Abstract
The rice–fish co-culture is an important model of carp farming in China; however, research on the dynamics and assembly of water bacterial communities in this system remains limited. Therefore, this study aimed to explore the dynamics and assembly of bacterial communities, as well as their correlation with environmental factors in paddy water. For these purposes, this study was divided into two groups: a rice–carp co-culture (WRC) group and a rice monoculture (WRM) group, with 20 rice fields in each group. After 60 days of farming, the concentrations of NH4+-N, NO2−-N, TN, and PO43− were significantly higher in the WRC group than those in the WRM group. Alpha diversity analysis showed that the Shannon index in the WRC group was significantly decreased compared with the WRM group. At the phylum level, the relative abundance of Actinobacteria significantly increased, while the relative abundance of Proteobacteria and Bacteroidetes significantly decreased in the paddy water of the WRC group. The neutral community model (NCM) indicated that a random process played a dominant role in the construction of bacterial communities in the two groups, and common carp cultivation increased migration rates, thereby affecting community assembly. The co-occurrence network displayed that common carp cultivation led to looser interactions between bacterial communities. In addition, the contents of nutrients significantly affected the abundance of bacteria in paddy water. In summary, carp cultivation decreased the diversity of bacteria and changed the relative abundance of dominant bacteria, thereby affecting the stability and assembly of bacterial communities.
Key Contribution:
This study focused on the dynamics and assembly of bacteria in paddy water. Rice–carp co-culture decreased the diversity of bacteria and accelerated their assembly towards stochastic evolution. Our study provides valuable insights into the dynamic alterations in the composition and assembly of bacterial communities in paddy water.
1. Introduction
Water quality is well understood to be critical for the well-being of aquatic animals [1]. Deteriorating water quality can lead to disease outbreaks in aquatic populations, drastically impacting their survival and productivity [2]. Aquaculture water pollution is primarily attributed to feed residues as well as the nitrogen and phosphorus waste excreted by the animals [3]. The persistent presence of these pollutants can lead to several environmental issues, including the eutrophication of water bodies [4,5]. The intensification of aquaculture has heightened the need for effective effluent treatment, which now represents a significant bottleneck in the industry’s expansion. Therefore, sustainable aquaculture practices are of the utmost importance for its development. Integrated rice–fish farming is considered a representative model of sustainable agriculture because it efficiently utilizes resources and reduces negative impacts on the environment [6]. It has become the second-largest freshwater aquaculture model in China, accounting for 11.77% of the total freshwater production in 2022 [7]. This mode works by using rice to absorb the nitrogen and phosphorus discharged by aquatic animals into the water, thereby preventing water quality deterioration. Additionally, rice can serve as a hidden shelter for aquatic animals, protecting them from natural predators [8]. Given these benefits, the Chinese government is promoting integrated rice–fish farming across the country [4]. In China, a diverse range of aquatic species has been reported to be farmed in rice paddies, including the red swamp crayfish (Procambarus clarkii), common carp (Cyprinus carpio), Chinese mitten crab (Eriocheir sinensis), bellamya, and bullfrogs [7]. Notably, the common carp is one of the most significant and widely cultured species in this system. The tradition of carp cultivation in paddy fields has a venerable history that stretches back about 2000 years in China [9].
The physicochemical indices of water, such as nutrient salt concentrations, dissolved oxygen (DO), and pH, are commonly employed to assess water quality in aquaculture [1]. However, the bacterial composition of water is frequently neglected. Aquaculture water harbors a significant bacterial population that plays crucial roles across various domains. For example, ammonia-oxidizing archaea and bacteria are involved in the nitrogen removal process in water bodies [10,11]. The genus Bacillus is capable of metabolizing organic carbon into CO2 in aquatic environments [12]. Moreover, certain bacterial strains, such as Bacteroides, Streptococcus, and Lactobacillus, function as probiotics in aquaculture systems [12,13]. Probiotics could enhance the growth performance, survival rate and health states of aquatic organisms, improve water quality, and inhibit the growth and reproduction of pathogenic microorganisms [14,15]. Additionally, the presence of pathogenic bacteria, including Vibrio, Aeromonas, Streptococcus, and Enterococcus, can pose risks to aquatic organisms [16]. In recent years, studies have reported on the bacterial diversity and community composition in water from integrated rice–crayfish systems as compared to rice monoculture systems [17]. Additionally, evidence has emerged highlighting disparities in soil bacterial diversity and community composition between these two cultivation approaches [18]. Notwithstanding these findings, research focusing on the water microorganisms within integrated rice–carp systems is notably scarce. Thus, in this study, the common carp, as a representative aquaculture species, was selected to investigate the differences in aquatic bacterial diversity and community composition between the integrated rice–fish system and the rice monoculture system.
2. Materials and Methods
2.1. Experiment Design and Sample Collection
Common carp (average weight of 105.33 ± 2.52 g) were randomly selected for the experiment from the Freshwater Fisheries Research Center farm (Jingjiang, China). This study was divided into two treatment groups: a rice–carp co-culture (WRC) group and a rice monoculture (WRM) group, with 20 rice fields in each group. Each rice field covered an area of 20 m2 (5 × 4 m). Rice seedlings of the “Nangeng 5055” variety were transplanted into all plots on June 20, and the rice was harvested in early November. The water level in paddy fields was maintained at depths ranging from 0.1 to 0.2 m. The carp were stocked in the rice fields at a density of 1.50 individuals/m2 (approximately 158.00 g/m2). The rearing period commenced on September 1 and concluded with the harvest on October 30. During the 60 days of farming, a commercial diet (Cargill Group Co., Ltd., Changzhou, China) was feed once daily at approximately 1% of fish body weight. The diet’s main nutritional components included 31.0% crude protein, 4.0% crude fat, and 18.0% crude ash. During the experimental period, dissolved oxygen levels ranged from 3.2 to 5.8 mg/L, pH levels were maintained between 6.80 and 7.60, and water temperatures varied from 18.5 to 30.5 °C.
Water samples were collected from 20 paddy fields of each group on October 30th utilizing the five-point sampling method [19]. For nutrient analysis, each sample was secured in a 500 mL polyethylene bottle and immediately stored in a portable icebox. For bacterial composition analysis, a 1000 mL water sample was filtrated using a 0.22 μm filter membrane. These collected microorganisms were then preserved at −80 °C for subsequent examination. A total of 20 samples were gathered for each group.
2.2. Detection of Nutrients in Water
Phosphate (PO43−) content was determined by the continuous flow analysis (CFA) and ammonium molybdate spectrophotometry [20]. Total nitrogen (TN) was detected by alkaline potassium persulfate digestion–UV spectrophotometry [21]. Ammonia (NH4+-N) was tested according to the Nessler’s reagent colorimetric method [22]. The spectrophotometric method was used to measure nitrite (NO2−-N) [23]. Nitrate (NO3−-N) was detected according to the spectrophotometric method with phenol disulfonic acid [24].
2.3. 16S rDNA Sequencing
Bacterial DNA in water was isolated using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). After quality assessment, PCR amplification was conducted using V4-V5 region primers: the forward primer 515F (5′-barcode-GTGCCAGCMGCCGCGG-3′) and the reverse primer 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). The PCR reaction mixture consisted of 4 μL of 5x FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer at 5 μM (forward and reverse), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. Amplicons were isolated from 2% agarose gels and subsequently purified utilizing the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). The purified PCR products were quantified with a Qubit® 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and used for the construction of a sequencing library. The resulting amplicon library was then sequenced on the Illumina platform (Shanghai Biozeron Biotechnology Co., Ltd., Shanghai, China).
After quality control analysis and the removal of low-quality reads, the clean reads were obtained. These reads were then processed to generate Amplicon Sequence Variants (ASVs) using QIIME2-dada2. To obtain species classification data for each ASV, we utilized the uclust algorithm to analyze the representative sequences and assign them taxonomically using the SILVA database. After ASV annotation, alpha indices, including the Shannon, Simpson, richness, and evenness indices were calculated using mother software (v.1.35.1), and the WRM and WRC groups were compared by Student’s t-test. Principal coordinates analysis (PCoA) utilizing weighted UniFrac distances was executed and visualized in the R software. Additionally, the Analysis of Similarities (ANOSIM) test was conducted via the “vegan” package (v 3.6.3) to ascertain the differences between different groups. Meanwhile, redundancy analysis (RDA) showed the relationship between microorganisms and the environment. Co-occurrence network analysis was utilized to examine the interrelationships among microorganisms and the structural composition of their communities. In addition, the FAPROTAX database was used to predict bacterial function in water. All analyses were completed using R v4.0.2.
2.4. Statistical Analysis
Data analysis in this study was performed using SPSS (version 25.0), with the results reported as the mean ± standard error of the mean (SEM). Differences in water quality parameters between the WRC and WRM groups were analyzed using a t-test. The level of significance was set at p < 0.05.
3. Results
3.1. Water Physicochemical Properties
The water nutrient salts in different cultural modes are shown in Figure 1. After 60 days of farming, the contents of NH4+-N, NO2−-N, TN, and PO43− in the WRC group were significantly higher than those in the WRM group (p < 0.05; Figure 1A,B,D,E). However, there was no significant difference in the content of NO3−-N between the WRC and WRM groups (p > 0.05; Figure 1C).
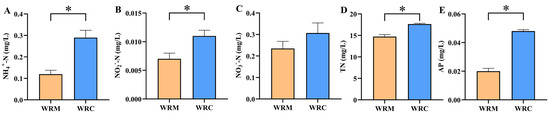
Figure 1.
Water quality parameters in different cultural modes. * p < 0.05. (A), ammonia (NH4+-N); (B), nitrite (NO2−-N); (C), nitrate (NO3−-N); (D), total nitrogen (TN); (E), total phosphorus (TP). WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
3.2. Alpha and Beta Diversities of Bacteria in Paddy Water
The alpha diversity indicated that the Shannon index in the WRC group was significantly decreased compared with the WRM group (p < 0.05; Figure 2B). Inversely, the Simpson index was markedly higher in the WRC group than that in the WRM group (p < 0.05; Figure 2C). Moreover, the species evenness of the WRM group was significantly raised compared with the WRC group (p < 0.05; Figure 2D). However, the species richness has no difference in each cultural mode (p > 0.05; Figure 2A). As for β-diversity, PCoA revealed that samples from the WRC and WRM groups formed distinct clusters (Figure 3A). Additionally, the ANOSIM test demonstrated a significant difference in the bacterial community composition between the WRC and WRM groups (R = 0.674, p = 0.001; Figure 3A).
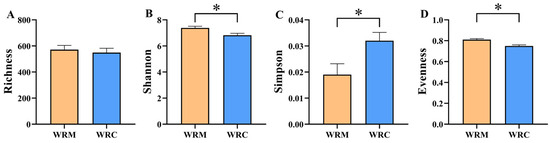
Figure 2.
Alpha diversity index in different cultural modes. * p < 0.05. (A), richness; (B), Shannon; (C), Simpson; (D), evenness. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
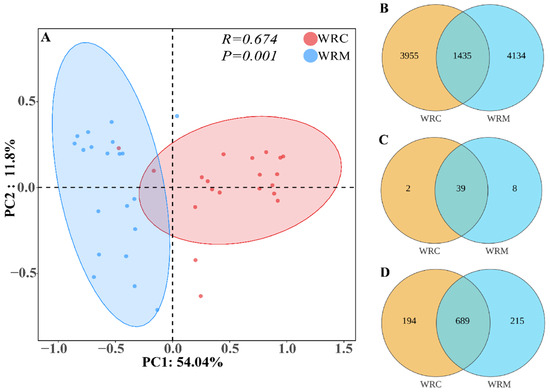
Figure 3.
Principal coordinates analysis (A) and Venn diagrams ((B), ASVs; (C), phylum; (D), genus) in different cultural modes. WRM, water rice monoculture; WRC, water rice–fish co-cultivation; PC, principal component.
Venn diagrams were used to visualize the number of common and unique ASVs between the WRM and WRC groups. A total of 9524 ASVs were identified. Of these, 1435 ASVs were common to both groups, constituting approximately 15.07% of the total ASVs detected (Figure 3B). At the phylum level, the WRC group exhibited 41 phyla, whereas the WRM group harbored 47 phyla. Notably, there was an overlap of 39 phyla between the two groups (Figure 3C). At the genus level, the WRC group contained 883 genera, compared to 904 genera in the WRM group. There was a considerable commonality with 689 genera shared between both groups (Figure 3D).
3.3. Bacterial Composition in Paddy Water
The bacterial composition at the phylum level is shown in Figure 4. The phylum-level composition of the predominant bacteria in both the WRM and WRC groups was similar, primarily consisting of Proteobacteria, Actinobacteria, and Bacteroidetes, together accounting for over 80% of the total bacteria. However, differences were observed in the relative abundance of these dominant phyla between the two groups. For example, Proteobacteria (52.91%) was the most prevalent phylum in the WRM group, while Actinobacteria (45.75%) exhibited the highest relative abundance in the WRC group. In particular, the differential analysis of the top six dominant bacteria revealed that the relative abundance of Proteobacteria and Bacteroidetes in the WRM group was significantly higher than that in the WRC group (p < 0.05; Figure 5A,C). Conversely, the relative abundance of Actinobacteria was markedly lower than that in the WRC group (p < 0.05; Figure 5B). Furthermore, there was no significant difference in the relative abundance of Firmicutes, Chloroflexi, and Cyanobacteria between the two groups (p > 0.05; Figure 5D–F).
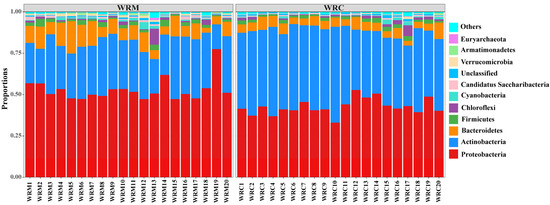
Figure 4.
Bacterial composition at the level of phylum in the WRM and WRC groups. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
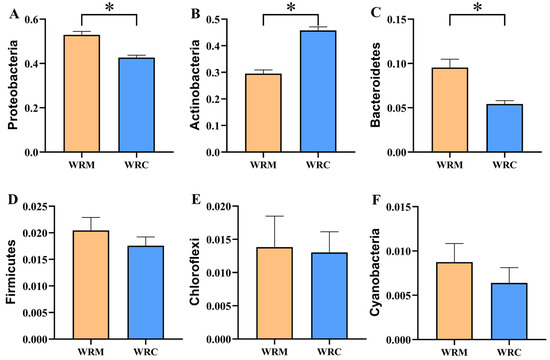
Figure 5.
The relative abundance of dominant phyla in the WRM and WRC groups. (A), Proteobacteria; (B), Actinobacteria; (C), Bacteroidetes; (D), Firmicutes; (E), Chloroflexi; (F), Cyanobacteria. * p < 0.05. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
At the genus level (Figure 6), the dominant bacteria in the WRM group were Rhodoluna (8.47%), Candidatus Planktophila (6.56%), Polynucleobacter (5.92%), Aurantimicrobium (5.33%), and Novosphingobium (4.86%), whereas the dominant bacteria in the WRC group were Rhodoluna (15.01%), Aurantimicrobium (14.52%), Polynucleobacter (5.41%), Candidatus Planktophila (4.46%), and Sideroxydans (3.56%). The differential analysis of the top six dominant bacteria showed that the relative abundances of Rhodoluna and Aurantimicrobium in the WRM group were significantly lower than those in the WRC group (p < 0.05; Figure 7A,B), while the relative abundance of Candidatus Planktophila was markedly higher than that in the WRC group (p < 0.05; Figure 7D) after 60 days of farming. In addition, the relative abundances of Polynucleobacter, Limnohabitans, and Cronobacter showed no difference (p > 0.05; Figure 7C,E,F).
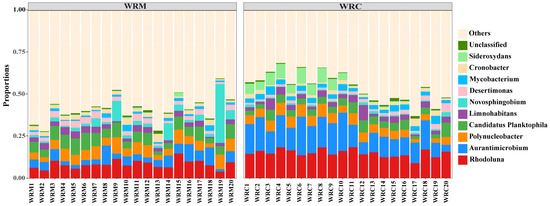
Figure 6.
Bacterial composition at the level of genus in the WRM and WRC groups. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
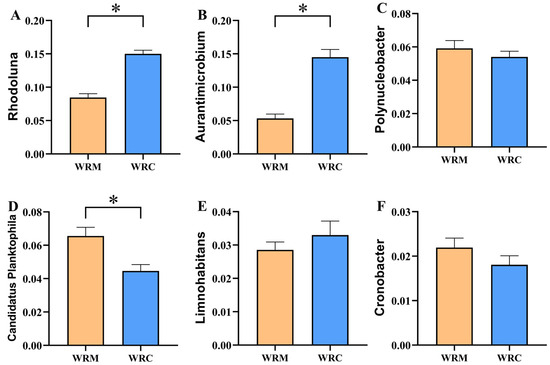
Figure 7.
The relative abundance of dominant genus in the WRM and WRC groups. (A), Rhodoluna; (B), Aurantimicrobium; (C), Polynucleobacter; (D), Candidatus Planktophila; (E), Limnohabitans; (F), Cronobacter. * p < 0.05. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
3.4. Assembly Processes and Co-Occurrence Patterns in the Water Bacterial Communities
A neutral community model (NCM) was used to identify the assembly processes of the bacteria (Figure 8). The R2 and m-values of the NCM for the WRM group were 0.821 and 0.012, respectively. The R2 and m-values of the NCM for the WRC group were 0.786 and 0.013, respectively. The results indicate that the assembly process of water bacterial communities was dominated by random process. In addition, the co-occurrence networks and topological parameters for the water bacterial communities in our study are illustrated in Figure 9. The co-occurrence network in the WRM group displayed 40 nodes and 188 edges, while the co-occurrence network in the WRC group included 33 nodes and 100 edges after 60 days of farming.
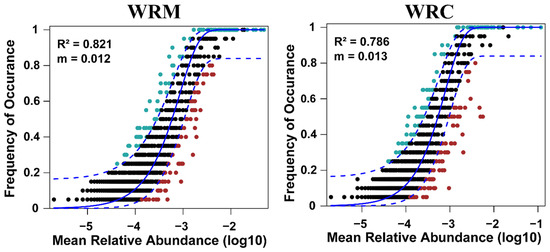
Figure 8.
Neutral community model in the water bacterial communities. The parameter “m” denotes immigration, and the value of “R2” represents the goodness of fit to the model. WRM, water rice monoculture; WRC, water rice–fish co-cultivation. The ASVs from each data were categorized into three partitions based on their frequency of occurrence relative to the predictions of the neutral community model: those occurring more frequently than (above partition, green dot), less frequently than (below partition, red dot), or within (neutral partition, black dot) the 95% confidence interval (blue solid line).
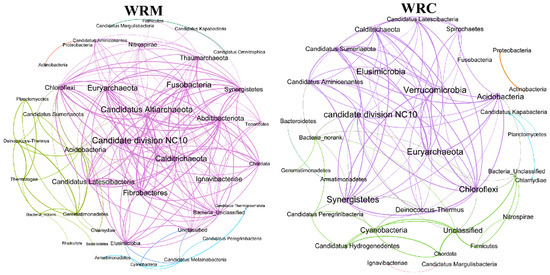
Figure 9.
Co-occurrence networks in the water bacterial communities. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
3.5. Function Prediction of Water Bacterial Communities
We selected twelve functions related to inorganic nitrogen conversion for differential analysis (Figure 10). Differential analysis showed that the relative abundance of nitrogen fixation in the WRM group was significantly lower than that in the WRC group (p < 0.05, Figure 10A). Nevertheless, the relative abundances of nitrate reduction, nitrous oxide denitrification, denitrification, nitrate denitrification, nitrite denitrification, nitrogen respiration, nitrate respiration, and nitrite respiration in the WRM group were markedly higher than those in the WRC group (p < 0.05, Figure 10B–I).
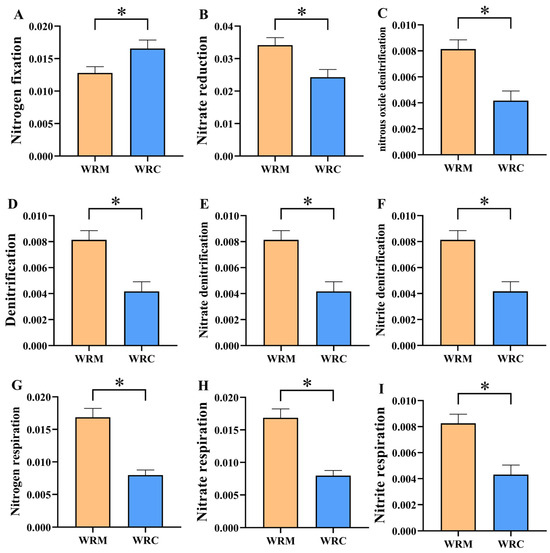
Figure 10.
Differential analysis of bacterial functions in paddy water between the WRM and WRC groups. * p < 0.05. (A), Nitrogen fixation; (B), Nitrate reduction; (C), Nitrous oxide denitrification; (D), Denitrification; (E), Nitrate denitrification; (F), Nitrite denitrification; (G), Nitrogen respiration; (H), Nitrate respiration; (I), Nitrite respiration. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
3.6. Relationship between Water Bacterial Community and Physicochemical Properties
RDA combines correspondence analysis and multiple regression on the basis of linear modes, which can intuitively see the relationship between samples and environmental factors. At the phylum level, the contribution rates of the X-axis and Y-axis were 96.21% and 3.46%, respectively, accounting for a total of 99.67% of the community changes (Figure 11). At the genus level, the contribution rates of the X-axis and Y-axis were 91.11% and 7.13%, respectively, accounting for a total of 98.24% of the community changes (Figure 11). In addition, all environmental factors had a significant positive correlation with Actinobacteria, Aurantimicrobium, and Rhodoluna (p < 0.05; Figure 12). However, ammonia, nitrite, and nitrate had a significant negative correlation with Bacteroidetes (p < 0.05; Figure 12). Ammonia, nitrite, TN, and PO43− were significantly negatively correlated with Protebacteria (p < 0.05; Figure 12). Ammonia, nitrite, and nitrate had a significant negative correlation with Candidatus Planktophila (p < 0.05; Figure 12).
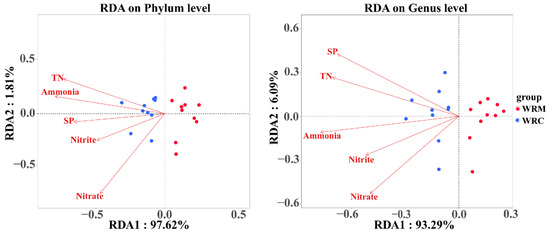
Figure 11.
Redundancy analysis (RDA) of water bacterial community composition and environmental factors. WRM, water rice monoculture; WRC, water rice–fish co-cultivation.
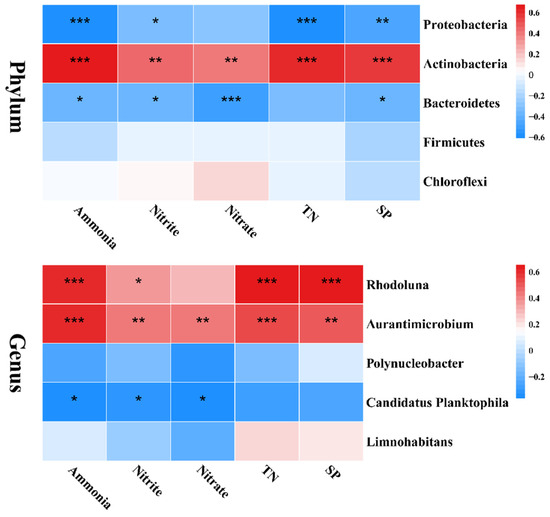
Figure 12.
The heatmap of redundancy analysis (RDA) at the phylum and genus levels. Asterisks denote statistically significant differences * p< 0.05; ** p < 0.01; *** p < 0.001.
4. Discussion
Water quality is crucial for fish growth and rice farming. In rice–fish ecosystems, the coexistence of these species provides mutual benefits [25]. Fish feces can improve soil fertility, increasing the nutrient availability for the rice [26]. Some studies have shown that there is no significant change in water parameters between the rice–fish co-culture and rice monoculture [27,28]. However, Li et al. observed that supplementary feeding in the rice–fish system contributed to heightened TN levels [29]. Similarly, Nayak et al. reported that concentrations of NO3−-N, ammonium nitrogen NH4+-N, and total phosphorus were significantly elevated in rice–fish ecosystems compared to rice monocultures, suggesting that fish activity, which disturbed the sediment, increased the nutrient levels in the water [30]. In line with these observations, we also found that the water quality parameters including NH4+-N, NO2−-N, TN, and PO43− were significantly higher in the WRC group than in the WRM group after 60 days of farming. We postulate that these elevated water quality parameters are the result of feed addition and carp activity. However, we found that there was no significant difference in the content of NO3−-N in the two groups. In aquaculture, this is a common phenomenon. For example, Peng et al. found that the content of nitrate and total nitrogen were significantly increased in the rice–fish co-culture system compared with the rice monoculture system in paddy water, while there was no significant difference in the content of nitrite, ammonia, and total phosphorus [31]. Wang et al. found that the content of total phosphorus was markedly increased in the rice–crab co-culture system compared with the rice monoculture system in paddy water, while there was no significant difference in the content of total nitrogen [32].
Bacterial communities are essential for maintaining the environmental health of aquaculture systems. They play a vital role in nutrient cycling, break down organic matter, and can even help to control pathogenic organisms. Numerous studies have revealed significant differences in bacterial community composition between integrated rice–fish farming systems and rice monoculture fields [26,33,34]. In our study, the composition of water bacterial communities in the WRM and WRC groups was found to be similar, with Proteobacteria, Actinobacteria, and Bacteroidetes identified as the dominant phyla. Nevertheless, there were significant differences in the relative abundance of these microorganisms between the two groups. Specifically, our findings indicated that the relative abundance of Actinobacteria in the WRC group was significantly higher than that in the WRM group. Actinobacteria are ubiquitous in both natural and anthropogenic environments, and they play a crucial role in the recycling of nutrients [35,36]. In addition, we found the relative abundances of Proteobacteria and Bacteroidetes in the WRC group were significantly lower than that in the WRM group. Proteobacteria has a high abundance in paddy soil [37,38]. It is not only related to sulfur, methane, and hydrogen oxidation, but also participates in nitrogen, phosphorus, and carbon cycling, degrading various environmental organic pollutants [39,40]. Bacteroidetes are the main bacteria in many environments, which can settle and decompose complex organic matter [41,42]. The bacterial community of Bacteroidetes can also efficiently utilize low-carbon substances [43]. Hence, the relative abundance of bacteria reflected that common carp cultivation may affect the nitrogen, phosphorus, and carbon cycling in paddy water. In addition, related studies have shown that rice growth can block light and inhibit the growth of phytoplankton [44]. Common carp feeding can also reduce phytoplankton [45]. Therefore, the abundance of bacteria, such as cyanobacteria, shows no significant difference between the two groups, although there is a significant difference in N and P concentrations in our experiment.
The assembly of bacterial communities exerts a profound influence on bacterial function [46]. Niche theory suggests that deterministic processes predominantly dictate the structure of bacterial communities [47]. In contrast, the neutral community model (NCM) offers an alternative perspective by evaluating these assemblies through a stochastic lens, thereby assessing the relative influence of random processes on community composition [48,49]. Among them, the value of “R2” represents the fit of the model [50]. A high R2 value indicates that the microbial community is greatly influenced by stochastic processes [51]. At the regional level, the NCM provided a better prediction of spatial community structures (mean R2 = 0.79) than deterministic factors [52]. In our study, we found that the R-squared values for the WRM and WRC groups were 0.821 and 0.786, respectively, indicating that random processes play a dominant role in the construction of bacterial communities in the paddy water. In NCM, random migration is the main determinant of community assembly [53]. Our results show that the migration rate of the WRC group was higher than that of the WRM group, indicating that common carp cultivation could enhance the mobility of bacterial communities and accelerate their assembly towards stochastic evolution. Further, our study found more nodes and edges in the WRM group than in the WRC group. In the co-occurrence network, edges and nodes represent the correlation and association between bacteria [54]. Hence, common carp cultivation has a higher fragmentation of the bacterial community and lower interactions, which may lead to a decrease in the stability of the bacterial community [55].
It has been reported that environmental factors directly affect the composition of the bacterial community [56,57,58]. Yang et al. (2017) found that Actinobacteria was positively correlated with the contents of NH4+-N, NO2−-N, NO3-N, TN, and TP [59]. Similarly, in this study, the contents of all environmental factors had a significant positive correlation with Actinobacteria after 60 days of farming. Some studies have shown that TN is negatively correlated with bacterial abundance, such as Bacteroidetes [60] and Proteobacteria [61,62], which is supported by our data. Meanwhile, our study found that ammonia, nitrite, and TN were significant positively correlated with Rhodoluna and Aurantimicrobium, which indicates that the differences in environmental factors significantly affected the relative abundance of dominant bacteria in the different cultural systems. In addition, the study showed that the relative abundances of nitrate reduction, nitrous oxide denitrification, denitrification, etc., in the WRM group were markedly higher than those in the WRC group. We speculate that common carp cultivation causes an increase in the concentrations of total nitrogen and nitrite. Therefore, the demand for bacteria to obtain nitrogen through other pathways is reduced, resulting in a decrease in the abundance of various functions related to nitrogen.
5. Conclusions
This study showed that common carp cultivation increased the contents of NH4+-N, NO2−-N, TN, and PO43− in paddy water. Meanwhile, common carp cultivation decreased the alpha diversity of bacteria in paddy water, mainly manifested in an increase in the Shannon index and a decrease in the Simpson index. Composition analysis showed that common carp cultivation markedly increased the relative abundance of Actinobacteria, Rhodoluna, and Aurantimicrobium, but decreased the relative abundance of Proteobacteria, Bacteroidetes, and Candidatus Planktophila. The neutral community model indicated that a random process played a dominant role in the construction of bacterial communities in the two groups, and common carp cultivation increased migration rates, thereby affecting community assembly and accelerating their assembly towards stochastic evolution in paddy water. In addition, the abundance of bacterial communities during our experiment were affected by environmental factors. Our study provides valuable insights into the dynamic alterations in the composition and assembly of bacterial communities in paddy water as a response to common carp cultivation.
Author Contributions
Conceptualization, writing—original draft preparation, software, W.D.; methodology, investigation, J.Y.; validation, formal analysis, Y.H., R.J. and L.Z.; resources, data curation, writing—review and editing, B.L.; visualization, supervision, project administration, funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Open Project of the Key Laboratory of Healthy Freshwater Aquaculture, Agriculture and Rural Affairs Ministry (ZJK202310), the earmarked fund for CARS (CARS-45), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2023TD64), and the National Key R&D Program of China (2019YFD0900305).
Institutional Review Board Statement
All animal use in this study was approved by the Animal Care and Use Ethics Committee of the Freshwater Fisheries (2020TD60, 10-08-2022), and all procedures were performed according to Jiangsu Laboratory’s Animal Management Guidelines (014000319/2008-00079).
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data of 16S rRNA used in this study have been submitted to the open database NCBI Sequence Read Archive (PRJNA1058949). All other data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- You, G.; Xu, B.; Su, H.; Zhang, S.; Pan, J.; Hou, X.; Li, J.; Ding, R. Evaluation of Aquaculture Water Quality Based on Improved Fuzzy Comprehensive Evaluation Method. Water 2021, 13, 1019. [Google Scholar] [CrossRef]
- Peter, M.R.; Jacob, K.E. Pond water quality and its relation to fish yield and disease occurrence in small-scale aquaculture in arid areas. Heliyon 2023, 9, e16753. [Google Scholar]
- Yu, J.; Kang, H.; Kong, Q. Research Progress of Aquaculture Tailwater Treatment Technology. Tianjin Agric. Sci. 2023, 29, 83–91. [Google Scholar]
- Liu, C.; Hu, N.; Song, W.; Chen, Q.; Zhu, L. Aquaculture Feeds Can Be Outlaws for Eutrophication When Hidden in Rice Fields? A Case Study in Qianjiang, China. Int. J. Environ. Res. Public Health 2019, 16, 4471. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Li, H.; Guo, X.; Chen, B.; Wang, M. A semi-continuous efficient strategy for removing phosphorus and nitrogen from eel aquaculture wastewater using the self-flocculating microalga Desmodesmus sp. PW1. J. Environ. Manag. 2023, 346, 118970. [Google Scholar] [CrossRef] [PubMed]
- Yi, S. Contingent Valuation of Sustainable Integrated Agriculture-Aquaculture Products: The Case of Rice–Fish Farming Systems in South Korea. Agronomy 2019, 9, 601. [Google Scholar] [CrossRef]
- Yu, X.; Hao, X.; Dang, Z.; Yang, L. Report on the Development of China’s Integrated Rice and Fishery Breeding Industry (2023). Chin. J. Aquac. 2023, 8, 19–26. [Google Scholar]
- Dong, Y.; Jia, R.; Hou, Y.R.; Diao, W.X.; Li, B.; Zhu, J. Effects of stocking density on the growth performance, mitophagy, endocytosis and metabolism of Cherax quadricarinatus in integrated rice-crayfish farming systems. Front. Physiol. 2022, 13, 1040712. [Google Scholar] [CrossRef] [PubMed]
- Xiang, A. Survey and New Inquiry on Fish Breeding in Rice Fields. China Hist. Mater. Sci. Technol. 1995, 16, 62–74. [Google Scholar]
- Lu, S.; Liao, M.; Xie, C.; He, X.; Li, D.; He, L.; Chen, J. Seasonal dynamics of ammonia-oxidizing microorganisms in freshwater aquaculture ponds. Ann. Microbiol. 2014, 65, 651–657. [Google Scholar] [CrossRef]
- Lu, S.; Liu, X.; Liu, C.; Cheng, G.; Zhou, R.; Li, Y. A Review of Ammonia-Oxidizing Archaea and Anaerobic Ammonia-Oxidizing Bacteria in the Aquaculture Pond Environment in China. Front. Microbiol. 2021, 12, 775794. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.J.; Amenyogbe, E.; Li, R.X.; Zhang, J.D.; Xie, R.T.; Wang, Z.L.; Chen, G.; Huang, J.S. The Microflora Structure in the Digestive Tract, Culture Water, and Feed of Hybrid Grouper (Epinephelus fuscoguttatus ♀ × E. polyphekadion ♂) Cultured in an Outdoor Pond Based on a High-Throughput Sequencing Technique. Aquac. Res. 2023, 2023, 9923362. [Google Scholar] [CrossRef]
- Eissa, E.S.H.; Baghdady, E.S.; Gaafar, A.Y.; El-Badawi, A.A.; Bazina, W.K.; Abd Al-Kareem, O.M.; Abd El-Hamed, N.N.B. Assessing the Influence of Dietary Pediococcus acidilactici Probiotic Supplementation in the Feed of European Sea Bass (Dicentrarchus labrax L.) (Linnaeus, 1758) on Farm Water Quality, Growth, Feed Utilization, Survival Rate, Body Composition, Blood Biochemical Parameters, and Intestinal Histology. Aquac. Nutr. 2022, 2022, 5841220. [Google Scholar] [CrossRef]
- Huang, H.H.; Li, C.Y. Adaptability of commercial probiotics to biofloc system: Influences on autochthonal bacterial community, water quality and growth performance of shrimp (Litopenaeus vannamei). Aquaculture 2024, 590, 740992. [Google Scholar] [CrossRef]
- Wuquan, L.; Zexu, L.; Minze, L.; Yuan, X.; Jianing, Z.; Yue, W.; Danqing, H.; Chengbo, S. Effects of sodium humate and probiotics on growth performance enzyme activity and microbial environment of Litopenaeus vannamei in high-density zero-water exchange systems. Front. Mar. Sci. 2022, 9, 989325. [Google Scholar]
- Bentzon-Tilia, M.; Sonnenschein, E.C.; Gram, L. Monitoring and managing microbes in aquaculture—Towards a sustainable industry. Microb. Biotechnol. 2016, 9, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wan, W.; Liu, B.; Xu, W.; Gu, Z. Effects of rice-crayfish integrated system on microbial diversity and community structure in paddy water. J. Huazhong Agric. Univ. 2022, 41, 141–151. [Google Scholar] [CrossRef]
- Arunrat, N.; Sansupa, C.; Kongsurakan, P.; Sereenonchai, S.; Hatano, R. Soil Microbial Diversity and Community Composition in Rice-Fish Co-Culture and Rice Monoculture Farming System. Biology 2022, 11, 1242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, G.; Gong, W.; Yu, E.; Xia, Y.; Li, Z.; Tian, J.; Xie, J. Study on Organic Carbon, Nitrogen, and Phosphorus Budgets of Zero-Water Exchange Ponds of Grass Carp, Bighead Carp, and Crucian Carp. Prog. Fish. Sci. 2022, 43, 188–198. [Google Scholar] [CrossRef]
- Huang, F.; Tang, Q.; Liang, P.; Xiao, L. Improvement of the ammonium molybdate spectrophotometric method for phosphorus monitoring in fresh water. J. Lake Sci. 2016, 28, 1404–1410. [Google Scholar]
- Smart, M.M.; Rada, R.G.; Donnermeyer, G.N. Determination of total nitrogen in sediments and plants using persulfate digestion. Water Res. 1983, 17, 1207–1211. [Google Scholar] [CrossRef]
- Wu, H.Z.; Cao, A. Preparation and Adding Methods of Nessler’s Reagent Having Effects on Determination of Water Quality Ammonia Nitrogen. Adv. Mater. Res. 2013, 726–731, 1362–1366. [Google Scholar] [CrossRef]
- Lo, H.S.; Lo, K.W.; Yeung, C.F.; Wong, C.Y. Rapid visual and spectrophotometric nitrite detection by cyclometalated ruthenium complex. Anal. Chim. Acta 2017, 990, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Noyes, H.A. Accurate Determination of Soil Nitrates by Phenol Disulfonic Acid Method. J. Ind. Eng. Chem. 2002, 11, 213–218. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Hang, Y.; Liu, X.; Li, P. Combination Modes and Ecological Effects of Planting-Breeding ecosystem in Rice Field. Chin. Agric. Sci. Bull. 2019, 35, 46–51. [Google Scholar]
- Herlambang, A.; Murwantoko, M.; Istiqomah, I. Dynamic change in bacterial communities in the integrated rice–fish farming system in Sleman, Yogyakarta, Indonesia. Aquac. Res. 2021, 52, 5566–5578. [Google Scholar] [CrossRef]
- Tsuruta, T.; Yamaguchi, M.; Abe, S.-I.; Iguchi, K. Effect of fish in rice-fish culture on the rice yield. Fish. Sci. 2010, 77, 95–106. [Google Scholar] [CrossRef]
- Zhijuan, N.; Feifan, L.; Wenwu, Z.; Gangchun, X.; Bo, L.; Yuyu, W.; Nailiang, S.; Jiawen, H.; Pao, X. Microbial community structure of the rice-carp co-culture systems in Hani Terraces. J. Fish. China 2020, 44, 469–479. [Google Scholar] [CrossRef]
- Li, F.; Gao, J.; Xu, Y.; Nie, Z.; Fang, J.; Zhou, Q.; Xu, G.; Shao, N.; Xu, D.; Xu, P.; et al. Biodiversity and sustainability of the integrated rice-fish system in Hani terraces, Yunnan province, China. Aquac. Rep. 2021, 20, 100763. [Google Scholar] [CrossRef]
- Nayak, P.K.; Nayak, A.K.; Panda, B.B.; Lal, B.; Gautam, P.; Poonam, A.; Shahid, M.; Tripathi, R.; Kumar, U.; Mohapatra, S.D.; et al. Ecological mechanism and diversity in rice based integrated farming system. Ecol. Indic. 2018, 91, 359–375. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, K.; Chen, C.; Xie, J. Effects of Carp (Cyprinus carpio) on Phytoplankton and Environmental Factors in Paddy Fields. J. Tianjin Agric. Univ. 2020, 27, 37–43. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Z.; Luo, L.; Zhang, R. Effects of Stocking Density on Growth Performance of Chinese Mitten Handed Crab (Erocheir sinensis) and Water Quality in a Rice-Crab Juveniles Co-culture System in Cold Regions. Chin. J. Fish. 2023, 36, 100–108. [Google Scholar]
- Zhao, X.-G.; Luo, H.; Liu, Q.-G.; Zhao, L.-J.; Cai, L.-R.; Dai, L.-L.; Zhang, Z. Influence of the cultured Odontobutis obscurus to the microbial community structure and diversity in rice-fish system. Freshw. Fish. 2017, 47, 8–14. [Google Scholar] [CrossRef]
- Jin, F. The Analysis of Microbial Community on Health Pelodiscus sinensis Breeding Aound Paddy and Penaeus vannamei Pond. Master’s Thesis, East China Normal University, Shanghai, China, 2013. [Google Scholar]
- Goodfellow, M. Ecology of Actinomycetes. Annu. Rev. Microbiol 1983, 37, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Henning, S.M.; Yang, J.; Shao, P.; Lee, R.P.; Huang, J.; Ly, A.; Hsu, M.; Lu, Q.Y.; Thames, G.; Heber, D.; et al. Health benefit of vegetable/fruit juice-based diet: Role of microbiome. Sci. Rep. 2017, 7, 2167. [Google Scholar] [CrossRef]
- Rudi, K.; Berg, F.; Gaustad, E.; Tannes, T.; Vatn, M. Ratios between Alpha-, Beta- and Gamma-proteobacteria in tap water determined by the ProteoQuant assay. Lett. Appl. Microbiol. 2010, 50, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Duan, J.; Abdoulaye, A.H.; Fu, Y.; Lin, Y.; Xie, J.; Cheng, J.; Jiang, D. Deciphering Bacterial Community of the Fallow and Paddy Soil Focusing on Possible Biocontrol Agents. Agronomy 2022, 12, 431. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Jia, R.; Sun, W.; Ding, H.; Li, B.; Zhu, J. Red Claw Crayfish Cherax quadricarinatus Cultivation Influences the Dynamics and Assembly of Benthic Bacterial Communities in Paddy Fields. Environments 2023, 10, 178. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, X.; Wang, N.; Yao, T. New subgroup of Bacteroidetes and diverse microorganisms in Tibetan plateau glacial ice provide a biological record of environmental conditions. FEMS Microbiol. Ecol. 2009, 67, 21–29. [Google Scholar] [CrossRef]
- Wolinska, A.; Kuzniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Blaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Zhong, L.-Q.; Wang, M.-H.; Zhang, S.-Y.; Jiang, H.-C.; Chen, X.-H.; Zhu, G.-W.; Bian, W.-J. Microbial community structure in a channel catfish pond in Nanjing, China. J. Agro-Environ. Sci. 2020, 39, 1594–1604. [Google Scholar]
- Li, L.; Liang, K.; Li, B.; Xu, H. Comparison of Water Quality Indicatiors and Environmental Organisms in Pond and Rice-fish Culture of Odontobutis obscura. Acta Hydrobiol. Sin. 2024, 48, 625–633. [Google Scholar]
- Peng, H. A Comparative Study of Rice-Fish Ecosystem and Rice-Monoculture Ecosystem. Master’s Thesis, Tianjin Agricultural University, Tianjin, China, 2019. [Google Scholar]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, Q.; Li, X.; Chao, X.; Liu, H.; Wei, L.; Ba, S. Deterministic processes dominate the geographic distribution pattern and community assembly of phytoplankton in typical plateau rivers. Biodivers. Sci. 2023, 31, 43–57. [Google Scholar] [CrossRef]
- Huang, X. Effects of Oyster Shell-Loading on Water Variables and Bacterial Community in Shrimop (Litopenaeus vannamei) Culture System. Ph.D. Thesis, Ningbo University, Ningbo, China, 2023. [Google Scholar]
- Wang, R.; Chen, H.; Zhu, Y.; Al-Masqari, Z.A.; Yan, M.; Wang, G.; Dong, P.; Gao, F.; Lu, T.; Zhang, D.; et al. Survival status of Penaeus vannamei is associated with the homeostasis and assembly process of the intestinal bacterial community. Aquaculture 2022, 558, 738398. [Google Scholar] [CrossRef]
- Ruyue, W.; Sai, X.; Yuxiang, Z.; Tao, Z.; Shijian, G. Denitrifying anaerobic methane-oxidizing bacteria in river networks of the Taihu Basin: Community dynamics and assembly process. Front. Microbiol. 2022, 13, 1074316. [Google Scholar]
- Xie, M.; Zhang, J.; Jia, J.; Wang, G.; Qin, Z.; Gao, H.; Li, H. Advances in the Assembly of Soil Microbial Communities in Natural and Farmland Ecosystems. Chin. J. Soil Sci. 2023, 54, 1503–1512. [Google Scholar] [CrossRef]
- Rogueta, A.; Laiglea, G.S.; Theriala, C.; Bressya, A.; Soulignac, F.; Catherine, A.; Lacroix, G.; Jardillier, L.; Bonhomme, C.; Lerch, T.Z.; et al. Neutral community model explains the bacterial community assembly in freshwater lakes. FEMS Microbiol. Ecol. 2015, 91, fiv125. [Google Scholar] [CrossRef]
- Wang, P.; Xu, M.; Yang, W.; Bai, K.; Liu, W.; Zhang, Z.; Ji, B.; Fan, Y.; Zhang, X. Nitrogen addition stimulate random migration of plant community in a semiarid steppe. Glob. Ecol. Conserv. 2021, 26, e01508. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, Z.; Waniek, J.J.; Niu, M.; Wang, Y.; Zhang, Z.; Zhou, M.; Zhang, R. The biogeography and co-occurrence network patterns of bacteria and microeukaryotes in the estuarine and coastal waters. Mar. Environ. Res. 2023, 184, 105873. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Bai, C.; Cai, J.; Dai, J.; Shao, K.; Tang, X.; Gao, G. Co-occurrence Network Reveals the Higher Fragmentation of the Bacterial Community in Kaidu River Than Its Tributaries in Northwestern China. Microbes Environ. 2018, 33, 127–134. [Google Scholar] [CrossRef]
- Ley, R.E.; Harris, J.K.; Wilcox, J.; Spear, J.R.; Miller, S.R.; Bebout, B.M.; Maresca, J.A.; Bryant, D.A.; Sogin, M.L.; Pace, N.R. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 2006, 72, 3685–3695. [Google Scholar] [CrossRef]
- Richter, D.; Matuła, J. Response of cyanobacteria and algae community from small water bodies to physicochemical parameters. Oceanol. Hydrobiol. Stud. 2012, 41, 18–28. [Google Scholar] [CrossRef]
- Yu, H.; Khan, A.U.; Subramanian, S.; Vesper, D.; Van Aken, B. Microbial Communities in Chesapeake & Ohio Canal National Historical Park and Their Function as Indicators of Water Quality. Geomicrobiol. J. 2019, 36, 673–682. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, G.-Z.; Yang, X.-N.; Wu, F.-P.; Zhao, W.; Zhang, H.-W.; Zhang, X. Microbial Community Structure and Diversity in Cellar Water by 16S rRNA High-throughput Sequencing. Huanjing Kexue 2017, 38, 1704–1716. [Google Scholar] [CrossRef]
- Li, Z.; Han, J.; Bai, H.; Peng, D.; Wang, L.; Bai, L. Effects of novel bioorganic fertilizer application on soil enzymes and bacterial community in multi-site rice paddies in China. AMB Express 2021, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Zheng, L.; Bai, Z.; Jia, A.; Wang, M. Long-term cultivation alter soil bacterial community in a forest-grassland transition zone. Front. Microbiol. 2022, 13, 1001781. [Google Scholar] [CrossRef]
- Zhang, Y.; Hou, Y.; Jia, R.; Li, B.; Zhu, J.; Ge, X. Nitrogen occurrence forms and bacterial community in sediment influenced by Bellamya purificata bioturbation. Front. Mar. Sci. 2022, 9, 1028716. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).