New Bayesian Posterior Approaches for Cytogenetic Partial Body Irradiation Inference
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Comparison of Approaches
3.2. Posterior Density Plots
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| D+R | Dicentric plus ring chromosome |
| FBI | Fraction of the body irradiated |
| HPDI | Highest posterior density interval |
| MLE | Maximum likelihood estimator |
| SD | Standard deviation |
| ZIP | Zero-inflated Poisson |
References
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis, 3rd ed.; CRC Texts in Statistical Science; Chapman & Hall: Boca Raton, FL, USA, 2013. [Google Scholar]
- Ainsbury, E.A.; Vinnikov, V.A.; Puig, P.; Higueras, M.; Maznyk, N.A.; Lloyd, D.C.; Rothkamm, K. Review of Bayesian Statistical Analysis Methods for Cytogenetic Radiation Biodosimetry, with a Practical Example. Radiat. Prot. Dosim. 2014, 162, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Suto, Y. A Bayesian hierarchical method to account for random effects in cytogenetic dosimetry based on calibration curves. Radiat. Environ. Biophys. 2014, 53, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Higueras, M.; Puig, P.; Ainsbury, E.A.; Rothkamm, K. A New Inverse Regression Model Applied to Radiation Biodosimetry. Proc. R. Soc. A 2015, 471, 20140588. [Google Scholar] [CrossRef] [PubMed]
- Higueras, M.; Puig, P.; Ainsbury, E.A.; Vinnikov, V.A.; Rothkamm, K. A new Bayesian Model Applied to Cytogenetic Partial Body Irradiation Estimation. Radiat. Prot. Dosim. 2016, 168, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Powojska, A.; Słonecka, I.; Fornalski, K. The Monte Carlo Method of Mixed Radiation Field Dose Assessment in the Cytogenetic Biodosimetry. Acta Phys. 2018, 134, 583–589. [Google Scholar] [CrossRef]
- Słonecka, I.; Łukasik, K.; Fornalski, K.W. Analytical and quasi-Bayesian methods as development of the iterative approach for mixed radiation biodosimetry. Radiat. Environ. Biophys. 2018, 57, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Słonecka, I.; Łukasik, K.; Fornalski, K.W. Simplified Bayesian method: Application in cytogenetic biological dosimetry of mixed η+γ radiation fields. Radiat. Environ. Biophys 2019, 58, 49–57. [Google Scholar] [CrossRef]
- Młynarczyk, D.; Puig, P.; Armero, C.; Gómez, R.V.; Barquinero, J.F.; Pujol, C.M. Radiation dose estimation with time-since-exposure uncertainty using the-H2AX biomarker. Sci. Rep. 2022, 12, 19877. [Google Scholar] [CrossRef] [PubMed]
- Młynarczyk, D.; Puig, P.; Barquinero, J.F.; Armero, C.; Gómez-Rubio, V. Comparative Analysis of the Yields of Dicentrics and Chromosomal Translocations. Int. J. Radiat. Biol. 2024, 100, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Dolphin, G.W. Biological Dosimetry with Particular Reference to Chromosome Aberration Analysis. A Review of Methods; IAEA: Vienna, Austria, 1969; pp. 215–224. [Google Scholar]
- Sasaki, M.; Miyata, H. Biological dosimetry in atom bomb survivors. Nature 1968, 220, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- International Atomic Energy Agency (IAEA). Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies; IAEA: Vienna, Austria, 2011. [Google Scholar]
- Pujol, M.; Barrios, L.; Puig, P.; Caballín, M.R.; Barquinero, J.F. A New Model for Biological Dose Assessment in Cases of Heterogeneous Exposures to Ionizing Radiation. Radiat. Res. 2016, 185, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.E.; Mendonça, J.C.G.; Barquinero, J.F.; Higueras, M.; Gonzalez, J.E.; Andrade, A.M.G.; Silva, L.M.; Nascimento, A.M.S.; Lima, J.C.F.; Silva, J.C.G.; et al. Comparative Study of Micronucleus Assays and Dicentric Plus Ring Chromosomes for Dose Assessment in Particular Cases of Partial-Body Exposure. Int. J. Radiat. Biol. 2019, 95, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Ainsbury, E.A.; Higueras, M.; Puig, P.; Einbeck, J.; Samaga, D.; Barquinero, J.F.; Barrios, L.; Brzozowska, B.; Fattibene, P.; Gregoire, E.; et al. Uncertainty of fast biological radiation dose assessment for emergency response scenarios. J. Radiat. Biol. 2017, 93, 127–135. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 27 February 2025).
- Kernighan, B.; Ritchie, D.M. The C Programming Language, 2nd ed.; Prentice Hall: Englewood Cliffs, NJ, USA, 1988. [Google Scholar]

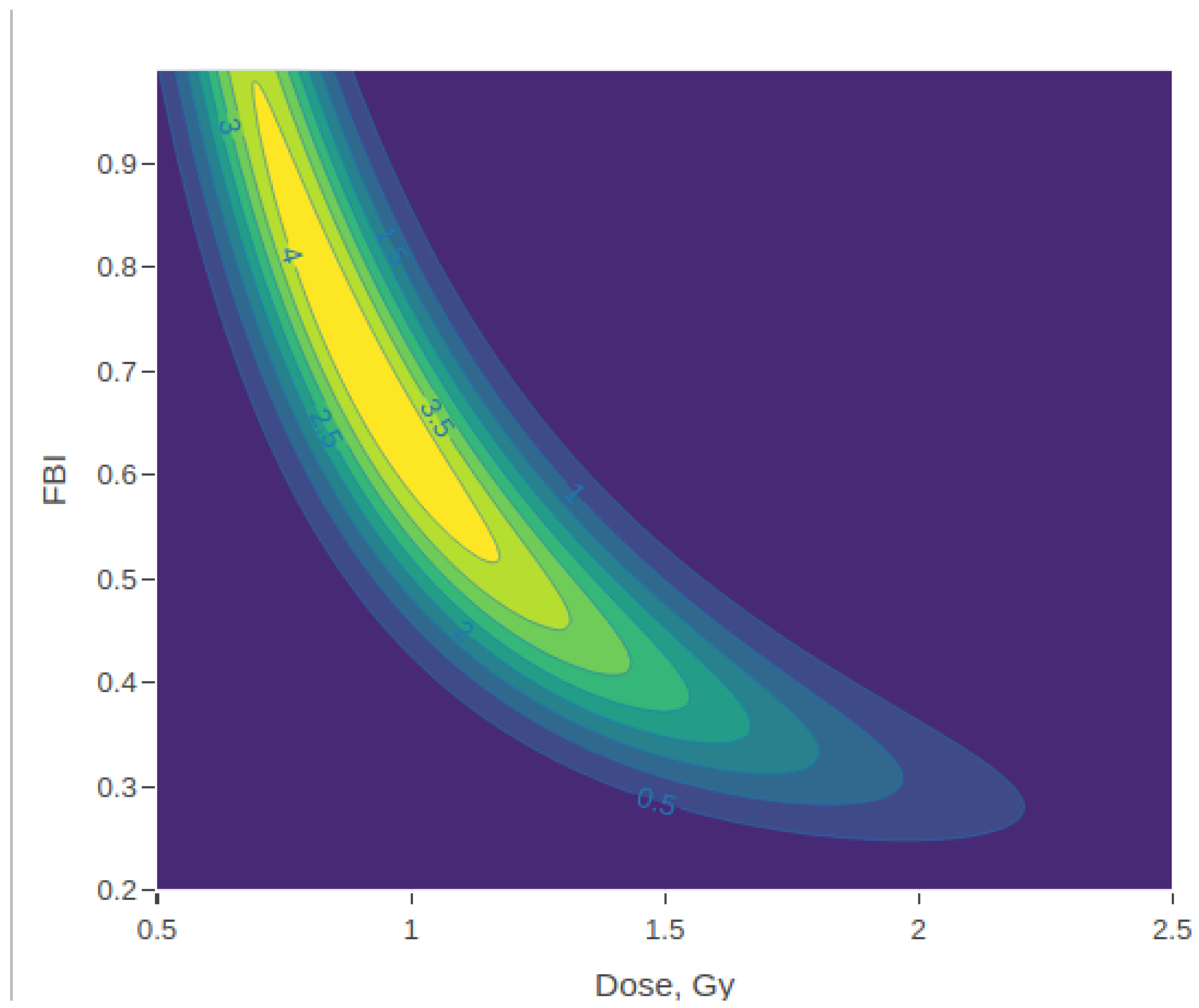
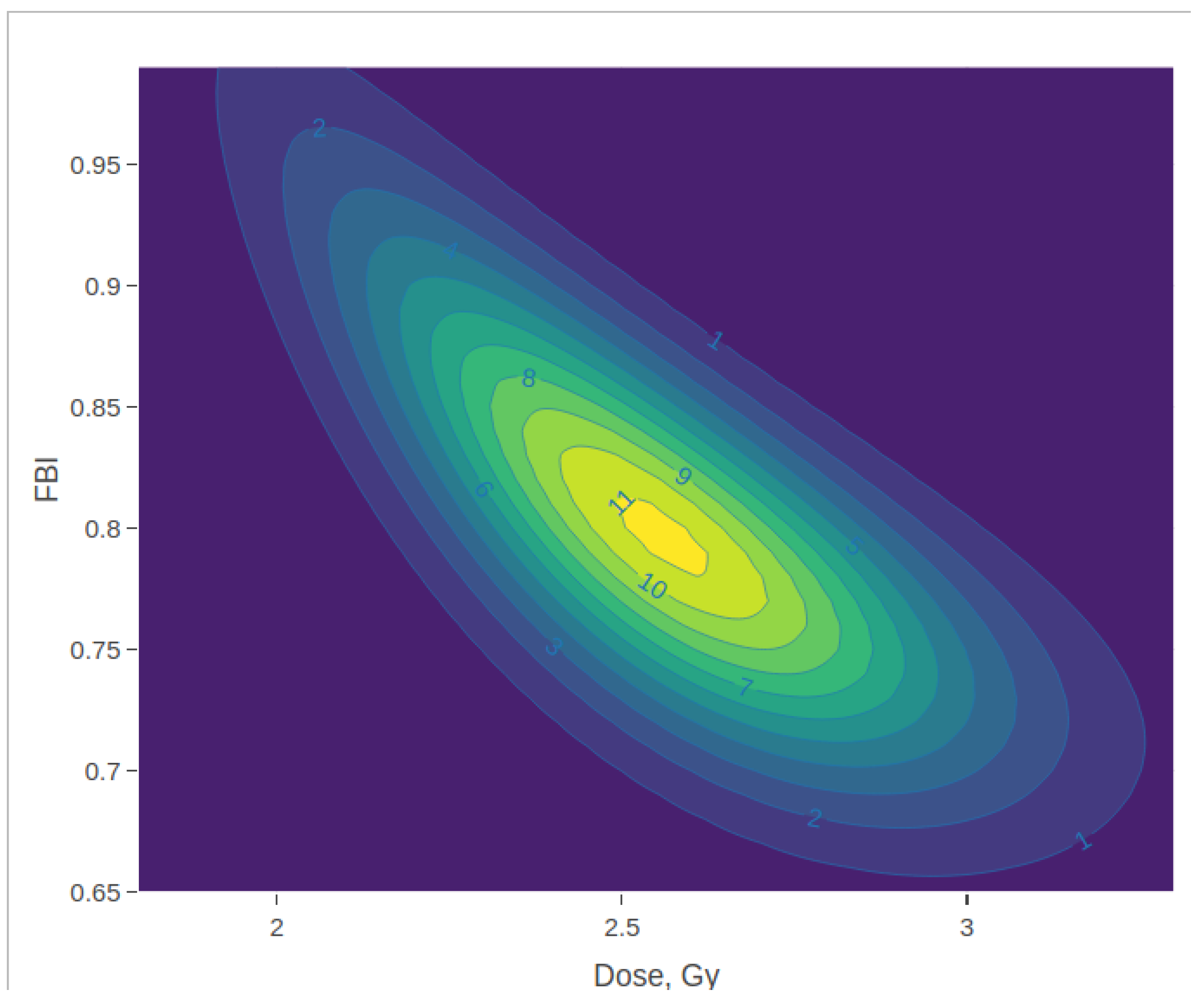
| Dose | FBI | Marginal Dose Posterior: | Marginal FBI Posterior: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mode | Expected | Median | SD | 95% HPDI | Mode | Expected | Median | SD | 95% HPDI | Correlation | |||
| 2.00 | 0.10 | 5.91 | 1.41 | 1.62 | 1.56 | 0.71 | (0.34, 2.99) | 0.11 | 0.14 | 0.12 | 0.07 | (0.05, 0.28) | −0.63 |
| 12.00 | 0.90 | 13.12 | 10.71 | 10.72 | 10.71 | 0.39 | (9.95, 11.49) | 0.91 | 0.91 | 0.91 | 0.03 | (0.86, 0.96) | 0.38 |
| Dose | FBI | Marginal Dose Posterior: | Marginal FBI Posterior: | |||||
|---|---|---|---|---|---|---|---|---|
| Mode | SD | 95% HPDI | Mode | SD | 95% HPDI | Correlation | ||
| 2.00 | 0.10 | 1.63 | 0.71 | (0.24, 3.02) | 0.11 | 0.04 | (0.04, 0.19) | −0.75 |
| 12.00 | 0.90 | 10.73 | 0.40 | (9.96, 11.51) | 0.91 | 0.03 | (0.84, 0.97) | 0.28 |
| Donor | Dose | FBI | Marginal Dose Posterior: | Marginal FBI Posterior: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mode | Expected | Median | SD | 95% HPDI | Mode | Expected | Median | SD | 95% HPDI | Correlation | ||||
| 1 | 0.00 | 1.00 | 10.12 | 0.03 | 0.68 | 0.23 | 1.10 | (0.00, 3.10) | 0.01 | 0.24 | 0.14 | 0.26 | (0.00, 0.81) | −0.47 |
| 1 | 0.50 | 0.25 | 8.18 | 0.20 | 0.85 | 0.48 | 0.96 | (0.00, 2.93) | 0.05 | 0.34 | 0.25 | 0.28 | (0.03, 1.00) | −0.61 |
| 1 | 0.50 | 0.50 | 7.27 | 0.25 | 0.75 | 0.50 | 0.72 | (0.04, 2.30) | 0.07 | 0.38 | 0.31 | 0.08 | (0.05, 1.00) | −0.67 |
| 1 | 0.50 | 1.00 | 3.31 | 0.68 | 0.92 | 0.82 | 0.36 | (0.41, 1.67) | 0.60 | 0.63 | 0.63 | 0.05 | (0.28, 1.00) | −0.83 |
| 1 | 1.00 | 0.25 | 6.17 | 0.63 | 1.29 | 1.16 | 0.68 | (0.30, 2.63) | 0.16 | 0.34 | 0.27 | 0.22 | (0.07, 0.83) | −0.76 |
| 1 | 1.00 | 0.50 | 4.21 | 0.84 | 1.13 | 1.05 | 0.39 | (0.55, 1.93) | 0.48 | 0.61 | 0.59 | 0.20 | (0.30, 1.00) | −0.84 |
| 1 | 1.00 | 1.00 | 2.90 | 1.19 | 1.29 | 1.25 | 0.20 | (0.96, 1.70) | 1.00 | 0.84 | 0.86 | 0.11 | (0.63, 1.00) | −0.82 |
| 1 | 2.00 | 0.25 | 5.11 | 1.20 | 1.46 | 1.39 | 0.54 | (0.57, 2.49) | 0.29 | 0.43 | 0.39 | 0.19 | (0.16, 0.85) | −0.81 |
| 1 | 2.00 | 0.50 | 3.48 | 1.47 | 1.61 | 1.57 | 0.25 | (1.19, 2.11) | 0.83 | 0.81 | 0.82 | 0.11 | (0.62, 1.00) | −0.84 |
| 1 | 2.00 | 1.00 | 3.55 | 2.21 | 2.23 | 2.23 | 0.15 | (1.95, 2.54) | 1.00 | 0.96 | 0.97 | 0.03 | (0.90, 1.00) | −0.49 |
| 1 | 4.00 | 0.25 | 4.27 | 1.82 | 1.88 | 1.86 | 0.38 | (1.15, 2.61) | 0.52 | 0.58 | 0.56 | 0.12 | (0.37, 0.85) | −0.83 |
| 1 | 4.00 | 0.50 | 4.41 | 2.54 | 2.57 | 2.56 | 0.30 | (2.00, 3.15) | 0.79 | 0.80 | 0.80 | 0.07 | (0.67, 0.95) | −0.75 |
| 1 | 4.00 | 1.00 | 5.27 | 3.74 | 3.75 | 3.75 | 0.22 | (3.33, 4.19) | 1.00 | 0.99 | 0.99 | 0.01 | (0.97, 1.00) | −0.11 |
| 2 | 0.00 | 1.00 | 10.12 | 0.03 | 0.68 | 0.23 | 1.10 | (0.00, 3.11) | 0.01 | 0.25 | 0.14 | 0.26 | (0.00, 0.82) | −0.47 |
| 2 | 0.50 | 0.25 | 9.49 | 0.26 | 0.95 | 0.58 | 0.98 | (0.01, 3.09) | 0.06 | 0.35 | 0.27 | 0.28 | (0.01, 0.90) | −0.62 |
| 2 | 0.50 | 0.50 | 7.13 | 0.66 | 1.47 | 1.32 | 0.82 | (0.26, 3.07) | 0.12 | 0.28 | 0.20 | 0.21 | (0.04, 0.76) | −0.72 |
| 2 | 0.50 | 1.00 | 3.07 | 0.73 | 0.93 | 0.85 | 0.31 | (0.48, 1.58) | 0.82 | 0.67 | 0.69 | 0.20 | (0.34, 1.00) | −0.84 |
| 2 | 1.00 | 0.25 | 4.88 | 0.40 | 0.82 | 0.64 | 0.57 | (0.15, 2.04) | 0.15 | 0.45 | 0.41 | 0.26 | (0.07, 0.94) | −0.75 |
| 2 | 1.00 | 0.50 | 3.26 | 0.70 | 0.93 | 0.84 | 0.35 | (0.43, 1.65) | 0.66 | 0.64 | 0.65 | 0.21 | (0.30, 1.00) | −0.83 |
| 2 | 1.00 | 1.00 | 2.81 | 1.02 | 1.12 | 1.08 | 0.21 | (0.77, 1.57) | 1.00 | 0.82 | 0.84 | 0.13 | (0.57, 1.00) | −0.81 |
| 2 | 2.00 | 0.25 | 3.07 | 0.79 | 0.97 | 0.90 | 0.30 | (0.52, 1.60) | 0.94 | 0.70 | 0.71 | 0.19 | (0.37, 1.00) | −0.83 |
| 2 | 2.00 | 0.50 | 3.71 | 1.30 | 1.48 | 1.44 | 0.30 | (0.99, 2.09) | 0.70 | 0.73 | 0.73 | 0.14 | (0.51, 1.00) | −0.85 |
| 2 | 2.00 | 1.00 | 3.81 | 2.21 | 2.28 | 2.26 | 0.23 | (1.87, 2.72) | 0.88 | 0.88 | 0.88 | 0.06 | (0.77, 1.00) | −0.78 |
| 2 | 4.00 | 0.25 | 5.38 | 2.24 | 2.29 | 2.27 | 0.51 | (1.29, 3.30) | 0.31 | 0.35 | 0.33 | 0.09 | (0.20, 0.53) | −0.70 |
| 2 | 4.00 | 0.50 | 4.74 | 2.95 | 2.96 | 2.95 | 0.30 | (2.38, 3.54) | 0.61 | 0.61 | 0.61 | 0.05 | (0.51, 0.71) | −0.52 |
| 2 | 4.00 | 1.00 | 5.17 | 3.68 | 3.70 | 3.69 | 0.21 | (3.28, 4.12) | 1.00 | 0.99 | 0.99 | 0.01 | (0.97, 1.00) | −0.09 |
| Donor | Dose | FBI | Marginal Dose Posterior: | Marginal FBI Posterior: | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mode | SD | 95% HPDI | Mode | SD | 95% HPDI | Correlation | |||
| 1 | 0.00 | 1.00 | 0.13 | 1.67 | (−3.14, 3.40) | 0.21 | 2.48 | (−4.65, 5.07) | −1.00 |
| 1 | 0.50 | 0.25 | 0.13 | 0.95 | (−1.73, 1.99) | 0.95 | 6.44 | (−11.66, 13.57) | −1.00 |
| 1 | 0.50 | 0.50 | 0.17 | 0.07 | (0.02, 0.31) | 1.00 | 0.04 | (0.92, 1.08) | −0.09 |
| 1 | 0.50 | 1.00 | 0.56 | 0.09 | (0.38, 0.74) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 1 | 1.00 | 0.25 | 1.39 | 0.79 | (−0.16, 2.94) | 0.21 | 0.14 | (−0.07, 0.50) | −0.92 |
| 1 | 1.00 | 0.50 | 0.92 | 0.54 | (−0.15, 1.99) | 0.69 | 0.49 | (−0.28, 1.65) | −0.97 |
| 1 | 1.00 | 1.00 | 1.09 | 0.09 | (0.92, 1.27) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 1 | 2.00 | 0.25 | 1.50 | 0.60 | (0.32, 2.67) | 0.35 | 0.16 | (0.04, 0.67) | −0.91 |
| 1 | 2.00 | 0.50 | 1.45 | 0.34 | (0.79, 2.11) | 0.89 | 0.22 | (0.47, 1.31) | −0.94 |
| 1 | 2.00 | 1.00 | 2.15 | 0.12 | (1.91, 2.38) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 1 | 4.00 | 0.25 | 1.89 | 0.39 | (1.12, 2.66) | 0.55 | 0.12 | (0.32, 0.77) | −0.86 |
| 1 | 4.00 | 0.50 | 2.56 | 0.31 | (1.96, 3.16) | 0.80 | 0.07 | (0.65, 0.94) | −0.77 |
| 1 | 4.00 | 1.00 | 3.71 | 0.21 | (3.30, 4.13) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 2 | 0.00 | 1.00 | 0.13 | 1.68 | (−3.16, 3.42) | 0.22 | 2.56 | (−4.80, 5.23) | −1.00 |
| 2 | 0.50 | 0.25 | 0.17 | 0.09 | (−0.02, 0.35) | 1.00 | 0.03 | (0.94, 1.06) | −0.06 |
| 2 | 0.50 | 0.50 | 1.63 | 0.91 | (−0.16, 3.42) | 0.15 | 0.10 | (−0.04, 0.34) | −0.88 |
| 2 | 0.50 | 1.00 | 0.62 | 0.09 | (0.44, 0.80) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 2 | 1.00 | 0.25 | 0.30 | 0.09 | (0.13, 0.47) | 1.00 | 0.00 | (0.99, 1.01) | −0.01 |
| 2 | 1.00 | 0.50 | 0.58 | 0.09 | (0.40, 0.76) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 2 | 1.00 | 1.00 | 0.92 | 0.09 | (0.74, 1.11) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 2 | 2.00 | 0.25 | 0.67 | 0.09 | (0.49, 0.86) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
| 2 | 2.00 | 0.50 | 1.36 | 0.39 | (0.60, 2.13) | 0.78 | 0.24 | (0.30, 1.25) | −0.95 |
| 2 | 2.00 | 1.00 | 2.22 | 0.26 | (1.70, 2.73) | 0.89 | 0.09 | (0.72, 1.06) | −0.87 |
| 2 | 4.00 | 0.25 | 2.32 | 0.51 | (1.32, 3.31) | 0.32 | 0.07 | (0.18, 0.46) | −0.70 |
| 2 | 4.00 | 0.50 | 2.95 | 0.30 | (2.37, 3.53) | 0.61 | 0.05 | (0.51, 0.71) | −0.51 |
| 2 | 4.00 | 1.00 | 3.66 | 0.21 | (3.25, 4.07) | 1.00 | 0.00 | (1.00, 1.00) | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Higueras, M.; Carrillo, H. New Bayesian Posterior Approaches for Cytogenetic Partial Body Irradiation Inference. Radiation 2025, 5, 16. https://doi.org/10.3390/radiation5020016
Higueras M, Carrillo H. New Bayesian Posterior Approaches for Cytogenetic Partial Body Irradiation Inference. Radiation. 2025; 5(2):16. https://doi.org/10.3390/radiation5020016
Chicago/Turabian StyleHigueras, Manuel, and Hans Carrillo. 2025. "New Bayesian Posterior Approaches for Cytogenetic Partial Body Irradiation Inference" Radiation 5, no. 2: 16. https://doi.org/10.3390/radiation5020016
APA StyleHigueras, M., & Carrillo, H. (2025). New Bayesian Posterior Approaches for Cytogenetic Partial Body Irradiation Inference. Radiation, 5(2), 16. https://doi.org/10.3390/radiation5020016






