Haplotypes of Echinococcus granulosus sensu stricto in Chile and Their Comparison Through Sequences of the Mitochondrial cox1 Gene with Haplotypes from South America and Other Continents
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Hydatid Cysts
2.2. DNA Extraction and Cox1 Gene Amplification
2.3. Sequence Analysis and Construction of Phylogenetic Trees
2.4. Cox1 Sequence Comparisons with GenBank Deposited Sequences
2.5. Diversity and Genetic Structuring of the Population
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMOVA | Molecular Analysis of Variance |
| cox1 | cytochrome oxidase subunit I |
| FST | Fixation index |
| G1-G3 | E. granulosus sensu stricto (sheep strain and buffalo strain) |
| G4 | E. equinus (horse strain) |
| G5 | E. ortleppi (bovine strain) |
| G6-G7 | E. intermedius |
| G8-G10 | E. canadensis |
| Hd | haplotypic diversity |
| K | number of haplotypes |
| mtDNA | mitochondrial DNA |
| N | number of samples |
| s.l. | sensu lato |
| s.s. | sensu stricto |
| S | number of segregation sites |
| π | nucleotide diversity |
Appendix A
| Locality/Country | Dog (HF) | Sheep (HF) | Goats (HF) | Bovine (HF) | Human (HF) |
|---|---|---|---|---|---|
| South America | |||||
| La Araucanía (IX), Maule (VII), Coquimbo (IV), Chile | KX227122 (EgCLP20) KX227123 (EgCLP21) | KX227116 (EgCLO11) KX227120 (EgCLO17) KX227126 (EgCLO22) MN399400 (EgCLO27) EgCLOM01-6,8-23* EgCLOCo07* KX227131(EgCL36) KX227125 (EgCL37) KX227121 (EgCL38) KX227124 (EgCL39) KX227119 (EgCL40) | KX227118 (EgCLC14) EgCLCCo03* | JQ250806.1 (EgCLB28) KT968704.1 (EgCLB05) KT968703 (EgCLB06) AB777904 (EgCLB08) KX227116 (EgCLB10) KX227117 (EgCLB13) KX227118 (EgCLB15) KX227120 (EgCLB18) KX227130 (EgCLB24) EgCLBA01-16,19,20* EgCLBCo17,18,21* AF458872 (EgCLB30) | U50464 (EgCLH01) M84666 (EgCLH02) AB522646 (EgCLH03) JQ250806.1 (EgCLH04) AB777904 (EgCLH07) KX227116 ((EgCLH09) KX227117 (EgCLH12) KX227118 (EgCLH16) KX227127 (EgCLH23) KX227128 (EgCLH25) KX227129 (EgCLH26) EgCLHCo01* AF458874 (EgCLH19) AF458875 (EgCLH29) KX227136 (EgCL31) KX227135 (EgCL32) KX227134 (EgCL33) KX227133 (EgCL34) KX227132 (EgCL35) |
| Perú | KT001408 (EgPeP09) KT001398 (EgPeP10) KT001407 (EgPeP11) KT001406 (EgPeP12) KT001405 (EgPeP13) KT001404 (EgPeP14) KT001403 (EgPeP15) KT001402 (EgPeP16) KT001401 (EgPeP17) KT001400 (EgPeP18) KT001399 (EgPeP19) KT001397 (EgPeP20) KT001396 (EgPeP21) KT001395 (EgPeP22) | AB458672 (EgPeO01) AB470527 (EgPeO02) AB688621 (EgPeO03) GU233854 (EgPeO04) GU23395 (EgPeO06) | MN732663 (EgPeLL23) AB458674 (EgPeB24) AB458673 (EgPeB23) AB688620 (EgPeB24) AB688621 (EgPeO25) | GU233952 (EgPeH05) AB458675 (EgPeH07) GU233951 (EgPeH08) | |
| Bolivia | JQ250806 (EgBolB01) KX227118 (EgBolB02) MT072973 (EgBolB03) MT072974 (EgBolB04) MT072975 (EgBolB05) MT072976 (EgBolB06) MT072977 (EgBolB07) MT072978 (EgBolB08) MT072979 (EgBolB09) | ||||
| Argentina | AF458875 (EgArgO03) MT796074 (EgArg07) KC579445 (EgArgO09) KC579450 (EgArgO10) KC579447 (EgArgO13) FN564569 (EgArgO16) GU980907 (EgArgO27) GU980906 (EgArgO28) KC954601 (EgArgO30) | MT796487 (EgArgC24) | AF458873 (EgArgB05) KC579445 (EgArgB08) KC579449 (EgArgB11) KC579448 (EgArgB12) KC579446 (EgArgB14) KC579447 (EgArgB18) KC954602 (EgArgB29) | M84661 (EgArgH01) AF458875 (EgArgH02) AF458873 (EgArgH04) AF458872 (EgArgH06) KT719395 (EgArgH15) MT800802 (EgArgH17) MT800801 (EgArgH19) MT800800 (EgArgH20) MT800797 (EgArgH21) MT800796 (EgArgH22) MT800795 (EgArgH23) MT796079 (EgArgH26) | |
| Middle East | |||||
| Palestine | KC109640 (EgPaO01), KC109657 (EgPaO02), KC109651 (EgPaO03) | ||||
| Iran Saudi Arabia | JN604097 (EgIP08), JN604099 (EgIP09, JN604102 (EgIP10) | JQ250806 (EgIO05), JQ250808 (EgIO06), JQ250811 (EgIO07) MN720282 (EgASO01) | JQ250810 (EgIH01), JQ250812 (EgIH02), JQ250815 (EgIH03), AB677811 (EgIH04) MT073987 (EgIH11), MN807921 (EgITH12) | ||
| Europe | |||||
| Austria | JF513058 (EgAO01) JF513060 (EgAO03) JF513061 ( (EgAO05) | JF513058 (EgAH02) JF513060 (EgAH04) JF513061 (EgAH06) AJ508018 (EgAH07) AJ508019 (EgAH08) AJ508021 (EgAH09) AJ508028 (EgAH10) | |||
| Bulgaria | JF513058 (EgBO01) JF513063 (EgBO02) JF513065 (EgBO03) JF513058 (EgBH07) JF513063 (EgBH08) JF513065 (EgBH09) | ||||
| Hungry | JF513058 (EgHO01) JF513061 (EgHO02) JF513063 (EgHO03) JF513064 (EgHO04) JF513065 (EgHO05) JF513067 (EgHO06) | ||||
| Italiy | JF513058 (EgIO01) F513059 (EgIO02) JF513060 (EgIO03) JF513062 (EgIO04) DQ062857 (EgIO06) | DQ062857 (EgIB05) | JF513058 (EgIH08) JF513060 (EgIH09) JF513062 (EgIH10) | ||
| Portugal | JF513058 (EgPO01) JF513060 (EgPO02) JF513079 (EgPO03) HF947595 (EgPO04) HF947592 (EgPO05) HF947558 (EgPO06) HF947585 (EgPO07) HF947574 (EgPO08) HF947579 (EgPO09) HF947557 (EgPO10) HF947566 (EgPO11) | ||||
| Romania | JF513058 (EgRO01) JF513061 (EgRO02) JF513063 (EgRO03) JF513072 (EgRO04) JF513079 (EgRO05) | JF513058 (EgRH06) JF513061 (EgRH07) JF513063 (EgRH08) JF513079 (EgRH09) JF520817 (EgRH010) JF520818 (EgRH011) AY686564 (EgRH012) AY686565 (EgRH013) | |||
| Greece | DQ856467 (EgGO01) | ||||
| UK | KT001398 (EgUKP01) KT001397 (EgUKP02) | ||||
| Asia | |||||
| Türkiye | JF513058 (EgTO01) JF513060 (EgTO02) JF513062 (EgTO03) JF513067 (EgTO04) JF513071 (EgTO05) JF513079 (EgTO06) JF775380 (EgTO07) EU929083 (EgTO08) | JF513058 (EgTH09) JF513060 (EgTH10) JF513062 (EgTH11) JF513067 (EgTH12) JF513079 (EgTH13) JF775379 (EgTH14) GU951512 (EgTH15) GU951513 (EgTH16) MT537165 (EgTH17) MN990735 (EgTH18) KX874714 (EgTH19) KX874725 (EgTH20) KX874719 (EgTH21) | |||
| China | DQ356881 (EgCHP01) DQ356882 (EgCHP02) | AB491414 (EgCHO03) AB491418 (EgCHO04) AB491421 (EgCHO05) AB491424 (EgCHO06) AB491425 (EgCHO07) AB491432 (EgCHO08) AB491438 (EgCHO09) AB491449 (EgCHO10) AB491454 (EgCHO11) AB688612 (EgCHO12) MG674403 (EgCHO31) MG674404 (EgCHO34) MG674406 (EgCHO40) MG674412 (EgCHO43) MG674416 (EgCHO46) MG674418 (EgCHO49) | MG674403 (EgCHY32) MG674404 (EgCHY35) MG674406 (EgCHY41) MG674412 (EgCHY44) MG674416 (EgCHY47) MG674418 (EgCHY50) | AB688602 (EgCHH13) AB688611 (EgCHH14) AB688613 (EgCHH15) AB688619 (EgCHH16) AB491419 (EgCHH17) AB491420 (EgCHH18) AB491422 (EgCHH19) AB491423 (EgCHH20) AB491428 (EgCHH21) AB491431 (EgCHH22) AB491434 (EgCHH23) AB491437 (EgCHH24) AB491439 (EgCHH25) AB491447 (EgCHH26) AB491451 (EgCHH27) AB491453 (EgCHH28) AB491455 (EgCHH29) MG674403 (EgCHH30) MG674404 (EgCHH33) MG674406 (EgCHH39) MG674412 (EgCHH42) MG674416 (EgCHH45) MG674418 (EgCHH48) | |
| Iraq, Kurdistan | MF004305 (EgKO01) MF004277 (EgKO04) MF004283 (EgKO07) MF004285 (EgKO10) MF004279 (EgKO13) MF004273 (EgKO14) MF004288 (EgKO15) | MF004305 (EgKC02) MF004277 (EgKC05) MF004283 (EgKC08) MF004285 (EgKC11) MF004278 (EgKO16) MF004287 (EgKO17) MF004293 (EgKO18) MF004306 (EgKO19) | MF004305 (EgKB03) MF004277 (EgKB06) MF004283 (EgKB09) MF004285 (EgKC12) MF004280 (EgKB20) MF004291 (EgKB21) | ||
| Kyrgyztan | MN787554 (EgKirH01) MN787559 (EgKirH02) MN787560 (EgKirH03) | ||||
| Oceania | |||||
| Australia | KT001398 (EgAusP05) KT001395 (EgAusP05) KT968706 (EgAusD07) KT968707 (EgAusM08) KU697314 (EgAusD14) JQ250806 (EgAusM16) JQ250809 (EgAusM17) | AJ508005 (EgAusO01) AJ508006 (EgAusO02) AJ508009 (EgAusO03) AJ508010 (EgAusO04) KT968705 (EgAusO09) KT968704 (EgAusO10) KT968703 (EgAusO11) KT968708 (EgAusO12) KT968702 (EgAusO13) AB522646 (EgAusO15) AB688591 (EgAusO18) | |||
| Africa | |||||
| Libya | KT001407 (EgLP01) KT001405 (EgLP02) KT001398 (EgLP03) | HM636641 (EgLH04) | |||
| Tunisia | KM014635 (EgTuP03) KM014636 (EgTuP04) KM014638 (EgTuP06) KM014640 (EgTuP08) KM001399 (EgTuP13) KT001401 (EgTuP14) KT001402 (EgTuP15) KT001403 (EgTuP16) KT001399 (EgTuP17) KT0014610 (EgTuP32) | KM014641 (EgTuO09) KM014644 (EgTuO12) KT0014628 (EgTuO20) KT0014627 (EgTuO21) KT0014614 (EgTuO28) KT0014613 (EgTuO29) | KT0014624 (EgTuC22) KT0014616 (EgTuC26) | KM014634 (EgTuB01) KM014633 (EgTuCam02) KM014639 (EgTuB07) KM014642 (EgTuB10) KT0014620 (EgTuB24 KT0014617 (EgTuB25) KT0014615 (EgTuB27) KT0014612 (EgTuB30) KT0014611 (EgTuCe31) KT0014606 (EgTuCe33) KT0014607 (EgTuCe34) | KM014637 (EgTuH05) KM014643 (EgTuH11) KT0014631 (EgTuH18) KT0014629 (EgTuH19) KT0014621 (EgTuH23) |
| Kenya | KM01398 (EgKP01) |
| Haplotypes | 1 | 96 | 101 | 134 | 137 | 182 | 197 | 218 | 251 | 253 | 348 | 359 | 368 | 371 | 383 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EGO1-JQ250806 | C | T | G | C | G | G | C | G | A | A | A | T | G | A | G | T |
| EG1A-AF458871 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1B-AF458872 | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| EG1C-AF458873 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| EG1D-AF458874 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EG1E-AF458875 | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | |
| EgCLCCoO3 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBCo17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBCo18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCIBCo21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH Co01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA03 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA04 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA07 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA10 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA12 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLBA20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM01 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM03 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM05 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOCo07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM08 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM10 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM16 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . |
| EgCLOM17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM21 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . |
| EgCLOM22 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLOM23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Haplotypes | 1 | 2 | 26 | 32 | 36 | 38 | 39 | 59 | 60 | 66 | 74 | 79 | 80 | 89 | 96 | 101 | 116 | 128 | 131 | 134 | 137 | 146 | 159 | 167 | 170 | 182 | 197 | 218 | 251 | 260 | 269 | 271 | 280 | 287 | 290 | 299 | 306 | 320 | 332 | 338 | 341 | 344 | 348 | 349 | 371 | 378 | 383 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EG01-JQ250806 | C | T | T | T | A | A | A | T | A | G | T | C | G | C | T | G | T | G | T | C | G | T | A | G | G | G | C | G | A | G | G | G | T | T | G | A | G | G | T | A | T | G | A | T | A | A | G | T |
| EG1A AF458871 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1B AF458872 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| EG1D AF458874 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1E AF458875 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG02 M84662 | - | - | . | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EG03 M84663 | - | - | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EG6 M84666 | - | - | . | . | G | T | . | G | . | T | G | T | T | T | . | . | . | A | . | T | T | A | . | A | A | T | T | T | G | A | T | A | C | . | T | G | A | T | . | C | C | T | . | . | G | G | A | - |
| EU178104 | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH01 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH02 | - | - | . | . | G | T | . | G | . | T | G | T | T | T | . | . | . | A | . | T | T | A | . | A | A | T | T | T | G | A | T | A | C | . | T | G | A | T | . | C | C | T | . | . | G | G | A | - |
| EgCLH04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB28 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH03 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLO11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH12 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB13 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLC14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLO17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLP20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLP21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLO22 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH23 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLB24 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH25 | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCLH26 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgClH19 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgClH29 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgClB30 | - | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| EgCL31 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL32 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL33 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL34 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL35 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL36 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL37 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL38 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL39 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCL40 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBolB08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgBolB09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH01 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| EgArgH04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . |
| EgArgB05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . |
| EgArgH07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgB08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgO09 | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgO10 | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgB11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| EgArgB12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgO13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgB14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EsArgO16 | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgB18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH22 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgH23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgArgC24 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgH25 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgH26 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgO27 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgO28 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgB29 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgArgO30 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgPeH07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgPeP09 | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP12 | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP16 | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP21 | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeP22 | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeLL23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeO02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgPeB24(2) | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | - |
| EgPeB23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgPeO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgPeO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeB24 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . |
| EgPeO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeH08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeH05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPeO25 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Haplotypes | 1 | 26 | 29 | 63 | 66 | 74 | 79 | 80 | 89 | 96 | 109 | 130 | 134 | 146 | 182 | 197 | 206 | 214 | 224 | 227 | 230 | 280 | 329 | 338 | 371 | 377 | 378 | 379 | 383 | 385 | 386 | 388 | 391 | 398 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EG01-JQ250806 | C | T | A | A | G | T | C | G | C | T | T | G | C | T | G | C | T | T | T | T | T | T | G | A | A | T | A | T | G | A | T | A | C | T | T |
| EG1A AF458871 | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1C AF458873 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . |
| EG1D AF458874 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG2 M84662 | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG3 M84663 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EU178103 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EU178104 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAO03 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH04 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAO05 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH06 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH07 | . | C | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH08 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH09 | . | . | G | . | . | . | T | . | T | . | C | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAH10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBH07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO02 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBH08 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBH09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO04 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO05 | . | . | . | . | . | . | . | . | T | . | . | . | . | C | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgBO06 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | T | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO02 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgHO06 | . | C | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItH08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItO02 | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItO03 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItH09 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItO04 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItH10 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItB05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgItP07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO02 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO03 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | A | T | T | A | G | A | A | . |
| EgPO08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPO11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRO02 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH07 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH08 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRO05 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH09 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH010 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH011 | . | . | . | G | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH012 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgRH013 | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgGO01 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . |
| EgUKP01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgUKP02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Haplotypes | 1 | 2 | 9 | 11 | 13 | 15 | 17 | 19 | 26 | 29 | 38 | 42 | 44 | 47 | 79 | 80 | 89 | 91 | 96 | 134 | 146 | 197 | 230 | 249 | 268 | 269 | 280 | 282 | 287 | 291 | 299 | 319 | 329 | 332 | 335 | 344 | 348 | 362 | 365 | 371 | 377 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EG01-JQ250806 | C | T | T | T | T | T | G | T | T | A | A | A | T | T | C | G | C | A | T | C | T | C | T | T | C | G | T | A | T | G | A | T | G | T | G | G | A | T | T | A | T | T |
| EG1A-AF458871 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1B-AF458872 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| EG1C-AF458873 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| EG1D-AF458874 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG2-M84662 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG3-M84663 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKirH01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKirH02 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKirH03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO04 | . | . | . | . | . | . | . | . | C | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH12 | . | . | . | . | . | . | . | . | C | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . |
| EgTO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTO08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH15 | . | . | . | C | A | G | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTH16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHP01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHP02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO08 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH14 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | T | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH18 | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| EgCHH21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH22 | . | . | . | . | . | . | . | . | . | . | G | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH24 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH25 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH26 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH27 | . | . | . | . | . | . | . | . | . | . | . | G | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH28 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH29 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKC02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKC05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKC08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKC11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO14 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO17 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | G | . |
| EgKO18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKO19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKB21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH30 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO31 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHY32 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH33 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO34 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHY35 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH36 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO37 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHY38 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH39 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO40 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHY41 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH42 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | C | C | C | . | C | . | C | . | . | . | . |
| EgCHO43 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | C | C | C | . | C | . | C | . | . | . | . |
| EgCHY44 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | C | . | C | C | C | . | C | . | C | . | . | . | . |
| EgCHH45 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHO46 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHY47 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgCHH48 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| EgCHO49 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| EgCHY50 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| Haplotypes | 1 | 79 | 80 | 96 | 101 | 128 | 131 | 134 | 137 | 197 | 218 | 239 | 306 | 310 | 311 | 320 | 383 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EG01-JQ250806 | C | C | G | T | G | G | T | C | G | C | G | G | G | C | T | G | G | T |
| EG1A-AF458871 | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1E-AF458875 | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . |
| EU178104 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPaO02 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPaO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgPaO03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIH01 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIH02 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIH03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIH04 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIO05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIO06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIO07 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgIP08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - |
| EgIP09 | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | - |
| EgIP10 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | - |
| EgIH11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgITH12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgASO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Haplotypes | 1 | 2 | 32 | 39 | 46 | 63 | 66 | 74 | 79 | 80 | 89 | 96 | 101 | 116 | 125 | 128 | 131 | 134 | 146 | 149 | 151 | 175 | 182 | 197 | 227 | 245 | 251 | 263 | 269 | 280 | 287 | 311 | 335 | 347 | 348 | 359 | 368 | 371 | 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EG01-JQ250806 | C | T | T | A | A | A | G | T | C | G | C | T | G | T | T | G | T | C | T | T | A | T | G | C | T | T | A | G | G | T | T | T | G | T | A | T | G | A | T |
| EG1-M84661 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | - |
| EG1A-AF458871 | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1B-AF458872 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . |
| EG1C-AF458873 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . |
| EG1D-AF458874 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG1E-AF458875 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EG2-M84662 | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EG3-M84663 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EF393619 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . |
| EF595654 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EU178103 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EU178104 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAusD07(2) | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAusO01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAusO02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAO03 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | G | . | . | . | A | . | . | . | . | . | C | . | . | . | . | . | . | . | . |
| EgAusO04 | . | . | . | . | T | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAusP05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAusP06 | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAusD07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus08 | . | . | . | . | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAus09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus13 | . | . | . | . | . | G | . | . | . | . | T | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAus14 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAus15 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| EgAus16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus17 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgAus18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgLP01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgLP02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . |
| EgLP03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuCam02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP03 | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP04 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH05 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP06 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB07 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP08 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| egTuO09 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB10 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH11 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuO12 | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . |
| EgTuP13 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP14 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP15 | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP17 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH18 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH19 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuO20 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuC22 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuH23 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB24 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB25 | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . |
| EgTuC26 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . |
| EgTuB27 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . |
| EgTuO28 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuO29 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuB30 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuC31 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuP32 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuC33 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgTuC34 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| EgKP01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
References
- Lymbery, A.J. Phylogenetic Pattern, Evolutionary Processes and Species Delimitation in the Genus Echinococcus. In Advances in Parasitology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 95, pp. 111–145. ISBN 978-0-12-811471-1. [Google Scholar]
- Nakao, M.; Lavikainen, A.; Yanagida, T.; Ito, A. Phylogenetic Systematics of the Genus Echinococcus (Cestoda: Taeniidae). Int. J. Parasitol. 2013, 43, 1017–1029. [Google Scholar] [CrossRef]
- Bowles, J.; Blair, D.; Mcmanus, D. Genetic Variants within the Genus Echinococcus Identified by Mitochondrial DNA Sequencing. Mol. Biochem. Parasitol. 1992, 54, 165–173. [Google Scholar] [CrossRef]
- Knapp, J.; Nakao, M.; Yanagida, T.; Okamoto, M.; Saarma, U.; Lavikainen, A.; Ito, A. Phylogenetic Relationships within Echinococcus and Taenia Tapeworms (Cestoda: Taeniidae): An Inference from Nuclear Protein-Coding Genes. Mol. Phylogenet. Evol. 2011, 61, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Yanagida, T.; Okamoto, M.; Knapp, J.; Nkouawa, A.; Sako, Y.; Ito, A. State-of-the-Art Echinococcus and Taenia: Phylogenetic Taxonomy of Human-Pathogenic Tapeworms and Its Application to Molecular Diagnosis. Infect. Genet. Evol. 2010, 10, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Yanagida, T.; Konyaev, S.; Lavikainen, A.; Odnokurtsev, V.A.; Zaikov, V.A.; Ito, A. Mitochondrial Phylogeny of the Genus Echinococcus (Cestoda: Taeniidae) with Emphasis on Relationships among Echinococcus canadensis Genotypes. Parasitology 2013, 140, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, T.; Lavikainen, A.; Hoberg, E.P.; Konyaev, S.; Ito, A.; Sato, M.O.; Zaikov, V.A.; Beckmen, K.; Nakao, M. Specific Status of Echinococcus canadensis (Cestoda: Taeniidae) Inferred from Nuclear and Mitochondrial Gene Sequences. Int. J. Parasitol. 2017, 47, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.A. The Molecular Epidemiology of Echinococcus Infections. Pathogens 2020, 9, 453. [Google Scholar] [CrossRef]
- Alvarez Rojas, C.A.; Ebi, D.; Gauci, C.G.; Scheerlinck, J.P.; Wassermann, M.; Jenkins, D.J.; Lightowlers, M.W.; Romig, T. Microdiversity of Echinococcus granulosus sensu stricto in Australia. Parasitology 2016, 143, 1026–1033. [Google Scholar] [CrossRef]
- Lightowlers, M.W. Cysticercosis and Echinococcosis. In One Health: The Human-Animal-Environment Interfaces in Emerging Infectious Diseases; Mackenzie, J.S., Jeggo, M., Daszak, P., Richt, J.A., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 365, pp. 315–335. ISBN 978-3-642-36888-2. [Google Scholar]
- Lymbery, A.J.; Constantine, C.C.; Thompson, R.C.A. Self-Fertilization Without Genomic or Population Structuring in A Parasitic Tapeworm. Evolution 1997, 51, 289–294. [Google Scholar] [CrossRef]
- Casulli, A.; Interisano, M.; Sreter, T.; Chitimia, L.; Kirkova, Z.; La Rosa, G.; Pozio, E. Genetic Variability of Echinococcus granulosus sensu stricto in Europe Inferred by Mitochondrial DNA Sequences. Infect. Genet. Evol. 2012, 12, 377–383. [Google Scholar] [CrossRef]
- Wang, J.; Wang, N.; Hu, D.; Zhong, X.; Wang, S.; Gu, X.; Peng, X.; Yang, G. Genetic Diversity of Echinococcus granulosus in Southwest China Determined by the Mitochondrial NADH Dehydrogenase Subunit 2 Gene. Sci. World J. 2014, 2014, 1–8. [Google Scholar] [CrossRef][Green Version]
- Han, X.; Jian, Y.; Zhang, X.; Ma, L.; Zhu, W.; Cai, Q.; Wu, S.; Wang, X.; Shi, B. Genetic Characterization of Echinococcus Isolates from Various Intermediate Hosts in the Qinghai-Tibetan Plateau Area, China. Parasitology 2019, 146, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Muqaddas, H.; Mehmood, N.; Arshad, M. Genetic Variability and Diversity of Echinococcus granulosus sensu lato in Human Isolates of Pakistan Based on Cox1 Mt-DNA Sequences (366bp). Acta Trop. 2020, 207, 105470. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Xiao, N.; Okamoto, M.; Yanagida, T.; Sako, Y.; Ito, A. Geographic Pattern of Genetic Variation in the Fox Tapeworm Echinococcus multilocularis. Parasitol. Int. 2009, 58, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Kamenetzky, L.; Gutierrez, A.M.; Canova, S.G.; Haag, K.L.; Guarnera, E.A.; Parra, A.; García, G.E.; Rosenzvit, M.C. Several Strains of Echinococcus granulosus Infect Livestock and Humans in Argentina. Infect. Genet. Evol. 2002, 2, 129–136. [Google Scholar] [CrossRef]
- Haag, K.L.; Ayala, F.J.; Kamenetzky, L.; Gutierrez, A.M.; Rosenzvit, M. Livestock Trade History, Geography, and Parasite Strains: The Mitochondrial Genetic Structure of Echinococcus granulosus in Argentina. J. Parasitol. 2004, 90, 234–239. [Google Scholar] [CrossRef]
- Alvarez Rojas, C.A.; Ebi, D.; Paredes, R.; Acosta-Jamett, G.; Urriola, N.; Roa, J.C.; Manterola, C.; Cortes, S.; Romig, T.; Scheerlinck, J.-P.; et al. High Intraspecific Variability of Echinococcus granulosus sensu stricto in Chile. Parasitol. Int. 2017, 66, 112–115. [Google Scholar] [CrossRef]
- Cortés, A.S.; Valle, B.C. Hidatidosis Humana: Generalidades y Situación Epidemiológica En Chile Según Egresos Hospitalarios y Notificación Obligatoria Entre Los Años 2001 y 2005. Rev. Chil. Infectol. 2010, 27, 329–335. [Google Scholar] [CrossRef]
- Hidalgo, C.; Stoore, C.; Pereira, I.; Paredes, R.; Alvarez Rojas, C.A. Multiple Haplotypes of Echinococcus granulosus sensu stricto in Single Naturally Infected Intermediate Hosts. Parasitol. Res. 2020, 119, 763–770. [Google Scholar] [CrossRef]
- Acosta-Jamett, G.; Cleaveland, S.; De C Bronsvoort, B.M.; Cunningham, A.; Bradshaw, H.; Craig, P. Echinococcus granulosus Infection in Foxes in Coquimbo District, Chile. Arch. Med. Vet. 2015, 47, 409–413. [Google Scholar] [CrossRef]
- Espinoza, S.; Salas, A.M.; Vargas, A.; Freire, V.; Diaz, E.; Sánchez, G.; Venegas, J. Detection of the G3 Genotype of Echinococcus granulosus from Hydatid Cysts of Chilean Cattle Using COX1and ND1mitochondrial Markers. Parasitol. Res. 2014, 113, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Manterola, C.; Benavente, F.; Melo, A.; Vial, M.; Roa, J.C. Description of Echinococcus granulosus Genotypes in Human Hydatidosis in a Region of Southern Chile. Parasitol. Int. 2008, 57, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, F.; Stoore, C.; Horlacher, P.; Jiménez, M.; Hidalgo, C.; Alvarez Rojas, C.A.; Figueiredo Barros, G.; Bunselmeyer Ferreira, H.; Hernández, M.; Cabrera, G.; et al. First Description of Echinococcus ortleppi and Cystic Echinococcosis Infection Status in Chile. PLoS ONE 2018, 13, e0197620. [Google Scholar] [CrossRef] [PubMed]
- Vural, G.; Baca, A.U.; Gauci, C.G.; Bagci, O.; Gicik, Y.; Lightowlers, M.W. Variability in the Echinococcus granulosus Cytochrome C Oxidase 1 Mitochondrial Gene Sequence from Livestock in Turkey and a Re-Appraisal of the G1–3 Genotype Cluster. Vet. Parasitol. 2008, 154, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, T.; Mohammadzadeh, T.; Kamhawi, S.; Nakao, M.; Sadjjadi, S.M.; Hijjawi, N.; Abdel-Hafez, S.K.; Sako, Y.; Okamoto, M.; Ito, A. Genetic Polymorphisms of Echinococcus granulosus sensu stricto in the Middle East. Parasitol. Int. 2012, 61, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; McMANUS, D.P.; Schantz, P.M.; Craig, P.S.; Ito, A. A Molecular Phylogeny of the Genus Echinococcus Inferred from Complete Mitochondrial Genomes. Parasitology 2006, 134, 713–722. [Google Scholar] [CrossRef]
- Boufana, B.; Lett, W.S.; Lahmar, S.; Buishi, I.; Bodell, A.J.; Varcasia, A.; Casulli, A.; Beeching, N.J.; Campbell, F.; Terlizzo, M.; et al. Echinococcus equinus and Echinococcus granulosus sensu stricto from the United Kingdom: Genetic Diversity and Haplotypic Variation. Int. J. Parasitol. 2015, 45, 161–166. [Google Scholar] [CrossRef]
- Moro, P.L.; Nakao, M.; Ito, A.; Schantz, P.M.; Cavero, C.; Cabrera, L. Molecular Identification of Echinococcus Isolates from Peru. Parasitol. Int. 2009, 58, 184–186. [Google Scholar] [CrossRef]
- Sánchez, E.; Cáceres, O.; Náquira, C.; Garcia, D.; Patiño, G.; Silvia, H.; Volotão, A.C.; Fernandes, O. Molecular Characterization of Echinococcus granulosus from Peru by Sequencing of the Mitochondrial Cytochrome C Oxidase Subunit 1 Gene. Mem. Inst. Oswaldo Cruz 2010, 105, 806–810. [Google Scholar] [CrossRef][Green Version]
- Adwan, G.; Adwan, K.; Bdir, S.; Abuseir, S. Molecular Characterization of Echinococcus granulosus Isolated from Sheep in Palestine. Exp. Parasitol. 2013, 134, 195–199. [Google Scholar] [CrossRef]
- AL-Mutairi, N.M.; Taha, H.A.; Nigm, A.H. Molecular Characterization of Echinococcus granulosus in Livestock of Al-Madinah (Saudi Arabia). J. Helminthol. 2020, 94, e157. [Google Scholar] [CrossRef]
- Barazesh, A.; Sarkari, B.; Shahabi, S.; Halidi, A.G.; Ekici, A.; Aydemir, S.; Mahami-Oskouei, M. Genetic Diversity of Echinococcus granulosus Isolated from Humans: A Comparative Study in Two Cystic Echinococcosis Endemic Areas, Turkey and Iran. BioMed Res. Int. 2020, 2020, 3054195. [Google Scholar] [CrossRef] [PubMed]
- Parsa, F.; Fasihi Harandi, M.; Rostami, S.; Sharbatkhori, M. Genotyping Echinococcus granulosus from Dogs from Western Iran. Exp. Parasitol. 2012, 132, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Pezeshki, A.; Akhlaghi, L.; Sharbatkhori, M.; Razmjou, E.; Oormazdi, H.; Mohebali, M.; Meamar, A.R. Genotyping of Echinococcus granulosus from Domestic Animals and Humans from Ardabil Province, Northwest Iran. J. Helminthol. 2013, 87, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, Y.; Sadjjadi, S.M.; Nikoupour Dailami, H.; Jafari, S.H.; Anbardar, M.H.; Khosravi, M.B. Cystic Echinococcosis/Hydatid Cyst Coinfection with HIV: A Report from Shiraz, Iran. Can. J. Gastroenterol. Hepatol. 2021, 2021, 8844104. [Google Scholar] [CrossRef]
- Bart, J.M.; Abdukader, M.; Zhang, Y.L.; Lin, R.Y.; Wang, Y.H.; Nakao, M.; Ito, A.; Craig, P.S.; Piarroux, R.; Vuitton, D.A.; et al. Genotyping of Human Cystic Echinococcosis in Xinjiang, PR China. Parasitology 2006, 133, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Beato, S.; Parreira, R.; Roque, C.; Gonçalves, M.; Silva, L.; Maurelli, M.P.; Cringoli, G.; Grácio, M.A. Echinococcus Granulosus in Portugal: The First Report of the G7 Genotype in Cattle. Vet. Parasitol. 2013, 198, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Obwaller, A.; Schneider, R.; Walochnik, J.; Gollackner, B.; Deutz, A.; Janitschke, K.; Aspöck, H.; Auer, H. Echinococcus granulosus Strain Differentiation Based on Sequence Heterogeneity in Mitochondrial Genes of Cytochrome c Oxidase-1 and NADH Dehydrogenase-1. Parasitology 2004, 128, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Šnábel, V.; Altintas, N.; D’Amelio, S.; Nakao, M.; Romig, T.; Yolasigmaz, A.; Gunes, K.; Turk, M.; Busi, M.; Hüttner, M.; et al. Cystic Echinococcosis in Turkey: Genetic Variability and First Record of the Pig Strain (G7) in the Country. Parasitol. Res. 2009, 105, 145–154. [Google Scholar] [CrossRef]
- Varcasia, A.; Canu, S.; Lightowlers, M.W.; Scala, A.; Garippa, G. Molecular Characterization of Echinococcus granulosus Strains in Sardinia. Parasitol. Res. 2006, 98, 273–277. [Google Scholar] [CrossRef]
- Varcasia, A.; Canu, S.; Kogkos, A.; Pipia, A.P.; Scala, A.; Garippa, G.; Seimenis, A. Molecular Characterization of Echinococcus granulosus in Sheep and Goats of Peloponnesus, Greece. Parasitol. Res. 2007, 101, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.I.; Meerkhan, A.A.; Boufana, B.; Hama, A.A.; Ahmed, B.D.; Mero, W.M.S.; Orsten, S.; Interisano, M.; Pozio, E.; Casulli, A. Two Haplotype Clusters of Echinococcus granulosus sensu stricto in Northern Iraq (Kurdistan Region) Support the Hypothesis of a Parasite Cradle in the Middle East. Acta Trop. 2017, 172, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Alvarez Rojas, C.A.; Kronenberg, P.A.; Aitbaev, S.; Omorov, R.A.; Abdykerimov, K.K.; Paternoster, G.; Müllhaupt, B.; Torgerson, P.; Deplazes, P. Genetic Diversity of Echinococcus multilocularis and Echinococcus granulosus sensu lato in Kyrgyzstan: The A2 Haplotype of E. multilocularis Is the Predominant Variant Infecting Humans. PLoS Negl. Trop. Dis. 2020, 14, e0008242. [Google Scholar] [CrossRef] [PubMed]
- Beyhan, Y.E.; Çobanoğlu, U.; Çelik, S.; Yılmaz, H.; Halidi, A.G. Molecular Characterization of Human Lung and Liver Cystic Echinococcosis Isolates in Van Province, Turkey. Acta Trop. 2020, 206, 105451. [Google Scholar] [CrossRef]
- Simsek, S.; Kaplan, M.; Ozercan, I.H. A Comprehensive Molecular Survey of Echinococcus Granulosus in Formalin-Fixed Paraffin-Embedded Tissues in Human Isolates in Turkey. Parasitol. Res. 2011, 109, 411–416. [Google Scholar] [CrossRef]
- Abushhewa, M.H.; Abushhiwa, M.H.S.; Nolan, M.J.; Jex, A.R.; Campbell, B.E.; Jabbar, A.; Gasser, R.B. Genetic Classification of Echinococcus granulosus Cysts from Humans, Cattle and Camels in Libya Using Mutation Scanning-Based Analysis of Mitochondrial Loci. Mol. Cell. Probes 2010, 24, 346–351. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical Tests of Neutrality of Mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Hudson, R.R. A New Statistic for Detecting Genetic Differentiation. Genetics 2000, 155, 2011–2014. [Google Scholar] [CrossRef]
- Alvarez Rojas, C.A.; Romig, T.; Lightowlers, M.W. Echinococcus granulosus sensu lato Genotypes Infecting Humans—Review of Current Knowledge. Int. J. Parasitol. 2014, 44, 9–18. [Google Scholar] [CrossRef]
- Laurimäe, T.; Kinkar, L.; Andresiuk, V.; Haag, K.L.; Ponce-Gordo, F.; Acosta-Jamett, G.; Garate, T.; Gonzàlez, L.M.; Saarma, U. Genetic Diversity and Phylogeography of Highly Zoonotic Echinococcus granulosus Genotype G1 in the Americas (Argentina, Brazil, Chile and Mexico) Based on 8279 Bp of mtDNA. Infect. Genet. Evol. 2016, 45, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Konyaev, S.V.; Yanagida, T.; Nakao, M.; Ingovatova, G.M.; Shoykhet, Y.N.; Bondarev, A.Y.; Odnokurtsev, V.A.; Loskutova, K.S.; Lukmanova, G.I.; Dokuchaev, N.E.; et al. Genetic Diversity of Echinococcus spp. in Russia. Parasitology 2013, 140, 1637–1647. [Google Scholar] [CrossRef]
- Ali, V.; Martinez, E.; Duran, P.; Seláez, M.A.; Barragan, M.; Nogales, P.; Peña, Y.; Lillo, A.; Castañares, M.; Claros, Y.; et al. Echinococcus granulosus sensu stricto, Echinococcus ortleppi and E. intermedius (G7) Are Present in Bolivia. Parasitology 2020, 147, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Romig, T.; Ebi, D.; Wassermann, M. Taxonomy and Molecular Epidemiology of Echinococcus granulosus sensu lato. Vet. Parasitol. 2015, 213, 76–84. [Google Scholar] [CrossRef]



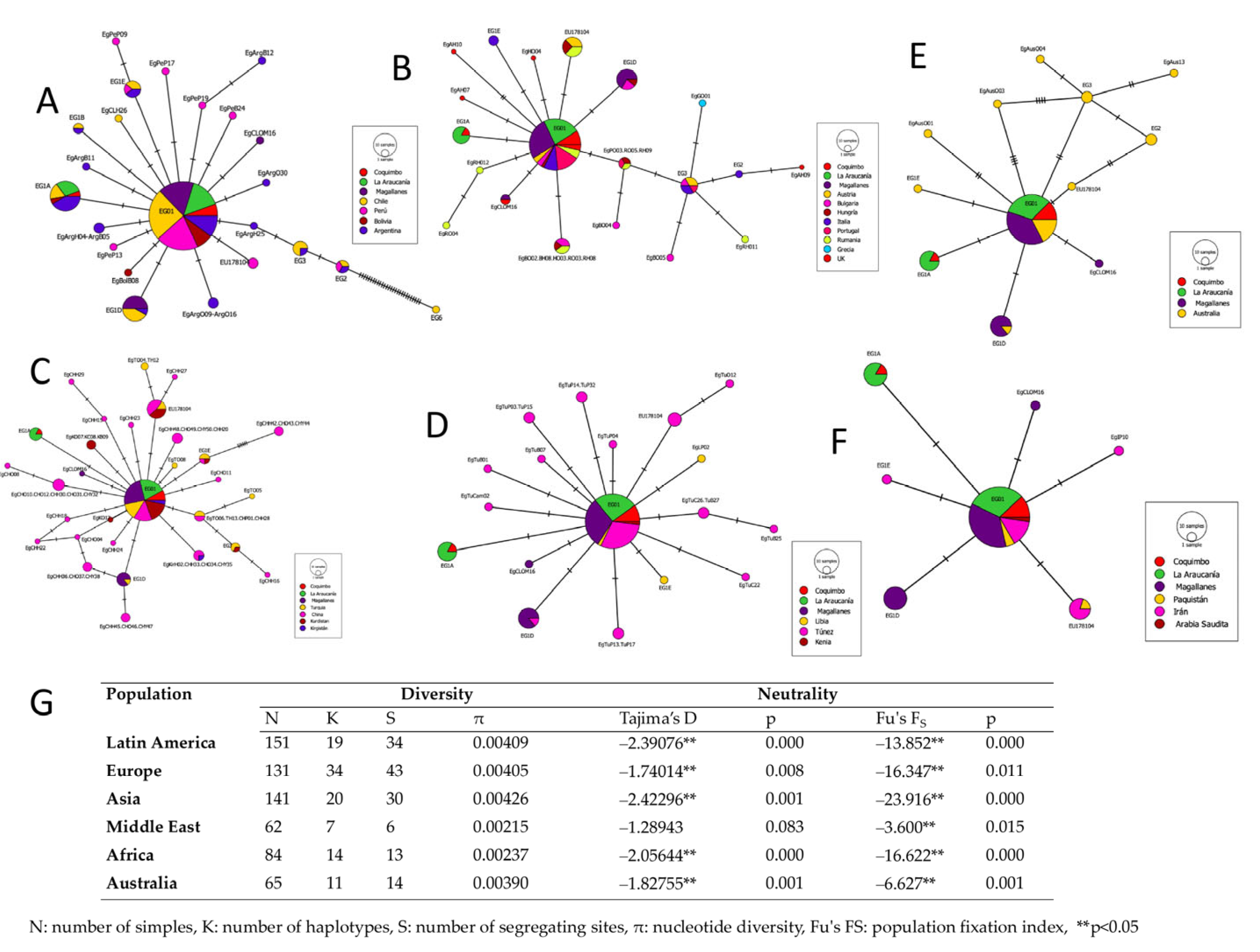
| Region | City/Locality | Number of Isolates | Host Speccies (Number of Isolates) |
|---|---|---|---|
| Coquimbo | Ovalle. Peña Blanca. Monte Patria. Illapel | 6 | goats (1). sheep (1). cattle (3). humans (1) |
| La Araucanía | Santa Cruz. Nueva Imperial. San José. Cunco. Los Maitenes. Puyehue. El Carmen. Carahue | 18 | cattle (18) |
| Magallanes | Magallanes | 22 | sheep (22) |
| Total | 46 | goats (1). sheep (23). cattle (21). humans (1) |
| Region | N | S | K | H | π | Tajima’s D | p | Fu’s FS | p |
|---|---|---|---|---|---|---|---|---|---|
| Coquimbo | 6 | 1 | 2 | 0.333 ± 0.04630 | 0.00084 | −0.93302 * | 0.061 | −0.003 * | 0.502 |
| La Araucanía | 17 | 1 | 2 | 0.425 ± 0.425 | 0.00107 | 0.8695 * | 0.780 | 1.039 | 0.801 |
| Magallanes | 22 | 2 | 3 | 0.481 ± 0.00875 | 0.00128 | −0.17406 * | 0.386 | −0.121 * | 0.414 |
| Total | 45 | 3 | 4 | 0.461 ± 0.00637 | 0.00141 | −0.52134 * | 0.326 | −0.856 * | 0.279 |
| Coquimbo | La Araucanía | Magallanes | |
|---|---|---|---|
| Coquimbo | ----------- | 0.59809 | 0.6823 |
| La Araucanía | −0.08761 | ---------- | 0.62728 ** |
| Magallanes | 0.03341 | 0.10703 ** | -------- |
| Source of Variation | D.f. | Sum of the Squares | Components of Variance | Percentage of Variation |
|---|---|---|---|---|
| Between populations | 2 | 3.285 | 0.062 | 20.340 |
| Within the populations | 43 | 10.226 | 0.243 | 79.659 |
| Total | 45 | 13.511 | 0.306 |
| Population | Number of Sequences Analyzed | Diversity Index | Neutrality Index | |||||
|---|---|---|---|---|---|---|---|---|
| Hn | hd ± SD | nd ± SD | Tajima’s D | p | Fu’s FS | p | ||
| Chile (this study) | 46 | 4 | 0.461 ± 0.00637 | 0.00181 ± 0.00036 | −0.54123 | 0.326 | −0.856 | 0.279 |
| Chile | 85 | 9 | 0.542 ± 0.059 | 0.00461 ± 0.00203 | −2.36563 ** | 0.000 | −2.083 | 0.201 |
| Argentina | 30 | 13 | 0.869 ± 0.042 | 0.0.00458 ± 0.00063 | −1.71486 ** | 0.010 | −8.639 ** | 0.000 |
| Peru | 27 | 9 | 0.561 ± 0.114 | 0.00251 ± 0.00073 | −1.99558 ** | 0.002 | −6.421 ** | 0.000 |
| Bolivia | 9 | 3 | 0.417 ± 0.191 | 0.00121 ± 0.00060 | −1.36240 ** | 0.016 | −1.081 ** | 0.022 |
| Austria | 10 | 6 | 0.889 ± 0.075 | 0.00766 ± 0.00161 | −0.70843 | 0.263 | −1.102 | 0.205 |
| Bulgaria | 9 | 6 | 0.917 ± 0.073 | 0.00607 ± 0.00104 | 0.62496 | 0.752 | −2.034 | 0.052 |
| Hungary | 8 | 6 | 0.893 ± 0.111 | 0.00411 ± 0.00095 | −117,532 | 0.111 | −3.589 ** | 0.000 |
| Italy | 10 | 4 | 0.733 ± 0.120 | 0.00428 ± 0.00102 | 0.20350 | 0.652 | −0.045 | 0.482 |
| Portugal | 11 | 3 | 0.345 ± 0.172 | 0.00146 ± 0.00078 | −0.77815 | 0.211 | −0.659 | 0.126 |
| Romania | 13 | 7 | 0.897 ± 0.054 | 0.00525 ± 0.00100 | −0.77485 | 0.215 | −2.640 | 0.015 |
| UK | 2 | 1 | 0.00000 | 0.00000 | ------ | ------ | ------ | ------ |
| Palestine | 3 | 1 | 0.00000 | 0.00000 | ------ | ------ | ------ | ------ |
| Iran | 12 | 4 | 0.682 ± 0.102 | 0.00224 ± 0.00050 | −0.57864 | 0.285 | −1.048 | 0.104 |
| Kiyrgyzstan | 3 | 2 | 0.667 ± 0.314 | 0.00182 ± 0.00086 | ----- | ------ | ------ | ------ |
| Türkiye | 21 | 5 | 0.424 ± 0.131 | 0.00221 ± 0.00077 | −165,358 | 0.176 | −3.127 ** | 0.004 |
| China | 50 | 21 | 0.935 ± 0.017 | 0.00729 ± 0.00106 | −1.63535 ** | 0.032 | −12.892 ** | 0.000 |
| Kurdistan | 21 | 6 | 0.724 ± 0.078 | 0.00304 ± 0.00065 | −1.38535 ** | 0.040 | −1.852 ** | 0.017 |
| Australia | 19 | 10 | 0.825 ± 0.084 | 0.00623 ± 0.00112 | −142,204 | 0.065 | −4.186 ** | 0.008 |
| Libya | 3 | 3 | 1.00 ± 0.272 | 0.00364 ± 0.00121 | ------ | ------ | ------ | ------ |
| Tunisia | 14 | 5 | 0.800 ± 0.068 | 0.00326 ± 0.00049 | −2.01165 ** | 0.001 | −12.237 ** | 0.000 |
| Latin America | 151 | 19 | 0.578 ± 0.00206 | 0.000409 ± 0.00371 | −2.39076 ** | 0.000 | −13.852 ** | 0.000 |
| Europe | 61 | 19 | 0.832 ± 0.041 | 0.00493 ± 0.00357 | −1.52138 ** | 0.036 | −13.824 ** | 0.142 |
| Middle East | 16 | 4 | 0.617 ± 0.00927 | 0.00204 ± 0.00168 | −0.62771 | 0.268 | −1.019 | 0.000 |
| Asia | 95 | 28 | 0.880 ± 0.00065 | 0.00581 ± 0.00477 | −1.95889 ** | 0.003 | −23.448 ** | 0.000 |
| Africa | 38 | 17 | 0.819 ± 0.00382 | 0.00307 ± 0.00358 | −2.19849 ** | 0.002 | −17.437 ** | 0.000 |
| total | 380 | 26 | 0.36539 ± 0.031 | 0.00321 ± 0.00054 | −2.42296 ** | 0.000 | −35.985 ** | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urriola-Urriola, N.; Rossi-Vargas, G.; Nilo-Bustios, Y. Haplotypes of Echinococcus granulosus sensu stricto in Chile and Their Comparison Through Sequences of the Mitochondrial cox1 Gene with Haplotypes from South America and Other Continents. Parasitologia 2025, 5, 40. https://doi.org/10.3390/parasitologia5030040
Urriola-Urriola N, Rossi-Vargas G, Nilo-Bustios Y. Haplotypes of Echinococcus granulosus sensu stricto in Chile and Their Comparison Through Sequences of the Mitochondrial cox1 Gene with Haplotypes from South America and Other Continents. Parasitologia. 2025; 5(3):40. https://doi.org/10.3390/parasitologia5030040
Chicago/Turabian StyleUrriola-Urriola, Nicole, Gabriela Rossi-Vargas, and Yenny Nilo-Bustios. 2025. "Haplotypes of Echinococcus granulosus sensu stricto in Chile and Their Comparison Through Sequences of the Mitochondrial cox1 Gene with Haplotypes from South America and Other Continents" Parasitologia 5, no. 3: 40. https://doi.org/10.3390/parasitologia5030040
APA StyleUrriola-Urriola, N., Rossi-Vargas, G., & Nilo-Bustios, Y. (2025). Haplotypes of Echinococcus granulosus sensu stricto in Chile and Their Comparison Through Sequences of the Mitochondrial cox1 Gene with Haplotypes from South America and Other Continents. Parasitologia, 5(3), 40. https://doi.org/10.3390/parasitologia5030040






