Using Drones to Monitor Broad-Leaved Orchids (Dactylorhiza majalis) in High-Nature-Value Grassland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
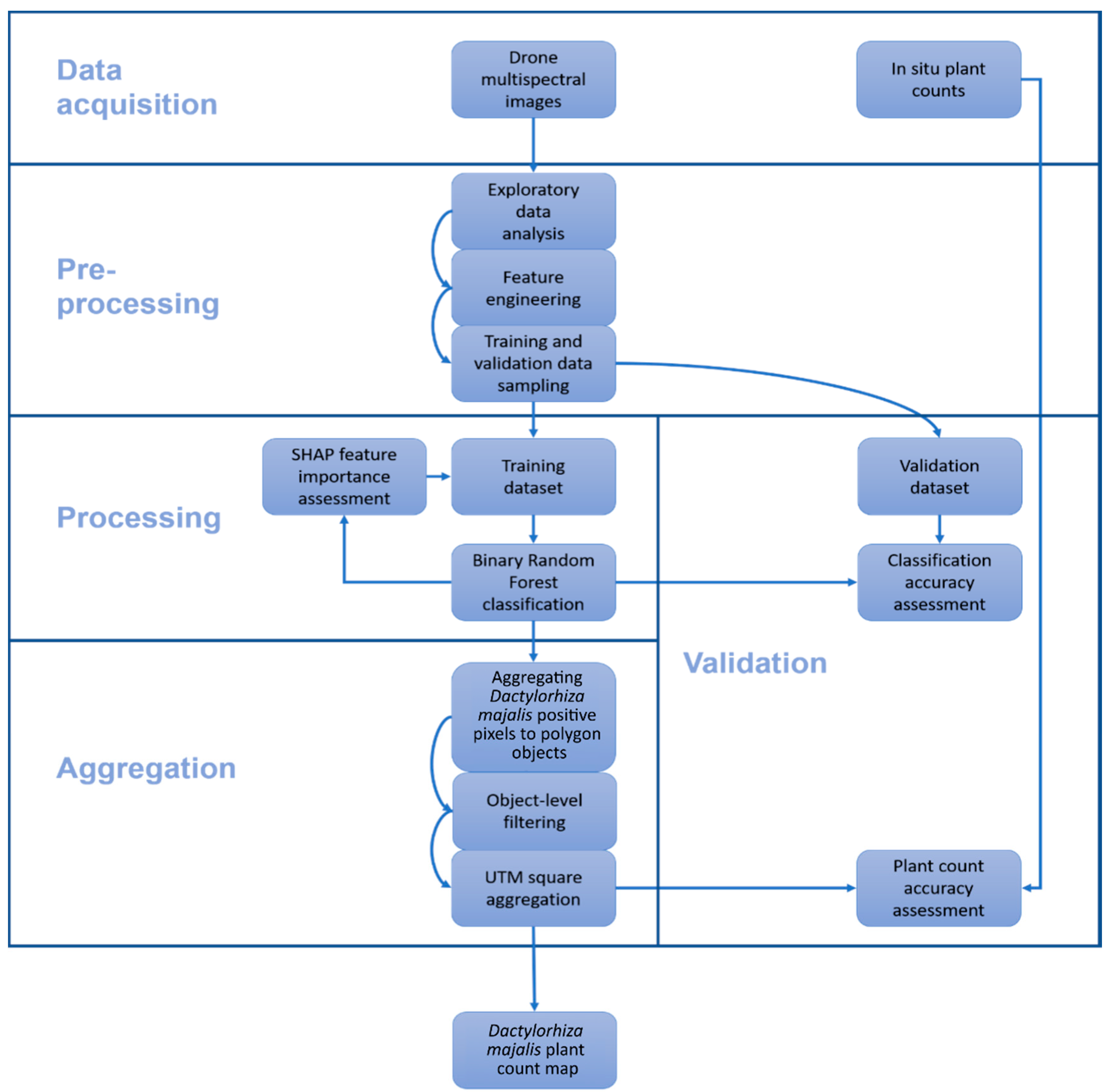
2.2. Data (Pre-)Processing and Analysis
2.3. Drone Data
2.4. In Situ Data
2.5. Labeling a Reference Dataset for Model Training and Validation
2.6. Magenta Vegetation Index—Main Ideas and Practical Implementation
2.7. Random Forest Classification
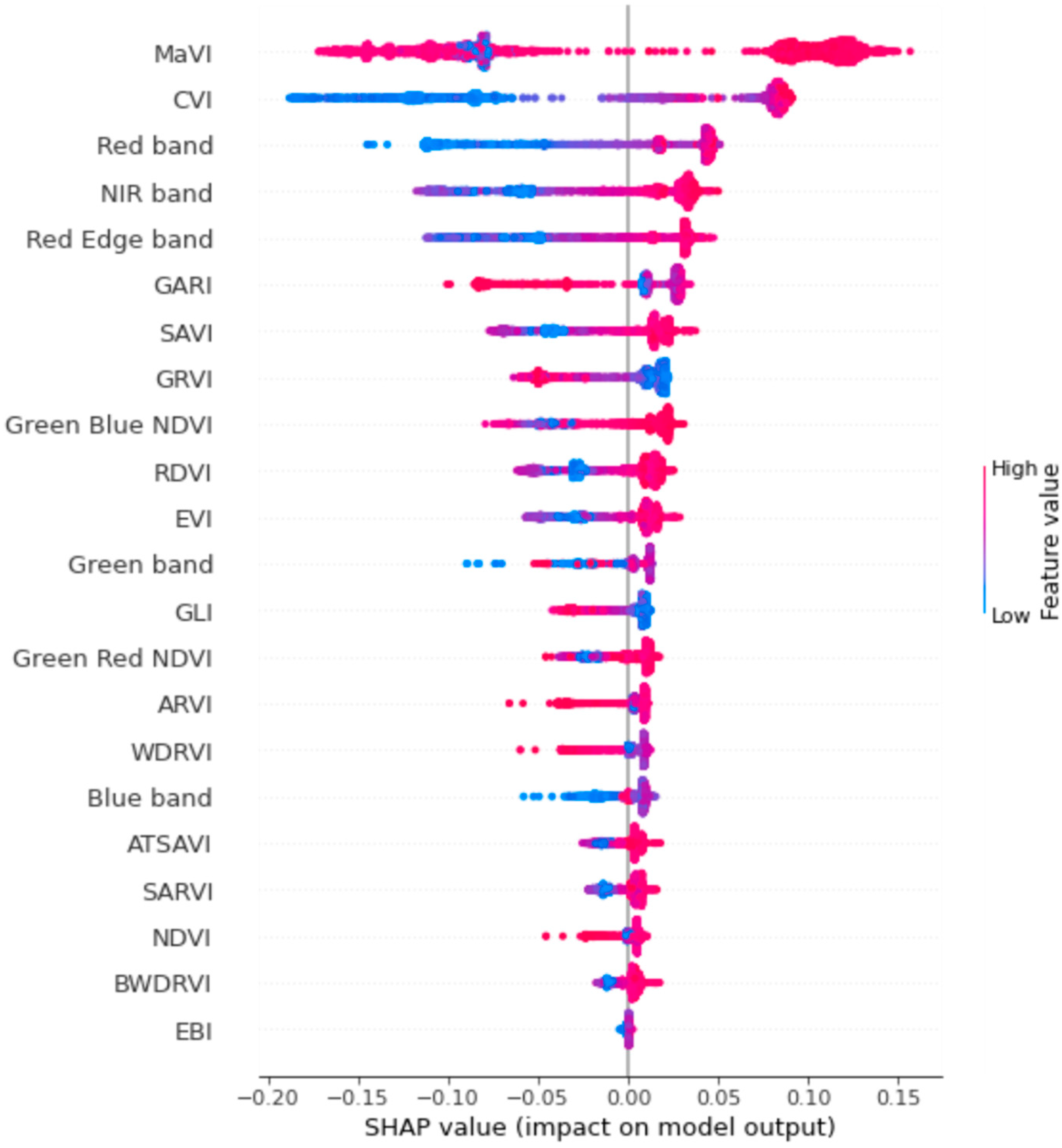
2.8. Feature Selection and Model Interpretation
2.9. Remote Sensing Plant Count Methodology
- Polygonize DM positive pixel clusters (i.e., neighboring pixels) to vector image objects;
- Calculate a filter threshold for each image object on the basis of the most descriptive feature of the image classification;
- Remove all pixels below the threshold from the remote sensing plant count.
3. Results
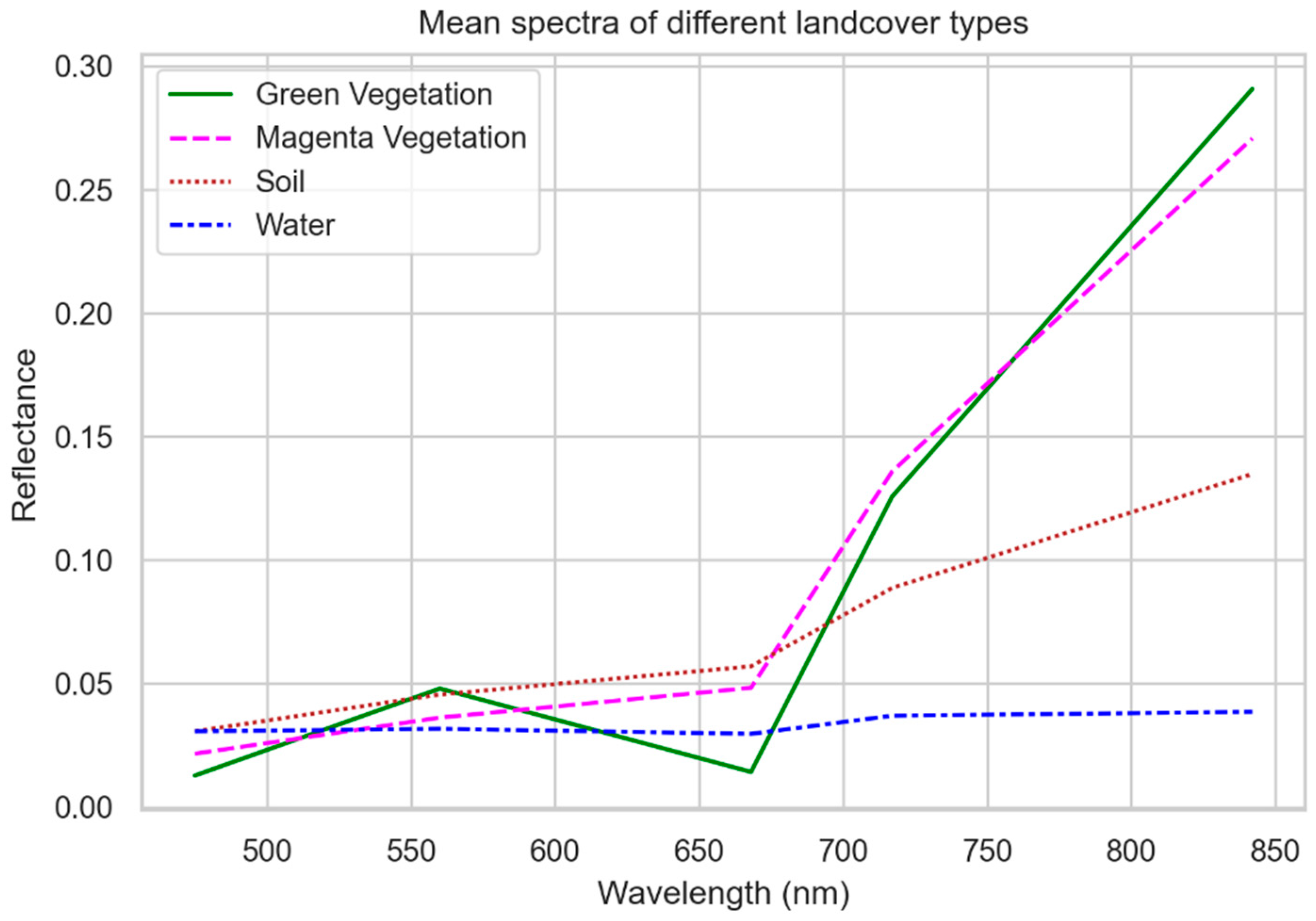
3.1. Ambiguity in the Drone Dataset
- Mixed pixel phenomena, due to (1) a DM individual located at the common boundary of multiple pixels, (2) multiple DM individuals in direct proximity and partly occupy multiple neighboring pixels, or (3) DM individuals which did not grow perfectly straight and, therefore, appeared in neighboring pixels;
- Adjacency effects, i.e., the magenta flowers spectrally superimpose the neighboring pixels;
- Motion blur caused by camera movement during exposure;
- Keystone effect of the camera, which may cause a slight cross-track displacement.
3.2. Classification Results before Feature Selection
3.3. Feature Selection and Predictive Performance of the MaVI
3.4. Classification Result after Feature Selection
3.5. Remote Sensing Plant Count Accuracy Assessment
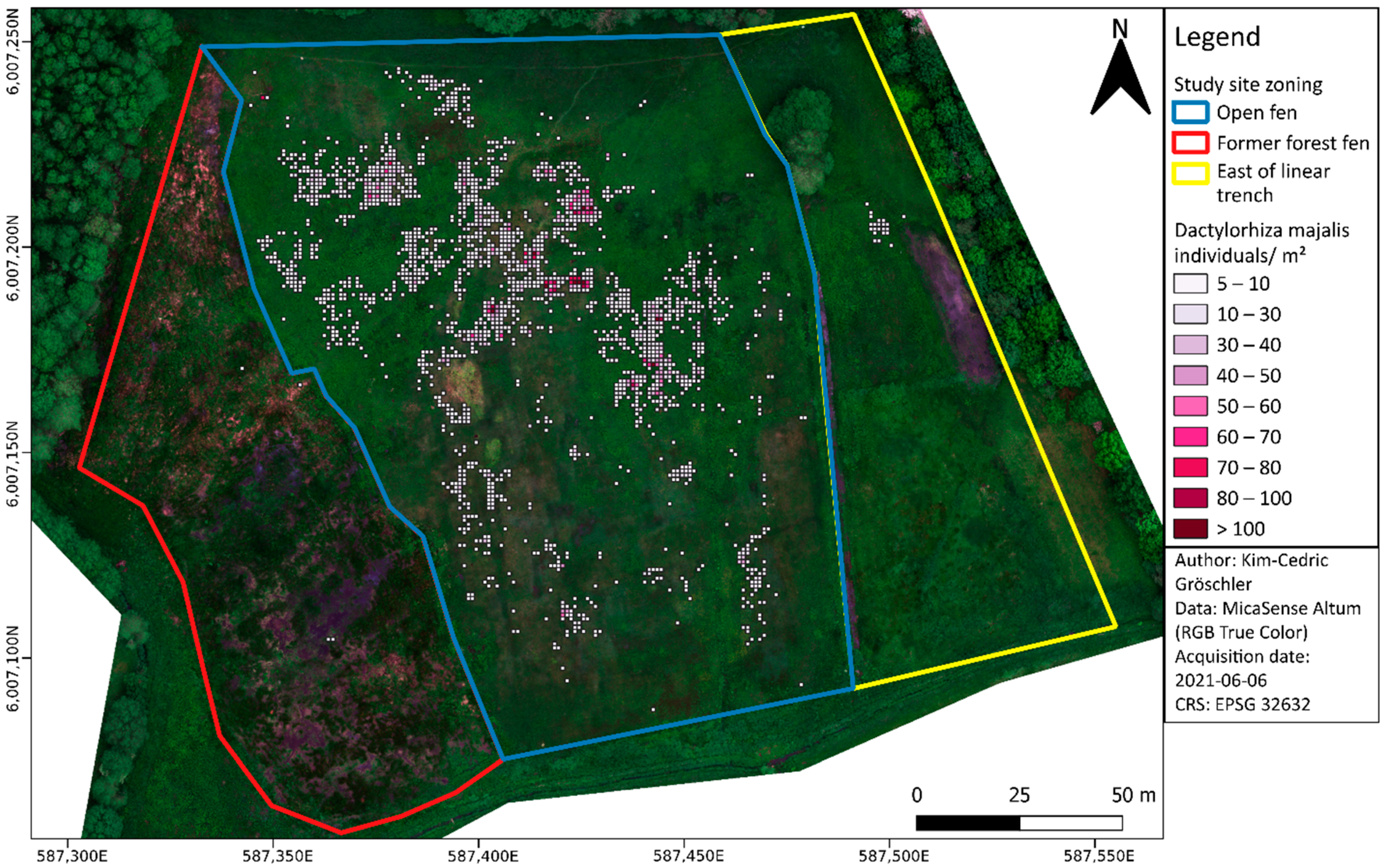
3.6. Assessing the Spatial Distribution and Abundance of Dactylorhiza majalis
3.7. Relevance to Nature Conservation and Management
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dullau, S.; Richter, F.; Adert, N.; Meyer, M.H.; Hensen, H.; Tischew, S. Handlungsempfehlung zur Populationsstärkung und Wiederansiedlung von Dactylorhiza majalis am Beispiel des Biosphärenreservat Karstlandschaft Südharz; Hochschule Anhalt: Bernburg, Germany, 2019. [Google Scholar] [CrossRef]
- Lohr, M.; Margenburg, B. Das Breitblättrige Knabenkraut Dactylorhiza majalis–Orchidee des Jahres 2020. J. Eur. Orchid. 2020, 52, 287–323. [Google Scholar]
- Gregor, T.; Saurwein, H.-P. Wer erhält das Großblättrige Knabenkraut (Dactylorhiza majalis). Beitr. Naturkunde Osthess. 2010, 47, 3–6. [Google Scholar]
- Messlinger, U.; Pape, T.; Wolf, S. Erhaltungsstrategien für das Breitblättrige Knabenkraut (Dactylorhiza majalis) in Stadt und Landkreis Ansbach. Regnitz Flora 2018, 9, 82–106. [Google Scholar]
- Wotavová, K.; Balounová, Z.; Kindlmann, P. Factors Affecting Persistence of Terrestrial Orchids in Wet Meadows and Implications for Their Conservation in a Changing Agricultural Landscape. Biol. Conserv. 2004, 118, 271–279. [Google Scholar] [CrossRef]
- Reinhard, H.R.; Gölz, P.; Peter, R.; Wildermuth, H. Die Orchideen der Schweiz und Angrenzender Gebiete; Fotorotar AG: Egg, Switzerland, 1991. [Google Scholar] [CrossRef]
- Pettorelli, N.; Safi, K.; Turner, W. Satellite Remote Sensing, Biodiversity Research and Conservation of the Future. Philos. Trans. R. Soc. B 2014, 369, 20130190. [Google Scholar] [CrossRef]
- Horton, R.; Cano, E.; Bulanon, D.; Fallahi, E. Peach Flower Monitoring Using Aerial Multispectral Imaging. J. Imaging 2017, 3, 2. [Google Scholar] [CrossRef]
- Fang, S.; Tang, W.; Peng, Y.; Gong, Y.; Dai, C.; Chai, R.; Liu, K. Remote Estimation of Vegetation Fraction and Flower Fraction in Oilseed Rape with Unmanned Aerial Vehicle Data. Remote Sens. 2016, 8, 416. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Rahman, E.; Makori, D.; Landmann, T.; Piiroinen, R.; Gasim, S.; Pellikka, P.; Raina, S. The Utility of AISA Eagle Hyperspectral Data and Random Forest Classifier for Flower Mapping. Remote Sens. 2015, 7, 13298–13318. [Google Scholar] [CrossRef] [Green Version]
- Hassan, N.; Numata, S.; Hosaka, T.; Hashim, M. Remote Detection of Flowering Somei Yoshino (Prunus × yedoensis ) in an Urban Park Using IKONOS Imagery: Comparison of Hard and Soft Classifiers. J. Appl. Remote Sens. 2015, 9, 096046. [Google Scholar] [CrossRef]
- Sulik, J.J.; Long, D.S. Spectral Indices for Yellow Canola Flowers. Int. J. Remote Sens. 2015, 36, 2751–2765. [Google Scholar] [CrossRef]
- Landmann, T.; Piiroinen, R.; Makori, D.M.; Abdel-Rahman, E.M.; Makau, S.; Pellikka, P.; Raina, S.K. Application of Hyperspectral Remote Sensing for Flower Mapping in African Savannas. Remote Sens. Environ. 2015, 166, 50–60. [Google Scholar] [CrossRef]
- Carl, C.; Landgraf, D.; van der Maaten-Theunissen, M.; Biber, P.; Pretzsch, H. Robinia Pseudoacacia L. Flower Analyzed by Using An Unmanned Aerial Vehicle (UAV). Remote Sens. 2017, 9, 1091. [Google Scholar] [CrossRef] [Green Version]
- Roosjen, P.; Suomalainen, J.; Bartholomeus, H.; Clevers, J. Hyperspectral Reflectance Anisotropy Measurements Using a Pushbroom Spectrometer on an Unmanned Aerial Vehicle—Results for Barley, Winter Wheat, and Potato. Remote Sens. 2016, 8, 909. [Google Scholar] [CrossRef] [Green Version]
- Severtson, D.; Callow, N.; Flower, K.; Neuhaus, A.; Olejnik, M.; Nansen, C. Unmanned Aerial Vehicle Canopy Reflectance Data Detects Potassium Deficiency and Green Peach Aphid Susceptibility in Canola. Precis. Agric. 2016, 17, 659–677. [Google Scholar] [CrossRef] [Green Version]
- Valente, J.; Sari, B.; Kooistra, L.; Kramer, H.; Mücher, S. Automated Crop Plant Counting from Very High-Resolution Aerial Imagery. Precis. Agric. 2020, 21, 1366–1384. [Google Scholar] [CrossRef]
- Shen, M.; Chen, J.; Zhu, X.; Tang, Y. Yellow Flowers Can Decrease NDVI and EVI Values: Evidence from a Field Experiment in an Alpine Meadow. Can. J. Remote Sens. 2009, 35, 8. [Google Scholar] [CrossRef]
- Shen, M.; Chen, J.; Zhu, X.; Tang, Y.; Chen, X. Do Flowers Affect Biomass Estimate Accuracy from NDVI and EVI? Int. J. Remote Sens. 2010, 31, 2139–2149. [Google Scholar] [CrossRef]
- Verma, K.S.; Saxena, R.K.; Hajare, T.N.; Kharche, V.K.; Kumari, P.A. Spectral Response of Gram Varieties under Variable Soil Conditions. Int. J. Remote Sens. 2002, 23, 313–324. [Google Scholar] [CrossRef]
- Chen, B.; Jin, Y.; Brown, P. An Enhanced Bloom Index for Quantifying Floral Phenology Using Multi-Scale Remote Sensing Observations. ISPRS J. Photogramm. Remote Sens. 2019, 156, 108–120. [Google Scholar] [CrossRef]
- Seer, F.K.; Schrautzer, J. Status, Future Prospects, and Management Recommendations for Alkaline Fens in an Agricultural Landscape: A Comprehensive Survey. J. Nat. Conserv. 2014, 22, 358–368. [Google Scholar] [CrossRef]
- Schrautzer, J.; Trepel, M. Niedermoore im Östlichen Hügelland-Lehmkuhlener Stauung. Tuexenia Mitt. Florist. Soziol. Arb. 2014, 7, 47–49. [Google Scholar]
- MELUND. Erhaltungsziele für das Gesetzlich Geschützte Gebiet von Gemeinschaftlicher Bedeutung DE-1728-303 “Lehmkuhlener Stauung”; Amtsblatt für Schleswig Holstein; Ministerium für Energiewende, Landwirtschaft, Umwelt, Natur und Digitalisierung (MELUND): Kiel, Germany, 2016; p. 1033. [Google Scholar]
- Wingtra AG. Wingtra One—Technical Specifications; Wingtra AG: Zurich, Switzerland, 2021; Available online: https://wingtra.com/wp-content/uploads/Wingtra-Technical-Specifications.pdf (accessed on 12 July 2022).
- MicaSense, Inc. MicaSense Altum—Specifications; MicaSense, Inc.: Seattle, WA, USA, 2020; Available online: https://1w2yci3p7wwa1k9jjd1jygxd-wpengine.netdna-ssl.com/wp-content/uploads/2022/03/Altum-PT-Specification-Table-Download.pdf (accessed on 12 July 2022).
- Tremp, H. Aufnahme und Analyse Vegetationsökologischer Daten-Kapitel 3: Datenaufnahme, 1st ed.; utb GmbH: Stuttgart, Germany, 2005; ISBN 978-3-8385-8299-3. [Google Scholar]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of Machine-Learning Classification in Remote Sensing: An Applied Review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, Y.J.; Tanre, D. Atmospherically Resistant Vegetation Index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Frederic, B.; Guyot, G. Potential and Limitations of Vegetation Indices for LAI and APAR Assessment. Remote Sens. Environ. 1991, 104, 88–95. [Google Scholar]
- Hancock, D.W.; Dougherty, C.T. Relationships between Blue- and Red-based Vegetation Indices and Leaf Area and Yield of Alfalfa. Crop Sci. 2007, 47, 2547–2556. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A Broad-Band Leaf Chlorophyll Vegetation Index at the Canopy Scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Huete, A.; Justice, C.; van Leeuwen, W. MODIS VEGETATION INDEX (MOD 13) ALGORITHM THEORETICAL BASIS DOCUMENT, VERSION 3. 1999. Available online: https://modis.gsfc.nasa.gov/data/atbd/atbd_mod13.pdf (accessed on 12 July 2022).
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Gobron, N.; Pinty, B.; Verstraete, M.M.; Widlowski, J.-L. Advanced Vegetation Indices Optimized for Up-Coming Sensors: Design, Performance, and Applications. IEEE Trans. Geosci. Remote Sens. 2000, 38, 2489–2505. [Google Scholar] [CrossRef]
- Wang, B.; Huang, J.; Tang, Y.; Wang, X. New Vegetation Index and Its Application in Estimating Leaf Area Index of Rice. Rice Sci. 2007, 14, 195–203. [Google Scholar] [CrossRef]
- Motohka, T.; Nasahara, K.; Hiroyuki, O.; Satoshi, T. Applicability of Green-Red Vegetation Index for Remote Sensing of Vegetation Phenology. Remote Sens. 2010, 2, 2369. [Google Scholar] [CrossRef] [Green Version]
- Rouse, J.W.; Haas, R.H.; Deering, D.W.; Schell, J.A.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation. [Great Plains Corridor]. 1973. Available online: https://ntrs.nasa.gov/citations/19730017588 (accessed on 12 July 2022).
- Roujean, J.-L.; Breon, F.-M. Estimating PAR Absorbed by Vegetation from Bidirectional Reflectance Measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide Dynamic Range Vegetation Index for Remote Quantification of Biophysical Characteristics of Vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Belgiu, M.; Drăguţ, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Shapley, L.S. 17. A Value for n-Person Games. In Contributions to the Theory of Games (AM-28); Kuhn, H.W., Tucker, A.W., Eds.; Princeton University Press: Princeton, NJ, USA, 1953; Volume II, pp. 307–318. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Guo, Q.; Li, W.; Liu, D.; Chen, J. A Framework for Supervised Image Classification with Incomplete Training Samples. Photogramm. Eng. Remote Sens. 2012, 78, 595–604. [Google Scholar] [CrossRef]
- Stehman, S.V. Sampling Designs for Accuracy Assessment of Land Cover. Int. J. Remote Sens. 2009, 30, 5243–5272. [Google Scholar] [CrossRef]
- Waldner, F. The T Index: Measuring the Reliability of Accuracy Estimates Obtained from Non-Probability Samples. Remote Sens. 2020, 12, 2483. [Google Scholar] [CrossRef]



| Vegetation Index | Formula | Reference |
|---|---|---|
| Atmospherically resistant vegetation index (ARVI) | y = 1 | [29] |
| Adjusted transformed soil-adjusted vegetation index (ATSAVI) | a = 1.22, X = 0.08, b = 0.03 | [30] |
| Blue-wide dynamic range vegetation index (BWDRVI) | [31] | |
| Chlorophyll vegetation index (CVI) | [32] | |
| Enhanced bloom index (EBI) | ε = 1 | [22] |
| Enhanced vegetation index (EVI) | [33] | |
| Green atmospherically resistant vegetation index (GARI) | [34] | |
| Green leaf index (GLI) | [35] | |
| Green–blue normalized difference vegetation index (GBNDVI) | [36] | |
| Green–red normalized difference vegetation index (GRNDVI) | [36] | |
| Green–red vegetation index (GRVI) | [37] | |
| Normalized difference vegetation index (NDVI) | [38] | |
| Renormalized difference vegetation index (RDVI) | [39] | |
| Soil and atmospherically resistant vegetation index (SARVI) | L = 0.5 y = 1 | [30] |
| Adjusted transformed soil-adjusted vegetation index (SAVI) | L = 0.5 | [30] |
| Transformed soil-adjusted vegetation index (TSAVI) | s = 0.33 a = 0.5 X = 1.5 | [30] |
| Wide dynamic range vegetation index (WDRVI) | [40] |
| In Situ | Without Filter | ≥10% Percentile | ≥20% Percentile | ≥30% Percentile | ≥40% Percentile | ≥50% Percentile | ≥60% Percentile | ≥70% Percentile | ≥80% Percentile | ≥90% Percentile | Mean Filter |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 68 | 104 | 91 | 82 | 73 | 61 | 53 | 44 | 31 | 24 | 15 | 48 |
| 35 | 63 | 50 | 44 | 39 | 34 | 31 | 23 | 17 | 13 | 11 | 29 |
| 54 | 88 | 72 | 66 | 59 | 54 | 47 | 37 | 31 | 24 | 17 | 49 |
| 57 | 66 | 56 | 50 | 46 | 40 | 38 | 29 | 23 | 19 | 13 | 32 |
| 34 | 74 | 62 | 57 | 51 | 44 | 39 | 32 | 25 | 21 | 14 | 34 |
| 22 | 42 | 36 | 34 | 30 | 26 | 22 | 20 | 14 | 12 | 8 | 22 |
| 13 | 11 | 9 | 8 | 7 | 6 | 6 | 5 | 4 | 3 | 2 | 6 |
| 62 | 87 | 71 | 66 | 59 | 51 | 45 | 38 | 28 | 24 | 17 | 47 |
| 66 | 156 | 141 | 129 | 115 | 98 | 85 | 69 | 55 | 36 | 22 | 86 |
| 69 | 126 | 114 | 106 | 96 | 85 | 77 | 65 | 55 | 40 | 24 | 76 |
| Count Setting | RMSE |
|---|---|
| Without filter | 42 |
| ≥10% percentile | 31 |
| ≥20% percentile | 26 |
| ≥30% percentile | 19 |
| ≥40% percentile | 14 |
| ≥50% percentile | 12 |
| ≥60% percentile | 16 |
| ≥70% percentile | 23 |
| ≥80% percentile | 29 |
| ≥90% percentile | 37 |
| Mean filter | 13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gröschler, K.-C.; Oppelt, N. Using Drones to Monitor Broad-Leaved Orchids (Dactylorhiza majalis) in High-Nature-Value Grassland. Drones 2022, 6, 174. https://doi.org/10.3390/drones6070174
Gröschler K-C, Oppelt N. Using Drones to Monitor Broad-Leaved Orchids (Dactylorhiza majalis) in High-Nature-Value Grassland. Drones. 2022; 6(7):174. https://doi.org/10.3390/drones6070174
Chicago/Turabian StyleGröschler, Kim-Cedric, and Natascha Oppelt. 2022. "Using Drones to Monitor Broad-Leaved Orchids (Dactylorhiza majalis) in High-Nature-Value Grassland" Drones 6, no. 7: 174. https://doi.org/10.3390/drones6070174
APA StyleGröschler, K.-C., & Oppelt, N. (2022). Using Drones to Monitor Broad-Leaved Orchids (Dactylorhiza majalis) in High-Nature-Value Grassland. Drones, 6(7), 174. https://doi.org/10.3390/drones6070174






