Heme-Based Gas Sensors in Nature and Their Chemical and Biotechnological Applications
Abstract
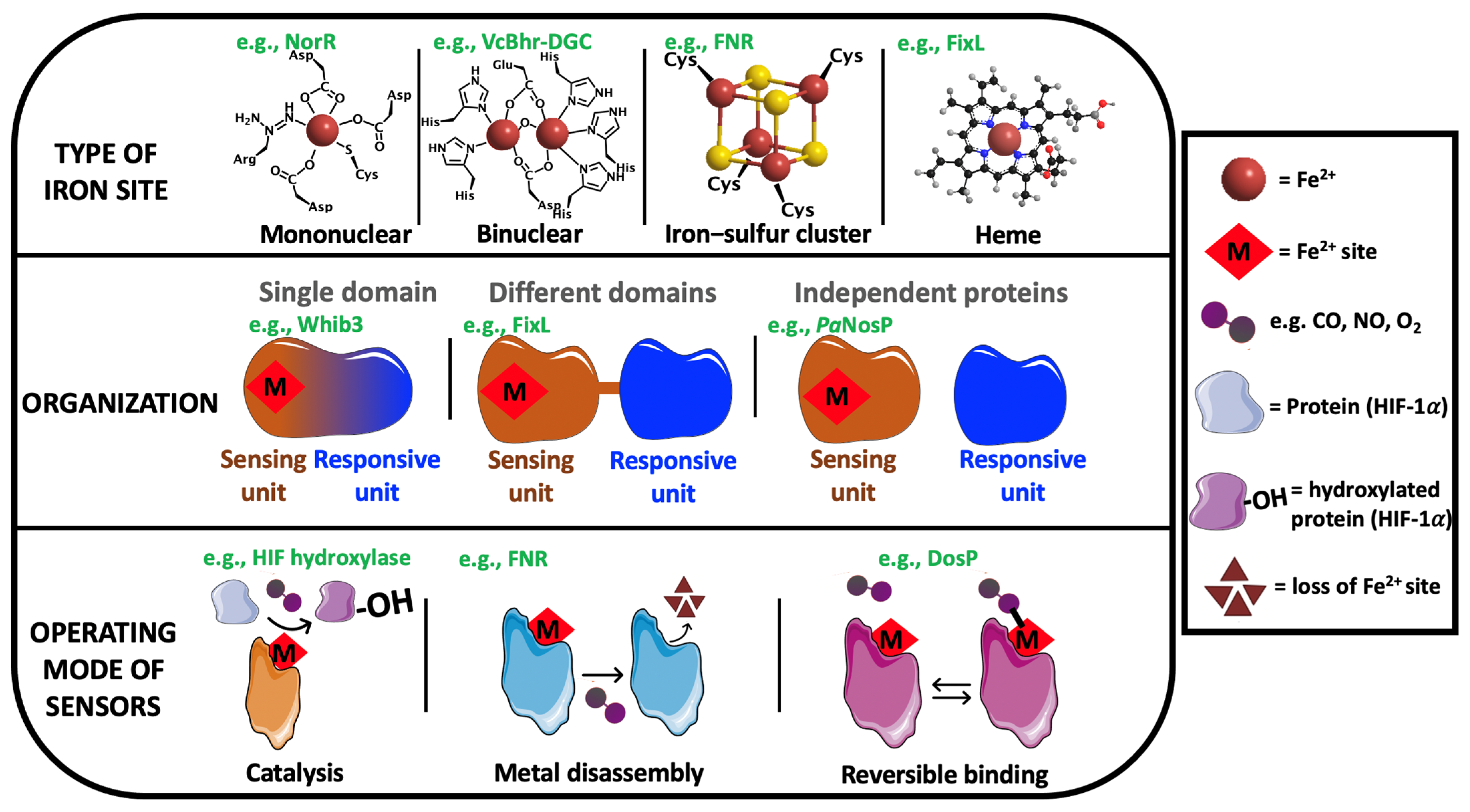
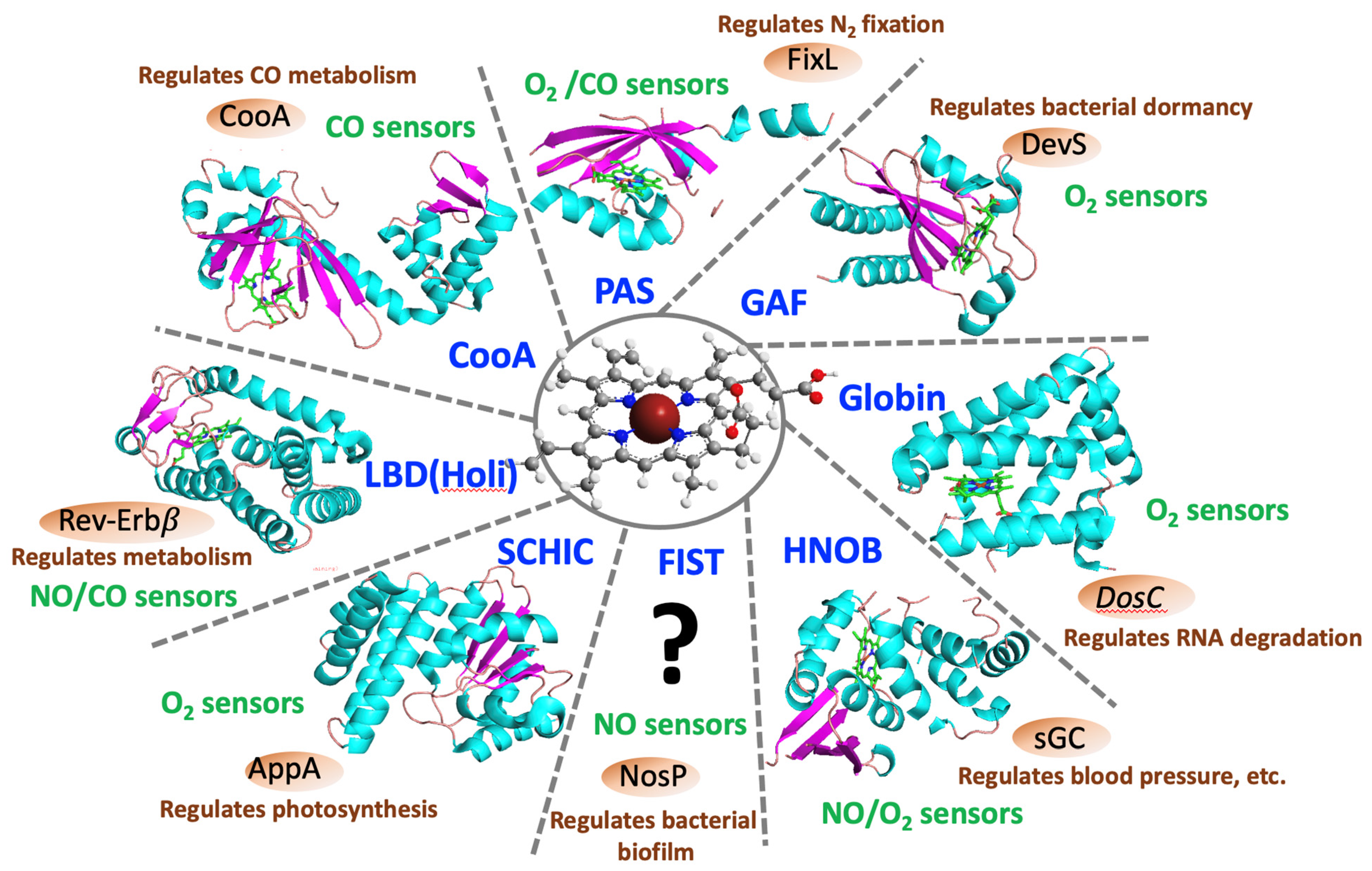
1. Introduction
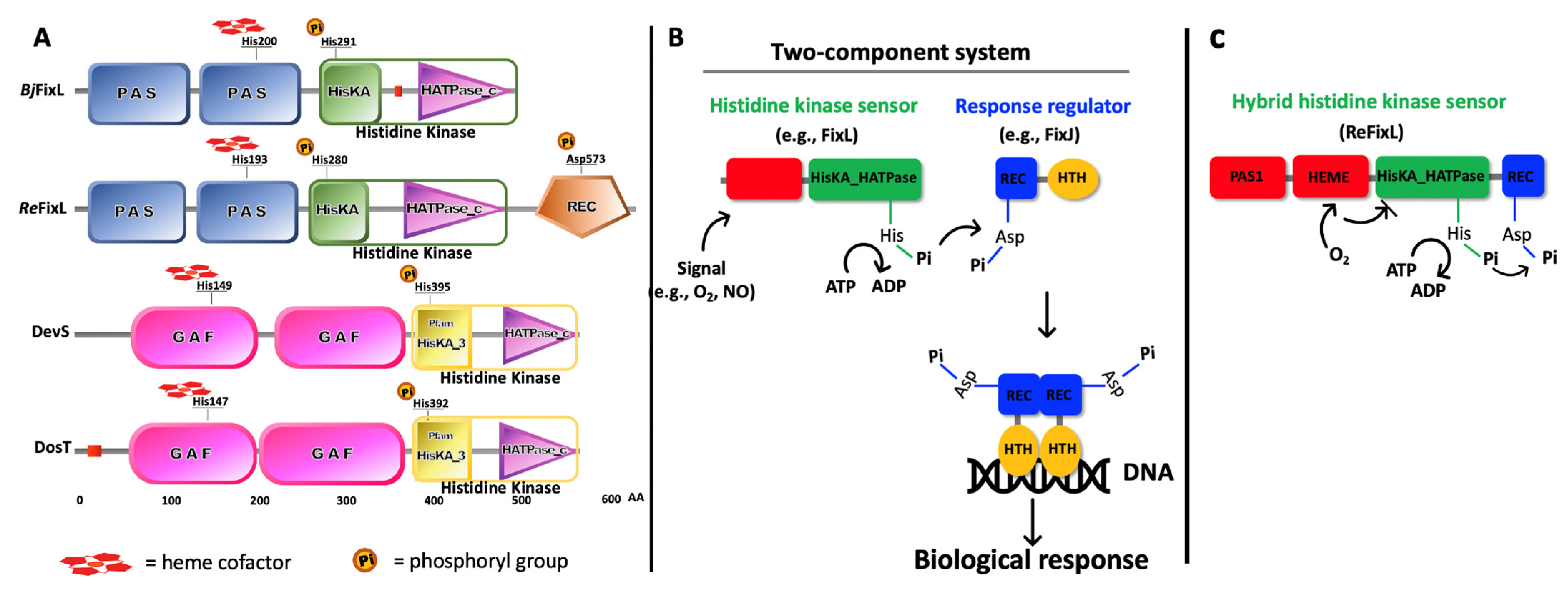
2. FixL—A Short Story
3. DevS and DosT—A Short History
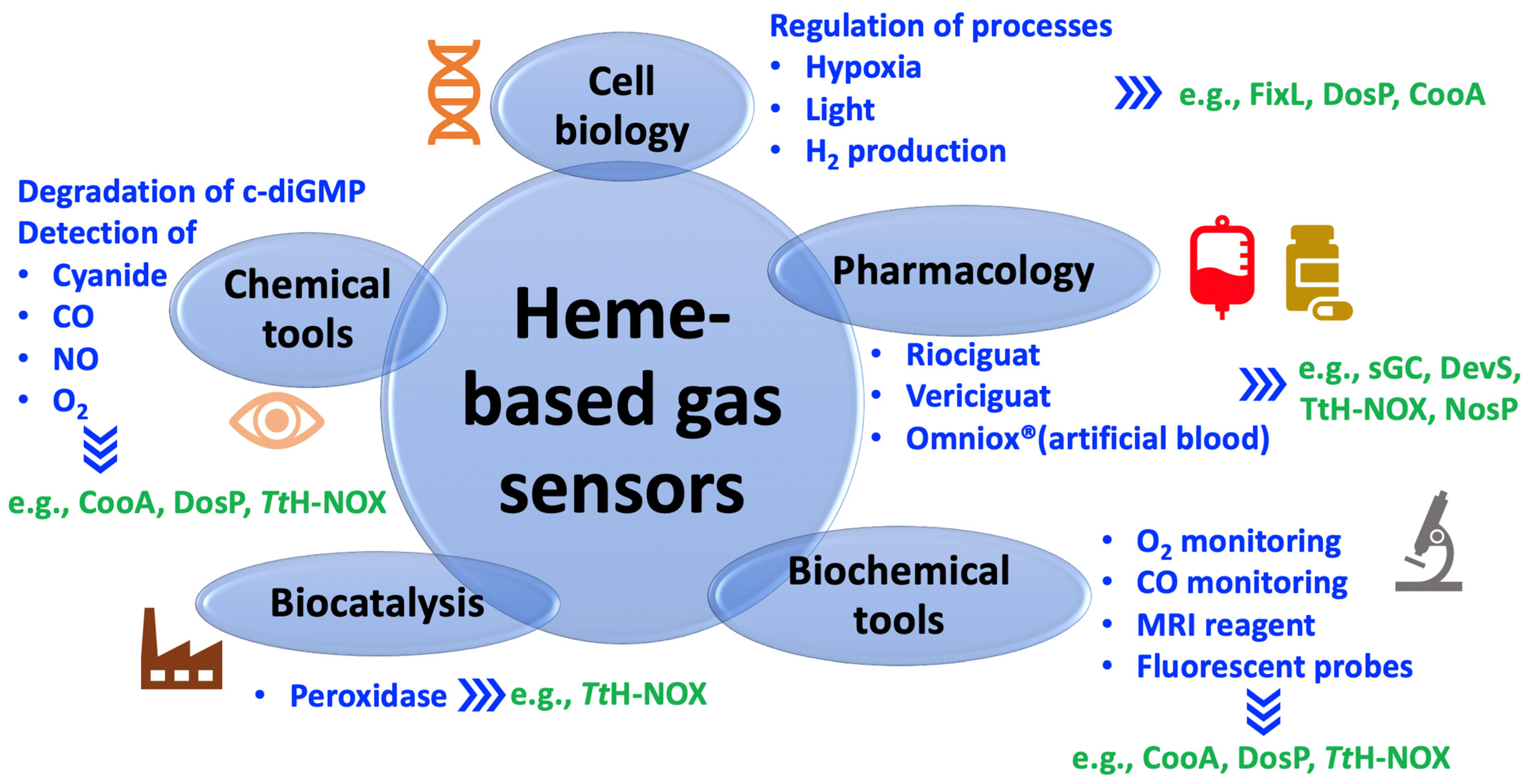
4. Diverse Potential Applications of Heme-Based Gas Sensors
4.1. Pharmacological Use
4.2. Chemical and Biological Tools
4.3. Systems Applied in Cell Biology
4.4. Biocatalysts
5. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BpeGReg | Bordetella pertussis globin-coupled regulator |
| CckA | Caulobacter crescentus histidine kinase protein A |
| c-di-GMP | Cyclic dimeric (3′,5′) guanosine monophosphate |
| cGMP | Cyclic guanosine monophosphate |
| CsH-NOX | H-NOX sensor from Caldanaerobacter subterraneus, formerly known as TtH-NOX |
| CskA | Chimeric sensory kinase A |
| CooA | CO oxidation activator protein |
| CORM-2 | CO-releasing molecule 2 |
| DevS | Differentially expressed in virulent strain sensor protein, the same protein as DosS |
| DevR | Differentially expressed in virulent strain response regulator protein, the same protein as DosR |
| DosC | Direct oxygen sensor cyclase protein |
| DosP | Direct oxygen sensor phosphodiesterase protein |
| DosS | Dormancy survival sensor protein |
| DosT | Dormancy survival sensor T protein |
| EcDOS | Escherichia coli direct oxygen sensor protein, the same protein as DosP |
| FIST | F-box and intracellular signal transduction proteins domain |
| FixL | Rhizobial nitrogen fixation gene L protein |
| FixJ | Nitrogen fixation gene J protein |
| FNR | Fumarate and nitrate reduction protein |
| GAF | cGMP-specific phosphodiesterases, adenylyl cyclase, and FhlA proteins domain |
| GTP | Guanosine-5′-triphosphate |
| HemAC-Lm | Heme-containing adenylate cyclase from Leishmania major |
| HemAT | Heme-based aerotactic transducer sensor protein |
| HIF | Hypoxia-inducible factor |
| HNOB | Heme NO-binding domain |
| HNOX | Heme NO- and oxygen-binding domain |
| HTH | Helix-turn-helix domain |
| HOLI | Ligand-binding domain of hormone receptors |
| Hpk2 | Histidine protein kinase 2 from Treponema denticola |
| YC-1 | 5-[1-(phenylmethyl)- 1H-indazol-3-yl]-2-furanmethanol |
| LBD | Ligand-binding domain of the nuclear receptor |
| LOV | Light–oxygen–voltage-sensing domain |
| MRI | Magnetic resonance imaging |
| Mtb | Mycobacterium tuberculosis |
| NifA | Oxygen-responsive nitrogen fixation sensor protein A |
| NorR | NO reductase regulatory protein |
| NosP | Nitric oxide-sensing protein |
| PAS | Per, period circadian protein, Arnt, aryl hydrocarbon receptor nuclear translocator protein, Sim, single-minded protein domain |
| pGpG | Linearized form of cyclic di-GMP, 5′-phosphoguanylyl- (3′ −> 5′)-guanosine |
| SAXS | Small-angle X-ray scattering |
| SCHIC | Sensor-containing heme instead of cobalamin domain |
| sGC | Soluble guanylate cyclase |
| ThkA | Thermotoga maritima histidine kinase A |
| TtH-NOX | H-NOX sensor from Thermoanaerobacter tengcongensis |
| VcBhr-DGC | Vibrio cholerae bacterial hemerythrin diguanylate cyclase sensor protein |
| Whib | regulator originally associated to the gene locus involved with the conversion of white multinucleoidal aseptate aerial hyphae into chains of mature grey uninucleoidal spores |
| Whib3 | NO and/or O2 sensor from Mycobacterium tuberculosis |
| YFP | Yellow fluorescent protein |
References
- Gonzaga de França Lopes, L.; Gouveia Júnior, F.S.; Karine Medeiros Holanda, A.; Maria Moreira de Carvalho, I.; Longhinotti, E.; Paulo, T.F.; Abreu, D.S.; Bernhardt, P.V.; Gilles-Gonzalez, M.-A.; Cirino Nogueira Diógenes, I.; et al. Bioinorganic systems responsive to the diatomic gases O2, NO, and CO: From biological sensors to therapy. Coord. Chem. Rev. 2021, 445, 214096. [Google Scholar] [CrossRef]
- Negrerie, M. Iron transitions during activation of allosteric heme proteins in cell signaling. Metallomics 2019, 11, 868–893. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Gonzalez, G. Heme-based sensors: Defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 2005, 99, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Huang, D.; Yan, F.; Stranava, M.; Bartosova, M.; Fojtikova, V.; Martinkova, M. Gaseous O2, NO, and CO in signal transduction: Structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 2015, 115, 6491–6533. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Gonzalez, M.-A.; Gonzalez, G. A Surfeit of Biological Heme-based Sensors. In The Smallest Biomolecules: Diatomics and Their Interactions with Heme Proteins; Ghosh, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Britt, R.D.; Rao, G.; Tao, L. Bioassembly of complex iron-sulfur enzymes: Hydrogenases and nitrogenases. Nat. Rev. Chem. 2020, 4, 542–549. [Google Scholar] [CrossRef] [PubMed]
- D’Autreaux, B.; Tucker, N.P.; Dixon, R.; Spiro, S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 2005, 437, 769–772. [Google Scholar] [CrossRef]
- Schaller, R.A.; Ali, S.K.; Klose, K.E.; Kurtz, D.M., Jr. A bacterial hemerythrin domain regulates the activity of a Vibrio cholerae diguanylate cyclase. Biochemistry 2012, 51, 8563–8570. [Google Scholar] [CrossRef]
- Crack, J.C.; Green, J.; Thomson, A.J.; Le Brun, N.E. Iron-sulfur clusters as biological sensors: The chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 2014, 47, 3196–3205. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Ditta, G.S.; Helinski, D.R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 1991, 350, 170–172. [Google Scholar] [CrossRef]
- Hossain, S.; Boon, E.M. Discovery of a Novel Nitric Oxide Binding Protein and Nitric-Oxide-Responsive Signaling Pathway in Pseudomonas aeruginosa. ACS Infect. Dis. 2017, 3, 454–461. [Google Scholar] [CrossRef]
- Taabazuing, C.Y.; Hangasky, J.A.; Knapp, M.J. Oxygen sensing strategies in mammals and bacteria. J. Inorg. Biochem. 2014, 133, 63–72. [Google Scholar] [CrossRef]
- Fandrey, J.; Schodel, J.; Eckardt, K.U.; Katschinski, D.M.; Wenger, R.H. Now a Nobel gas: Oxygen. Pflug. Arch. 2019, 471, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Crack, J.C.; Thomson, A.J.; Le Brun, N.E. Mass spectrometric identification of intermediates in the O2-driven [4Fe-4S] to [2Fe-2S] cluster conversion in FNR. Proc. Natl. Acad. Sci. USA 2017, 114, E3215–E3223. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Gonzalez, M.A.; Gonzalez, G.; Sousa, E.H.; Tuckerman, J. Oxygen-sensing histidine-protein kinases: Assays of ligand binding and turnover of response-regulator substrates. Methods Enzymol. 2008, 437, 173–189. [Google Scholar] [CrossRef]
- Fojtikova, V.; Stranava, M.; Vos, M.H.; Liebl, U.; Hranicek, J.; Kitanishi, K.; Shimizu, T.; Martinkova, M. Kinetic Analysis of a Globin-Coupled Histidine Kinase, AfGcHK: Effects of the Heme Iron Complex, Response Regulator, and Metal Cations on Autophosphorylation Activity. Biochemistry 2015, 54, 5017–5029. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.P.; Mittal, C.K.; Katsuki, S.; Murad, F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. USA 1977, 74, 3203–3207. [Google Scholar] [CrossRef]
- Patterson, D.C.; Ruiz, M.P.; Yoon, H.; Walker, J.A.; Armache, J.P.; Yennawar, N.H.; Weinert, E.E. Differential ligand-selective control of opposing enzymatic activities within a bifunctional c-di-GMP enzyme. Proc. Natl. Acad. Sci. USA 2021, 118, e2100657118. [Google Scholar] [CrossRef] [PubMed]
- Tuckerman, J.R.; Gonzalez, G.; Sousa, E.H.; Wan, X.; Saito, J.A.; Alam, M.; Gilles-Gonzalez, M.A. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 2009, 48, 9764–9774. [Google Scholar] [CrossRef]
- Shimizu, T.; Lengalova, A.; Martinek, V.; Martinkova, M. Heme: Emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 2019, 48, 5624–5657. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Sousa, E.H.S. Escherichia coli DosC and DosP: A role of c-di-GMP in compartmentalized sensing by degradosomes. Adv. Microb. Physiol. 2019, 75, 53–67. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 2011, 407, 633–639. [Google Scholar] [CrossRef]
- Shelver, D.; Kerby, R.L.; He, Y.; Roberts, G.P. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. USA 1997, 94, 11216–11220. [Google Scholar] [CrossRef]
- Schofield, C.J.; Ratcliffe, P.J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004, 5, 343–354. [Google Scholar] [CrossRef]
- Nunes, E.D.; Villela, A.D.; Basso, L.A.; Teixeira, E.H.; Andrade, A.L.; Vasconcelos, M.A.; Neto, L.G.D.; Gondim, A.C.S.; Diogenes, I.C.N.; Romo, A.I.B.; et al. Light-induced disruption of an acyl hydrazone link as a novel strategy for drug release and activation: Isoniazid as a proof-of-concept case. Inorg. Chem. Front. 2020, 7, 859–870. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, R.; Wu, J.X.; Chen, L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature 2019, 574, 206–210. [Google Scholar] [CrossRef]
- Wittenborn, E.C.; Marletta, M.A. Structural Perspectives on the Mechanism of Soluble Guanylate Cyclase Activation. Int. J. Mol. Sci. 2021, 22, 5439. [Google Scholar] [CrossRef]
- Horst, B.G.; Yokom, A.L.; Rosenberg, D.J.; Morris, K.L.; Hammel, M.; Hurley, J.H.; Marletta, M.A. Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy. Elife 2019, 8, e50634. [Google Scholar] [CrossRef]
- Lanzilotta, W.N.; Schuller, D.J.; Thorsteinsson, M.V.; Kerby, R.L.; Roberts, G.P.; Poulos, T.L. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 2000, 7, 876–880. [Google Scholar] [CrossRef]
- Williams, D.E.; Nisbett, L.M.; Bacon, B.; Boon, E. Bacterial Heme-Based Sensors of Nitric Oxide. Antioxid. Redox Signal. 2018, 29, 1872–1887. [Google Scholar] [CrossRef]
- Stuehr, D.J.; Misra, S.; Dai, Y.; Ghosh, A. Maturation, inactivation, and recovery mechanisms of soluble guanylyl cyclase. J. Biol. Chem. 2021, 296, 100336. [Google Scholar] [CrossRef]
- Friebe, A.; Sandner, P.; Schmidtko, A. cGMP: A unique 2nd messenger molecule—Recent developments in cGMP research and development. Naunyn-Schmiedebergs Arch. Pharmacol. 2020, 393, 287–302. [Google Scholar] [CrossRef]
- Sousa, E.H.S.; Gonzalez, G.; Gilles-Gonzalez, M.A. Soluble guanylate cyclase and its microbial relatives. In The Smallest Biomolecules: Perspectives on Heme-Diatomic Interactions; Ghosh, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 524–539. [Google Scholar]
- Bian, K.; Murad, F. What is next in nitric oxide research? From cardiovascular system to cancer biology. Nitric Oxide 2014, 43, 3–7. [Google Scholar] [CrossRef]
- Rutten, P.J.; Poole, P.S. Oxygen regulatory mechanisms of nitrogen fixation in rhizobia. Adv. Microb. Physiol. 2019, 75, 325–389. [Google Scholar] [CrossRef]
- Sousa, E.H.S.; Gilles-Gonzalez, M.A. Haem-Based Sensors of O2: Lessons and Perspectives. Adv. Microb. Physiol. 2017, 71, 235–257. [Google Scholar] [CrossRef]
- Ganesh, I.; Gwon, D.A.; Lee, J.W. Gas-Sensing Transcriptional Regulators. Biotechnol. J. 2020, 15, e1900345. [Google Scholar] [CrossRef]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef]
- Ledermann, R.; Schulte, C.C.M.; Poole, P.S. How Rhizobia Adapt to the Nodule Environment. J. Bacteriol. 2021, 203, e0053920. [Google Scholar] [CrossRef]
- Wright, G.S.A.; Saeki, A.; Hikima, T.; Nishizono, Y.; Hisano, T.; Kamaya, M.; Nukina, K.; Nishitani, H.; Nakamura, H.; Yamamoto, M.; et al. Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Caceres, A.I.; Sousa, E.H.; Tomchick, D.R.; Brautigam, C.; Gonzalez, C.; Machius, M. A proximal arginine R206 participates in switching of the Bradyrhizobium japonicum FixL oxygen sensor. J. Mol. Biol. 2006, 360, 80–89. [Google Scholar] [CrossRef]
- Balland, V.; Bouzhir-Sima, L.; Anxolabéhère-Mallart, E.; Boussac, A.; Vos, M.H.; Liebl, U.; Mattioli, T.A. Functional Implications of the Propionate 7−Arginine 220 Interaction in the FixLH Oxygen Sensor from Bradyrhizobium japonicum. Biochemistry 2006, 45, 2072–2084. [Google Scholar] [CrossRef]
- Sousa, E.H.; Gonzalez, G.; Gilles-Gonzalez, M.A. Oxygen blocks the reaction of the FixL-FixJ complex with ATP but does not influence binding of FixJ or ATP to FixL. Biochemistry 2005, 44, 15359–15365. [Google Scholar] [CrossRef]
- Key, J.; Moffat, K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry 2005, 44, 4627–4635. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Dioum, E.M.; Gilles-Gonzalez, M.A. Ligand and oxidation-state specific regulation of the heme-based oxygen sensor FixL from Sinorhizobium meliloti. Biochemistry 2002, 41, 6170–6177. [Google Scholar] [CrossRef]
- Hao, B.; Isaza, C.; Arndt, J.; Soltis, M.; Chan, M.K. Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 2002, 41, 12952–12958. [Google Scholar] [CrossRef]
- Gong, W.; Hao, B.; Mansy, S.S.; Gonzalez, G.; Gilles-Gonzalez, M.A.; Chan, M.K. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 1998, 95, 15177–15182. [Google Scholar] [CrossRef]
- Guimaraes, W.G.; Gondim, A.C.S.; Costa, P.; Gilles-Gonzalez, M.A.; Lopes, L.G.F.; Carepo, M.S.P.; Sousa, E.H.S. Insights into signal transduction by a hybrid FixL: Denaturation study of on and off states of a multi-domain oxygen sensor. J. Inorg. Biochem. 2017, 172, 129–137. [Google Scholar] [CrossRef]
- Honorio-Felicio, N.; Carepo, M.S.; Paulo, T.D.F.; de Franca Lopes, L.G.; Sousa, E.H.; Diogenes, I.C.; Bernhardt, P.V. The Heme-Based Oxygen Sensor Rhizobium etli FixL: Influence of Auxiliary Ligands on Heme Redox Potential and Implications on the Enzyme Activity. J. Inorg. Biochem. 2016, 164, 34–41. [Google Scholar] [CrossRef][Green Version]
- Yamawaki, T.; Ishikawa, H.; Mizuno, M.; Nakamura, H.; Shiro, Y.; Mizutani, Y. Regulatory Implications of Structural Changes in Tyr201 of the Oxygen Sensor Protein FixL. Biochemistry 2016, 55, 4027–4035. [Google Scholar] [CrossRef]
- Yamada, S.; Sugimoto, H.; Kobayashi, M.; Ohno, A.; Nakamura, H.; Shiro, Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure 2009, 17, 1333–1344. [Google Scholar] [CrossRef]
- Miksovska, J.; Suquet, C.; Satterlee, J.D.; Larsen, R.W. Characterization of conformational changes coupled to ligand photodissociation from the heme binding domain of FixL. Biochemistry 2005, 44, 10028–10036. [Google Scholar] [CrossRef]
- Dunham, C.M.; Dioum, E.M.; Tuckerman, J.R.; Gonzalez, G.; Scott, W.G.; Gilles-Gonzalez, M.A. A distal arginine in oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure. Biochemistry 2003, 42, 7701–7708. [Google Scholar] [CrossRef]
- Miyatake, H.; Mukai, M.; Adachi, S.; Nakamura, H.; Tamura, K.; Iizuka, T.; Shiro, Y.; Strange, R.W.; Hasnain, S.S. Iron coordination structures of oxygen sensor FixL characterized by Fe K-edge extended X-ray absorption fine structure and resonance raman spectroscopy. J. Biol. Chem. 1999, 274, 23176–23184. [Google Scholar] [CrossRef]
- Gong, W.; Hao, B.; Chan, M.K. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 2000, 39, 3955–3962. [Google Scholar] [CrossRef] [PubMed]
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Sousa, E.H.; Tuckerman, J.R.; Gondim, A.C.; Gonzalez, G.; Gilles-Gonzalez, M.A. Signal transduction and phosphoryl transfer by a FixL hybrid kinase with low oxygen affinity: Importance of the vicinal PAS domain and receiver aspartate. Biochemistry 2013, 52, 456–465. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Gonzalez, G.; Perutz, M.F.; Kiger, L.; Marden, M.C.; Poyart, C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 1994, 33, 8067–8073. [Google Scholar] [CrossRef]
- Sousa, E.H.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A. A memory of oxygen binding explains the dose response of the heme-based sensor FixL. Biochemistry 2007, 46, 6249–6257. [Google Scholar] [CrossRef]
- Vauquelin, G.; Charlton, S.J. Long-lasting target binding and rebinding as mechanisms to prolong in vivo drug action. Br. J. Pharmacol. 2010, 161, 488–508. [Google Scholar] [CrossRef]
- Nuernberger, P.; Lee, K.F.; Bonvalet, A.; Bouzhir-Sima, L.; Lambry, J.C.; Liebl, U.; Joffre, M.; Vos, M.H. Strong ligand-protein interactions revealed by ultrafast infrared spectroscopy of CO in the heme pocket of the oxygen sensor FixL. J. Am. Chem. Soc. 2011, 133, 17110–17113. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Srajer, V.; Pahl, R.; Moffat, K. Time-resolved crystallographic studies of the heme domain of the oxygen sensor FixL: Structural dynamics of ligand rebinding and their relation to signal transduction. Biochemistry 2007, 46, 4706–4715. [Google Scholar] [CrossRef]
- Yano, S.; Ishikawa, H.; Mizuno, M.; Nakamura, H.; Shiro, Y.; Mizutani, Y. Ultraviolet resonance Raman observations of the structural dynamics of rhizobial oxygen sensor FixL on ligand recognition. J. Phys. Chem. B 2013, 117, 15786–15791. [Google Scholar] [CrossRef]
- Yamawaki, T.; Mizuno, M.; Ishikawa, H.; Takemura, K.; Kitao, A.; Shiro, Y.; Mizutani, Y. Regulatory Switching by Concerted Motions on the Microsecond Time Scale of the Oxygen Sensor Protein FixL. J. Phys. Chem. B 2021. [Google Scholar] [CrossRef]
- Crosson, S.; McGrath, P.T.; Stephens, C.; McAdams, H.H.; Shapiro, L. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl. Acad. Sci. USA 2005, 102, 8018–8023. [Google Scholar] [CrossRef] [PubMed]
- Roset, M.S.; Almiron, M.A. FixL-like sensor FlbS of Brucella abortus binds haem and is necessary for survival within eukaryotic cells. FEBS Lett. 2013, 587, 3102–3107. [Google Scholar] [CrossRef] [PubMed]
- Murthy, U.M.; Wecker, M.S.; Posewitz, M.C.; Gilles-Gonzalez, M.A.; Ghirardi, M.L. Novel FixL homologues in Chlamydomonas reinhardtii bind heme and O(2). FEBS Lett. 2012, 586, 4282–4288. [Google Scholar] [CrossRef]
- Schaefers, M.M.; Liao, T.L.; Boisvert, N.M.; Roux, D.; Yoder-Himes, D.; Priebe, G.P. An Oxygen-Sensing Two-Component System in the Burkholderia cepacia Complex Regulates Biofilm, Intracellular Invasion, and Pathogenicity. PLoS Pathog. 2017, 13, e1006116. [Google Scholar] [CrossRef] [PubMed]
- Schaefers, M.M.; Wang, B.X.; Boisvert, N.M.; Martini, S.J.; Bonney, S.L.; Marshall, C.W.; Laub, M.T.; Cooper, V.S.; Priebe, G.P. Evolution towards Virulence in a Burkholderia Two-Component System. MBio 2021, 12, e0182321. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Tuberculosis Report; World Health Organization: Geneva, Switzerland, 2020.
- Campanico, A.; Harjivan, S.G.; Warner, D.F.; Moreira, R.; Lopes, F. Addressing Latent Tuberculosis: New Advances in Mimicking the Disease, Discovering Key Targets, and Designing Hit Compounds. Int. J. Mol. Sci. 2020, 21, 8854. [Google Scholar] [CrossRef] [PubMed]
- Sousa, E.H.S.; Diogenes, I.C.N.; Lopes, L.G.F.; Moura, J.J.G. Potential therapeutic approaches for a sleeping pathogen: Tuberculosis a case for bioinorganic chemistry. J. Biol. Inorg. Chem. 2020, 25, 685–704. [Google Scholar] [CrossRef]
- Dasgupta, N.; Kapur, V.; Singh, K.K.; Das, T.K.; Sachdeva, S.; Jyothisri, K.; Tyagi, J.S. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 2000, 80, 141–159. [Google Scholar] [CrossRef]
- Saini, D.K.; Malhotra, V.; Dey, D.; Pant, N.; Das, T.K.; Tyagi, J.S. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 2004, 150, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Saini, D.K.; Malhotra, V.; Tyagi, J.S. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 2004, 565, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Liao, R.P.; Wisedchaisri, G.; Hol, W.G.; Sherman, D.R. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 2004, 279, 23082–23087. [Google Scholar] [CrossRef]
- Gupta, V.K.; Kumar, M.M.; Singh, D.; Bisht, D.; Sharma, S. Drug targets in dormant Mycobacterium tuberculosis: Can the conquest against tuberculosis become a reality? Infect. Dis. 2018, 50, 81–94. [Google Scholar] [CrossRef]
- Sardiwal, S.; Kendall, S.L.; Movahedzadeh, F.; Rison, S.C.; Stoker, N.G.; Djordjevic, S. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 2005, 353, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Sousa, E.H.S.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.-A. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 2007, 16, 1708–1719. [Google Scholar] [CrossRef]
- Sousa, E.H.S.; Gonzalez, G.; Gilles-Gonzalez, M.A. Target DNA stabilizes Mycobacterium tuberculosis DevR/DosR phosphorylation by the full-length oxygen sensors DevS/DosS and DosT. FEBS J. 2017, 284, 3954–3967. [Google Scholar] [CrossRef]
- Kumar, A.; Toledo, J.C.; Patel, R.P.; Lancaster, J.R.; Steyn, A.J.C. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 2007, 104, 11568–11573. [Google Scholar] [CrossRef] [PubMed]
- Barreto, G.A.; Carepo, M.S.P.; Gondim, A.C.S.; Guimarães, W.G.; Lopes, L.G.F.; Bernhardt, P.V.; Paulo, T.F.; Sousa, E.H.S.; Diógenes, I.C.N. A spectroelectrochemical investigation of the heme-based sensor DevS from Mycobacterium tuberculosis: A redox versus oxygen sensor. FEBS J. 2019, 286, 4278–4293. [Google Scholar] [CrossRef]
- Honaker, R.W.; Leistikow, R.L.; Bartek, I.L.; Voskuil, M.I. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect. Immun. 2009, 77, 3258–3263. [Google Scholar] [CrossRef]
- Cho, H.Y.; Cho, H.J.; Kim, Y.M.; Oh, J.I.; Kang, B.S. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 13057–13067. [Google Scholar] [CrossRef]
- Ioanoviciu, A.; Meharenna, Y.T.; Poulos, T.L.; Ortiz de Montellano, P.R. DevS oxy complex stability identifies this heme protein as a gas sensor in Mycobacterium tuberculosis dormancy. Biochemistry 2009, 48, 5839–5848. [Google Scholar] [CrossRef]
- Podust, L.M.; Ioanoviciu, A.; Ortiz de Montellano, P.R. 2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry 2008, 47, 12523–12531. [Google Scholar] [CrossRef] [PubMed]
- Basudhar, D.; Madrona, Y.; Yukl, E.T.; Sivaramakrishnan, S.; Nishida, C.R.; Moenne-Loccoz, P.; Ortiz de Montellano, P.R. Distal Hydrogen-bonding Interactions in Ligand Sensing and Signaling by Mycobacterium tuberculosis DosS. J. Biol. Chem. 2016, 291, 16100–16111. [Google Scholar] [CrossRef]
- Yukl, E.T.; Ioanoviciu, A.; de Montellano, P.R.; Moenne-Loccoz, P. Interdomain interactions within the two-component heme-based sensor DevS from Mycobacterium tuberculosis. Biochemistry 2007, 46, 9728–9736. [Google Scholar] [CrossRef] [PubMed]
- Lobao, J.; Gondim, A.C.S.; Guimaraes, W.G.; Gilles-Gonzalez, M.A.; Lopes, L.G.F.; Sousa, E.H.S. Oxygen triggers signal transduction in the DevS (DosS) sensor of Mycobacterium tuberculosis by modulating the quaternary structure. FEBS J. 2019, 286, 479–494. [Google Scholar] [CrossRef]
- Skalova, T.; Lengalova, A.; Dohnalek, J.; Harlos, K.; Mihalcin, P.; Kolenko, P.; Stranava, M.; Blaha, J.; Shimizu, T.; Martinkova, M. Disruption of the dimerization interface of the sensing domain in the dimeric heme-based oxygen sensor AfGcHK abolishes bacterial signal transduction. J. Biol. Chem. 2020, 295, 1587–1597. [Google Scholar] [CrossRef]
- Burns, J.L.; Rivera, S.; Deer, D.D.; Joynt, S.C.; Dvorak, D.; Weinert, E.E. Oxygen and Bis(3′,5′)-cyclic Dimeric Guanosine Monophosphate Binding Control Oligomerization State Equilibria of Diguanylate Cyclase-Containing Globin Coupled Sensors. Biochemistry 2016, 55, 6642–6651. [Google Scholar] [CrossRef] [PubMed]
- Sousa, E.H.; Lopes, L.G.; Gonzalez, G.; Gilles-Gonzalez, M.A. Drug discovery targeting heme-based sensors and their coupled activities. J. Inorg. Biochem. 2017, 167, 12–20. [Google Scholar] [CrossRef]
- Sandner, P.; Zimmer, D.P.; Milne, G.T.; Follmann, M.; Hobbs, A.; Stasch, J.P. Soluble Guanylate Cyclase Stimulators and Activators. Handb. Exp. Pharmacol. 2021, 264, 355–394. [Google Scholar] [CrossRef] [PubMed]
- Muniz Carvalho, E.; Silva Sousa, E.H.; Bernardes-Génisson, V.; Gonzaga de França Lopes, L. When NO Is not Enough: Chemical Systems, Advances and Challenges in the Development of NO and HNO Donors for Old and Current Medical Issues. Eur. J. Inorg. Chem. 2021, 2021, 4316–4348. [Google Scholar] [CrossRef]
- Ko, F.N.; Wu, C.C.; Kuo, S.C.; Lee, F.Y.; Teng, C.M. YC-1, a novel activator of platelet guanylate cyclase. Blood 1994, 84, 4226–4233. [Google Scholar] [CrossRef]
- Sa, D.S.; Fernandes, A.F.; Silva, C.D.; Costa, P.P.; Fonteles, M.C.; Nascimento, N.R.; Lopes, L.G.; Sousa, E.H. Non-nitric oxide based metallovasodilators: Synthesis, reactivity and biological studies. Dalton Trans. 2015, 44, 13633–13640. [Google Scholar] [CrossRef]
- He, B.; Chen, Z. Molecular Targets for Small-Molecule Modulators of Circadian Clocks. Curr. Drug Metab. 2016, 17, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Ellermann, M.; Sperandio, V. Bacterial signaling as an antimicrobial target. Curr. Opin. Microbiol. 2020, 57, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Wille, J.; Coenye, T. Biofilm dispersion: The key to biofilm eradication or opening Pandora’s box? Biofilm 2020, 2, 100027. [Google Scholar] [CrossRef]
- Cai, Y.M.; Webb, J.S. Optimization of nitric oxide donors for investigating biofilm dispersal response in Pseudomonas aeruginosa clinical isolates. Appl. Microbiol. Biotechnol. 2020, 104, 8859–8869. [Google Scholar] [CrossRef] [PubMed]
- da Silva Filho, P.M.; Andrade, A.L.; Lopes, J.; Pinheiro, A.A.; de Vasconcelos, M.A.; Fonseca, S.; Lopes, L.G.F.; Sousa, E.H.S.; Teixeira, E.H.; Longhinotti, E. The biofilm inhibition activity of a NO donor nanosilica with enhanced antibiotics action. Int. J. Pharm. 2021, 610, 121220. [Google Scholar] [CrossRef]
- Howlin, R.P.; Cathie, K.; Hall-Stoodley, L.; Cornelius, V.; Duignan, C.; Allan, R.N.; Fernandez, B.O.; Barraud, N.; Bruce, K.D.; Jefferies, J.; et al. Low-Dose Nitric Oxide as Targeted Anti-biofilm Adjunctive Therapy to Treat Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. Mol. Ther. 2017, 25, 2104–2116. [Google Scholar] [CrossRef] [PubMed]
- Boce, M.; Tasse, M.; Mallet-Ladeira, S.; Pillet, F.; Da Silva, C.; Vicendo, P.; Lacroix, P.G.; Malfant, I.; Rols, M.P. Effect of trans(NO, OH)-[RuFT(Cl)(OH)NO](PF6) ruthenium nitrosyl complex on methicillin-resistant Staphylococcus epidermidis. Sci. Rep. 2019, 9, 4867. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Abramovitch, R.B. Inhibiting DosRST as a new approach to tuberculosis therapy. Future Med. Chem. 2020, 12, 457–467. [Google Scholar] [CrossRef]
- Gupta, R.K.; Thakur, T.S.; Desiraju, G.R.; Tyagi, J.S. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J. Med. Chem. 2009, 52, 6324–6334. [Google Scholar] [CrossRef]
- Kaur, K.; Taneja, N.K.; Dhingra, S.; Tyagi, J.S. DevR (DosR) mimetic peptides impair transcriptional regulation and survival of Mycobacterium tuberculosis under hypoxia by inhibiting the autokinase activity of DevS sensor kinase. BMC Microbiol. 2014, 14, 195. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dhingra, S.; Kaur, K.; Taneja, N.K.; Tyagi, J.S. DevR (DosR) binding peptide inhibits adaptation of Mycobacterium tuberculosis under hypoxia. FEMS Microbiol. Lett. 2012, 330, 66–71. [Google Scholar] [CrossRef]
- Zheng, H.; Colvin, C.J.; Johnson, B.K.; Kirchhoff, P.D.; Wilson, M.; Jorgensen-Muga, K.; Larsen, S.D.; Abramovitch, R.B. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 2017, 13, 218–225. [Google Scholar] [CrossRef]
- Zheng, H.; Williams, J.T.; Aleiwi, B.; Ellsworth, E.; Abramovitch, R.B. Inhibiting Mycobacterium tuberculosis DosRST Signaling by Targeting Response Regulator DNA Binding and Sensor Kinase Heme. ACS Chem. Biol. 2020, 15, 52–62. [Google Scholar] [CrossRef]
- Patel, Y.S.; Mistry, N.; Mehra, S. Repurposing artemisinin as an anti-mycobacterial agent in synergy with rifampicin. Tuberculosis 2019, 115, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.H. Novel Pharmacological Activity of Artesunate and Artemisinin: Their Potential as Anti-Tubercular Agents. J. Clin. Med. 2017, 6, 30. [Google Scholar] [CrossRef]
- Wan, X.; Tuckerman, J.R.; Saito, J.A.; Freitas, T.A.; Newhouse, J.S.; Denery, J.R.; Galperin, M.Y.; Gonzalez, G.; Gilles-Gonzalez, M.A.; Alam, M. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J. Mol. Biol. 2009, 388, 262–270. [Google Scholar] [CrossRef]
- Sarkar, J.; Miller, D.P.; Oliver, L.D.; Marconi, R.T. The Treponema denticola PAS Domain-Containing Histidine Kinase Hpk2 Is a Heme Binding Sensor of Oxygen Levels. J. Bacteriol. 2018, 200, e00116–e00118. [Google Scholar] [CrossRef]
- Sen Santara, S.; Roy, J.; Mukherjee, S.; Bose, M.; Saha, R.; Adak, S. Globin-coupled heme containing oxygen sensor soluble adenylate cyclase in Leishmania prevents cell death during hypoxia. Proc. Natl. Acad. Sci. USA 2013, 110, 16790–16795. [Google Scholar] [CrossRef]
- Abuchowski, A. SANGUINATE (PEGylated Carboxyhemoglobin Bovine): Mechanism of Action and Clinical Update. Artif. Organs 2017, 41, 346–350. [Google Scholar] [CrossRef]
- Azarov, I.; Wang, L.; Rose, J.J.; Xu, Q.; Huang, X.N.; Belanger, A.; Wang, Y.; Guo, L.; Liu, C.; Ucer, K.B.; et al. Five-coordinate H64Q neuroglobin as a ligand-trap antidote for carbon monoxide poisoning. Sci. Transl. Med. 2016, 8, 368ra173. [Google Scholar] [CrossRef]
- Hoops, H.E.; Manning, J.E.; Graham, T.L.; McCully, B.H.; McCurdy, S.L.; Ross, J.D. Selective aortic arch perfusion with fresh whole blood or HBOC-201 reverses hemorrhage-induced traumatic cardiac arrest in a lethal model of noncompressible torso hemorrhage. J. Trauma Acute Care Surg. 2019, 87, 263–273. [Google Scholar] [CrossRef]
- Alam, M.; Larsen, R. Heme Proteins hemAT-Hs and hemAT-Bs and Their Use in Medicine and Microsensors. U.S. Patent 7,129,329, 31 October 2006. [Google Scholar]
- Boehme, J.; Le Moan, N.; Kameny, R.J.; Loucks, A.; Johengen, M.J.; Lesneski, A.L.; Gong, W.; Goudy, B.D.; Davis, T.; Tanaka, K.; et al. Preservation of myocardial contractility during acute hypoxia with OMX-CV, a novel oxygen delivery biotherapeutic. PLoS Biol. 2018, 16, e2005924. [Google Scholar] [CrossRef]
- Itoh, T.; Matsuura, S.I.; Chuong, T.T.; Tanaike, O.; Hamakawa, S.; Shimizu, T. Successful Mesoporous Silica Encapsulation of Optimally Functional EcDOS (E. coli Direct Oxygen Sensor), a Heme-based O2-Sensing Phosphodiesterase. Anal. Sci. 2019, 35, 329–335. [Google Scholar] [CrossRef]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef]
- Opoku-Temeng, C.; Sintim, H.O. Targeting c-di-GMP Signaling, Biofilm Formation, and Bacterial Motility with Small Molecules. Methods Mol. Biol. 2017, 1657, 419–430. [Google Scholar] [CrossRef]
- Wang, J.; Karpus, J.; Zhao, B.S.; Luo, Z.; Chen, P.R.; He, C. A selective fluorescent probe for carbon monoxide imaging in living cells. Angew. Chem. Int. Ed. Engl. 2012, 51, 9652–9656. [Google Scholar] [CrossRef]
- Winter, M.B.; McLaurin, E.J.; Reece, S.Y.; Olea, C., Jr.; Nocera, D.G.; Marletta, M.A. Ru-porphyrin protein scaffolds for sensing O2. J. Am. Chem. Soc. 2010, 132, 5582–5583. [Google Scholar] [CrossRef]
- Nierth, A.; Marletta, M.A. Direct meso-Alkynylation of Metalloporphyrins Through Gold Catalysis for Hemoprotein Engineering. Angew. Chem. Int. Ed. 2014, 53, 2611–2614. [Google Scholar] [CrossRef]
- Lemon, C.M.; Marletta, M.A. Corrole-Substituted Fluorescent Heme Proteins. Inorg. Chem. 2021, 60, 2716–2729. [Google Scholar] [CrossRef] [PubMed]
- Fardeau, M.L.; Salinas, M.B.; L’Haridon, S.; Jeanthon, C.; Verhe, F.; Cayol, J.L.; Patel, B.K.C.; Garcia, J.L.; Ollivier, B. Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: Reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int. J. Syst. Evol. Microbiol. 2004, 54, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Boon, E.M. Engineering of the heme pocket of an H-NOX domain for direct cyanide detection and quantification. J. Am. Chem. Soc. 2010, 132, 11496–11503. [Google Scholar] [CrossRef]
- Winter, M.B.; Klemm, P.J.; Phillips-Piro, C.M.; Raymond, K.N.; Marletta, M.A. Porphyrin-substituted H-NOX proteins as high-relaxivity MRI contrast agents. Inorg. Chem. 2013, 52, 2277–2279. [Google Scholar] [CrossRef]
- Kumita, H.; Yamada, S.; Nakamura, H.; Shiro, Y. Chimeric sensory kinases containing O2 sensor domain of FixL and histidine kinase domain from thermophile. Biochim. Biophys. Acta 2003, 1646, 136–144. [Google Scholar] [CrossRef]
- Moglich, A.; Ayers, R.A.; Moffat, K. Design and signaling mechanism of light-regulated histidine kinases. J. Mol. Biol. 2009, 385, 1433–1444. [Google Scholar] [CrossRef]
- Iniesta, A.A.; Hillson, N.J.; Shapiro, L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. USA 2010, 107, 7012–7017. [Google Scholar] [CrossRef]
- Nomata, J.; Hisabori, T. Development of heme protein based oxygen sensing indicators. Sci. Rep. 2018, 8, 11849. [Google Scholar] [CrossRef]
- Ainala, S.K.; Seol, E.; Sekar, B.S.; Park, S. Improvement of carbon monoxide-dependent hydrogen production activity in Citrobacter amalonaticus Y19 by over-expressing the CO-sensing transcriptional activator, CooA. Int. J. Hydrog. Energ. 2014, 39, 10417–10425. [Google Scholar] [CrossRef]
- Arnold, F.H. Innovation by Evolution: Bringing New Chemistry to Life (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2019, 58, 14420–14426. [Google Scholar] [CrossRef]
- Leveson-Gower, R.B.; Mayer, C.; Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. 2019, 3, 687–705. [Google Scholar] [CrossRef]
- Aggrey-Fynn, J.E.; Surmeli, N.B. A novel thermophilic hemoprotein scaffold for rational design of biocatalysts. J. Biol. Inorg. Chem. 2018, 23, 1295–1307. [Google Scholar] [CrossRef]
- Ortmayer, M.; Lafite, P.; Menon, B.R.; Tralau, T.; Fisher, K.; Denkhaus, L.; Scrutton, N.S.; Rigby, S.E.; Munro, A.W.; Hay, S.; et al. An oxidative N-demethylase reveals PAS transition from ubiquitous sensor to enzyme. Nature 2016, 539, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.J.; Martin, N.I.; Marletta, M.A. An Escherichia coli expression-based method for heme substitution. Nat. Methods 2007, 4, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, N.; Shoji, O.; Watanabe, Y. Single-step reconstitution of apo-hemoproteins at the disruption stage of Escherichia coli cells. Chembiochem 2012, 13, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Lelyveld, V.S.; Brustad, E.; Arnold, F.H.; Jasanoff, A. Metal-substituted protein MRI contrast agents engineered for enhanced relaxivity and ligand sensitivity. J. Am. Chem. Soc. 2011, 133, 649–651. [Google Scholar] [CrossRef]
- Perkins, L.J.; Weaver, B.R.; Buller, A.R.; Burstyn, J.N. De novo biosynthesis of a nonnatural cobalt porphyrin cofactor in E. coli and incorporation into hemoproteins. Proc. Natl. Acad. Sci. USA 2021, 118, e2017625118. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gondim, A.C.S.; Guimarães, W.G.; Sousa, E.H.S. Heme-Based Gas Sensors in Nature and Their Chemical and Biotechnological Applications. BioChem 2022, 2, 43-63. https://doi.org/10.3390/biochem2010004
Gondim ACS, Guimarães WG, Sousa EHS. Heme-Based Gas Sensors in Nature and Their Chemical and Biotechnological Applications. BioChem. 2022; 2(1):43-63. https://doi.org/10.3390/biochem2010004
Chicago/Turabian StyleGondim, Ana Claudia Silva, Wellinson Gadelha Guimarães, and Eduardo Henrique Silva Sousa. 2022. "Heme-Based Gas Sensors in Nature and Their Chemical and Biotechnological Applications" BioChem 2, no. 1: 43-63. https://doi.org/10.3390/biochem2010004
APA StyleGondim, A. C. S., Guimarães, W. G., & Sousa, E. H. S. (2022). Heme-Based Gas Sensors in Nature and Their Chemical and Biotechnological Applications. BioChem, 2(1), 43-63. https://doi.org/10.3390/biochem2010004






