamine against rat prostate tumorigenesis. Cancer Res. 2008, 68, 4384–4391. [Google Scholar] [CrossRef] [PubMed]
- Mossine, V.V.; Kelley, S.P.; Mawhinney, T.P. Crystal structure of (E)-3-methoxy-N’-(1-(pyridin-2-yl)ethylidene) benzohydrazide, C15H15N3O2. Z. Kristallogr. New Crystal Struct. 2020, 235, 907–909. [Google Scholar] [CrossRef]
- Watanabe, K. Current chemotherapeutic approaches for hepatoblastoma. Int. J. Clin. Oncol. 2013, 18, 955–961. [Google Scholar] [CrossRef]
- Mossine, V.V.; Kelley, S.P.; Waters, J.K.; Mawhinney, T.P. Screening a small hydrazide-hydrazone combinatorial library for targeting the STAT3 in monocyte-macrophages with insulated reporter transposons. Med. Chem. Res. 2023, 32, 682–693. [Google Scholar] [CrossRef]
- Sunder, S.; Peet, N.P.; Barbuch, R.J. A reinvestigation of the cyclodesulfurization of thiosemicarbazides. J. Heterocycl. Chem. 1981, 18, 1601–1604. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Mossine, V.V.; Waters, J.K.; Gu, Z.; Sun, G.Y.; Mawhinney, T.P. Bidirectional responses of eight neuroinflammation-related transcriptional factors to 64 flavonoids in astrocytes with transposable insulated signaling pathway reporters. ACS Chem. Neurosci. 2022, 13, 613–623. [Google Scholar] [CrossRef]
- MacKenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCr J. 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Lopes, A.B.; Miguez, E.; Kümmerle, A.E.; Rumjanek, V.M.; Fraga, C.A.M.; Barreiro, E.J. Characterization of amide bond conformers for a novel heterocyclic template of N-acylhydrazone derivatives. Molecules 2013, 18, 11683–11704. [Google Scholar] [CrossRef]
- Kadyan, K.; Singh, R.; Sindhu, J.; Kumar, P.; Devi, M.; Lal, S.; Kumar, A.; Singh, D.; Kumar, H. Exploring the structural versatility and dynamic behavior of acyl/aroyl hydrazones: A comprehensive review. Top. Curr. Chem. 2025, 383, 18. [Google Scholar] [CrossRef]
- Chou, T.-C. The median-effect principle and the combination index for quantitation of synergism and antagonism. In Synergism and Antagonism in Chemotherapy; Chou, T.-C., Rideout, D.C., Eds.; Academic Press: San Diego, CA, USA, 1991; pp. 61–102. [Google Scholar]
- Socea, L.-I.; Barbuceanu, S.-F.; Pahontu, E.M.; Dumitru, A.-C.; Nitulescu, G.M.; Sfetea, R.C.; Apostol, T.-V. Acylhydrazones and their biological activity: A review. Molecules 2022, 27, 8719. [Google Scholar] [CrossRef] [PubMed]
- Frei, P.; Pang, L.; Silbermann, M.; Eris, D.; Muehlethaler, T.; Schwardt, O.; Ernst, B. Target-directed dynamic combinatorial chemistry: A study on potentials and pitfalls as exemplified on a bacterial target. Chem. Eur. J. 2017, 23, 11570–11577. [Google Scholar] [CrossRef]
- Elrayess, R.; Darwish, K.M.; Nafie, M.S.; El-Sayyed, G.S.; Said, M.M.; Yassen, A.S.A. Quinoline-hydrazone hybrids as dual mutant EGFR inhibitors with promising metallic nanoparticle loading: Rationalized design, synthesis, biological investigation and computational studies. New J. Chem. 2022, 46, 18207–18232. [Google Scholar] [CrossRef]
- Ezzoukhry, Z.; Louandre, C.; Trecherel, E.; Godin, C.; Chauffert, B.; Dupont, S.; Diouf, M.; Barbare, J.-C.; Maziere, J.-C.; Galmiche, A. EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int. J. Cancer 2012, 131, 2961–2969. [Google Scholar] [CrossRef]


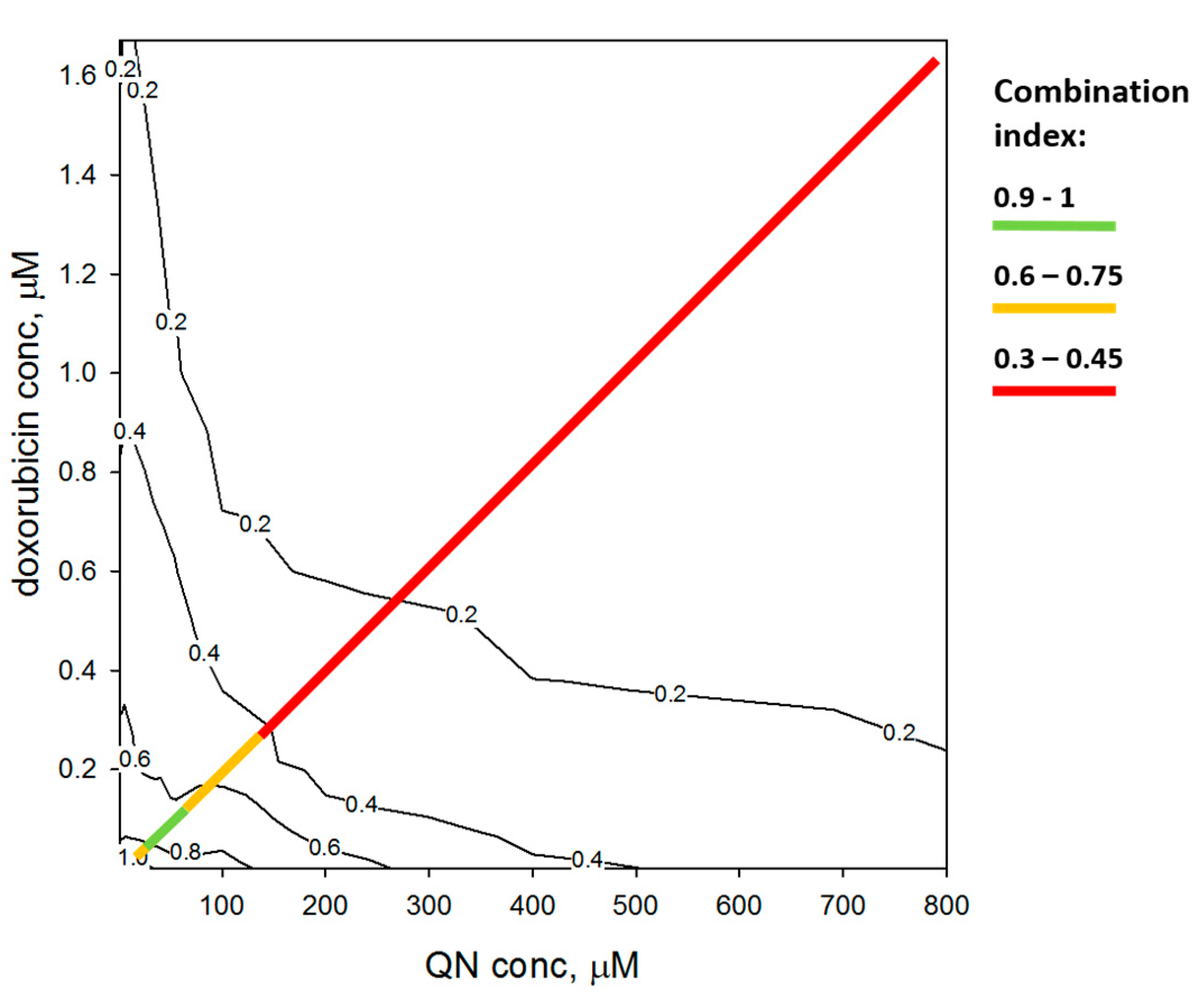
| Agent | Mechanism | Single Agent | Combination with QN | |||
|---|---|---|---|---|---|---|
| IC50, μM | r | IC50, μM | r | DRI | ||
| Doxorubicin | topoisomerase inh | 0.792 | 0.960 | 0.410 | 0.961 | 1.9 |
| Daunorubicin | topoisomerase inh | 0.650 | 0.954 | 0.319 | 0.966 | 2.0 |
| Epirubicin | topoisomerase inh | 3.00 | 0.980 | 1.16 | 0.941 | 2.6 |
| Idarubicin | topoisomerase inh | 0.213 | 0.935 | 0.155 | 0.953 | 1.4 |
| Mitoxanthrone | topoisomerase inh | 0.884 | 0.934 | 0.718 | 0.936 | 1.2 |
| Topotecan | topoisomerase inh | 4.47 | 0.947 | 2.27 | 0.967 | 2.0 |
| Etoposide | topoisomerase inh | 132 | 0.980 | 79.7 | 0.959 | 1.7 |
| Actinomycin D | topoisomerase inh | 0.015 | 0.990 | 0.0046 | 0.844 | 3.3 |
| Docetaxel | microtubule stabil | 0.145 | 0.953 | 0.070 | 0.909 | 2.1 |
| Vincristine | microtubule inh | 1.80 | 0.900 | 0.406 | 0.905 | 4.4 |
| Cisplatin | DNA crosslink | 1.59 | 0.954 | 2.41 | 0.941 | 0.7 |
| Carboplatin | DNA crosslink | 132 | 0.962 | 65.6 | 0.942 | 2.0 |
| Oxaliplatin | DNA crosslink | 17.4 | 0.982 | 24.3 | 0.975 | 0.7 |
| Mitomycin C | DNA crosslink | 4.26 | 0.993 | 2.23 | 0.987 | 1.9 |
| 5-Fluorouracil | antimetabolite | 137 | 0.929 | 140 | 0.983 | 1.0 |
| Sorafenib | multi-kinase inh | 13.2 | 0.876 | 7.55 | 0.948 | 1.7 |
| Vorinostat | HDAC inhibitor | 4.77 | 0.996 | 4.42 | 0.901 | 1.1 |
| QN | 418 | 0.998 | ||||
| Parameter | Value |
|---|---|
| GI absorption | High |
| BBB permeability | Low |
| P-glycoprotein substrate | No |
| CYP1A2 inhibitor | Yes |
| CYP2C19 inhibitor | Yes |
| CYP2C9 inhibitor | Yes |
| CYP2D6 inhibitor | No |
| CYP3A4 inhibitor | No |
| Log Kp (skin permeability) | −5.94 cm/s |
| Lipinski rule violations | 0 |
| PAINS alerts | 0 |
| Bioavailability score | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mossine, V.V.; Kelley, S.P.; Mawhinney, T.P. Quinaldehyde o-Nitrobenzoylhydrazone: Structure and Sensitization of HepG2 Cells to Anti-Cancer Drugs. Compounds 2025, 5, 24. https://doi.org/10.3390/compounds5030024
Mossine VV, Kelley SP, Mawhinney TP. Quinaldehyde o-Nitrobenzoylhydrazone: Structure and Sensitization of HepG2 Cells to Anti-Cancer Drugs. Compounds. 2025; 5(3):24. https://doi.org/10.3390/compounds5030024
Chicago/Turabian StyleMossine, Valeri V., Steven P. Kelley, and Thomas P. Mawhinney. 2025. "Quinaldehyde o-Nitrobenzoylhydrazone: Structure and Sensitization of HepG2 Cells to Anti-Cancer Drugs" Compounds 5, no. 3: 24. https://doi.org/10.3390/compounds5030024
APA StyleMossine, V. V., Kelley, S. P., & Mawhinney, T. P. (2025). Quinaldehyde o-Nitrobenzoylhydrazone: Structure and Sensitization of HepG2 Cells to Anti-Cancer Drugs. Compounds, 5(3), 24. https://doi.org/10.3390/compounds5030024






