Abstract
Temperature is an important factor in the physiological processes of aquatic organisms and can seriously affect several chemical and biological processes in their bodies, including respiratory metabolism, antioxidant capacity, immune capacity, and signal transduction. In this study, physiological, transcriptomic, and metabolomic analyses were used to investigate the response of Penaeus vannamei to cold stress. The results indicated that cold stress disrupted nucleotide metabolism and inhibited gluconeogenesis. However, shrimp exhibited response mechanisms in order to enhance cold tolerance, regulating changes in key genes and metabolites in amino acid and lipid metabolism to increase energy supply and protect cell membrane stability. In addition, the response included regulating Ca2+ pumps and ion channels to maintain intracellular ion homeostasis and osmotic balance. When the temperature dropped further, oxidative damage occurred due to overwhelming of the antioxidant defense system, and immune function was inhibited. This research provides some references regarding the molecular mechanisms involved in responding to cold stress and potential strategies to improve cold tolerance in P. vannamei; these are important references for studying the cold stress response of shrimp.
Key Contribution:
Cold stress results in impaired antioxidant and immune defense and inhibits gluconeogenesis in the hemocytes of P. vannamei. P. vannamei produces energy by regulating amino acid and lipid metabolism in response to cold stress.
1. Introduction
Penaeus vannamei, commonly known as whiteleg shrimp, has become one of the most productive aquaculture species in the world due to its significant economic value [1]. Water temperature changes are common stressors in aquaculture; among these stressors, low temperature is a serious environmental stress factor [2,3,4]. Low temperatures can affect shrimp health by suppressing the immune system and disrupting physiological processes [5,6,7]. It has been reported that P. vannamei can grow normally between 25 °C and 35 °C [8], stop feeding at temperatures lower than 18 °C [9], and die below 12 °C [10].
Biological phenomena are complex and diverse, with intricate regulation of gene expression. When conducting single-omics studies, the conclusions often fall short of providing a comprehensive understanding. Thus, single-omics approaches have inherent restrictions. To overcome these limitations, combined analysis of the transcriptome and metabolome emerges as a powerful tool for unraveling important biological phenomena by establishing connections between changes in gene expression, metabolic pathways, and physiological processes. For example, transcriptome and metabolome analysis revealed that natural astaxanthin had superior effects on color formation, gene expression, and metabolite profiles in the leopard coral grouper (Plectropomus leopardus) compared to synthetic astaxanthin [11]; amino acid, lipid, and carbohydrate metabolisms have been confirmed to be the major metabolic pathways affected by AgNP exposure in carp [12]; in juvenile hybrid sturgeon, glutamine supplementation alleviated the liver damage caused by soybean meal substitution, possibly through downregulating the elevated levels of metabolites or genes as well as related metabolic pathways [13]; amino acid metabolism plays a critical role in the metabolic changes of clams in response to acute hypoxia, and a variety of free amino acids may not only serve as potential osmolytes for osmotic regulation but also contribute to energy production, during acute hypoxia exposure [14]. To the best of our knowledge, there is still a lack of research through an integrated analysis of physiology, transcriptome, and metabolome on the response mechanism to cold stress of P. vannamei. The molecular mechanisms underlying the coregulation of genes and metabolites in the cold tolerance of P. vannamei remain unclear.
Therefore, it is of great significance to systematically understand the molecular mechanism of P. vannamei in response to cold stress. In this study, we performed physiological analysis on shrimp hemolymph’s serum, studied the changes in plasma metabolites under cold stress through metabonomics, and analyzed the gene expression characteristics of hemocytes under cold stress by transcriptome analysis. The results will provide some important insights into the molecular mechanisms of cold adaptation in P. vannamei and potential strategies to increase its cold tolerance, which are important references for studying the cold stress response of shrimp.
2. Materials and Methods
2.1. Experimental Shrimp Collection and Rearing
Healthy P. vannamei (weight: 9.06 ± 0.23 g, length: 10.0 ± 0.4 cm) were obtained from the Rudong base of the Institute of Oceanology and Marine Fisheries, Jiangsu, Nantong, China. Shrimp were temporarily cultured in an indoor tank for 7 days before the experiment. During this period, approximately 30% of the volume of filtered, precipitated, and fully aerated seawater (24 ± 0.5 °C, salinity: 13 ± 1.0) was replaced daily. Commercial feed was fed three times a day, and dead shrimp, feed wastes, and excreta were cleaned up in a timely manner.
2.2. Experimental Design and Sample Collection
A total of 300 shrimp were randomly selected and divided into three 1000 L PVC drums as three replicates (the conditions were 24 ± 0.5 °C, salinity: 13 ± 1.0), and all shrimp were adapted to the environment in the drum for two days before the experiment. The starting temperature of the experiment was set at 24 °C (control group, CK) and was automatically cooled by a chiller at a rate of 2 °C/2 h. Then, the cooling was stopped when the water temperature reached the predetermined temperature (22, 20, 18, 16, 14, and 12 °C) and remained stable for 22 h. During this period, these temperature points (24 °C/CK, 20 °C/T1, 18 °C/T2, 14 °C/T3, and 12 °C/T4) were set as sampling points. Shrimp were randomly selected from each temperature point and quickly anesthetized on ice (10–15 s). Hemolymph (150 µL) was obtained from the ventral sinus of one shrimp with a 1 mL sterile syringe. Hemolymph samples from three shrimp in each drum were collected at each point (CK, T1, T2, T3, T4) and then immediately centrifuged (5000 rpm/min (2291× g), 4 °C)) for 10 min to obtain supernatant (hemolymph’s serum) for physiological analysis [15]. Hemolymph samples from six shrimp in each drum were collected at each of the CK, T2, and T4 points, mixed with an equivalent volume of ice-cold ACD anticoagulant (85 mM sodium citrate, 62.2 mM citric acid, and 110 mM dextrose, pH 4.9) into a centrifuge tube, and then centrifuged (5000 rpm/min (2291× g), 4 °C) for 10 min to obtain sediment (hemocytes) and supernatant (plasma). The hemocytes were preserved for transcriptomics analysis, and the plasma was preserved for metabolomics analysis [16]. All samples for physiological, transcriptome, and metabolome analysis were promptly frozen in liquid nitrogen and preserved at −80 °C. The experimental process is shown in Figure 1.
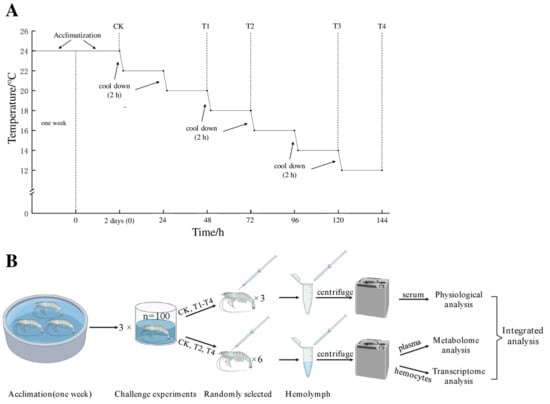
Figure 1.
Experimental process. Control check (CK/24 °C), treatment group 1 (T1/20 °C), treatment group 2 (T2/18 °C), treatment group 3 (T3/14 °C), treatment group 4 (T4/12 °C). (A): After acclimating, the starting temperature of the experiment was 24 °C, and samples collected at this temperature were named as CK (control group) before cooling. The temperature of each drum was automatically cooled by a chiller at a rate of 2 °C/2 h, the cooling was stopped when the water temperature reached the predetermined temperature (22, 20, 18, 16, 14, and 12 °C) and remained stable for 22 h, and then sampling was performed. (B): After centrifuging, serum from three shrimp in each drum were collected at CK, T1, T2, T3, and T4 for physiological analysis, and hemocytes and plasma from six shrimp in each drum were collected at CK, T2 and T4 for omics analysis.
2.3. Physiological Parameter Determination
To investigate the impact of cold on the energy reserves, oxidative damage, and immune capacity of P. vannamei, the levels of triglycerides (TG), malondialdehyde (MDA), and the enzyme activities of superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), acid phosphatase (ACP), and alkaline phosphatase (AKP) in serum were determined with assay kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Duncan’s multiple comparison procedure and one-way ANOVA were performed using the SPSS software (version 20.0; IBM, Armonk, NY, USA); p < 0.05 was considered statistically significant.
2.4. Transcriptome Analysis
Total RNA extraction from the hemocytes was performed utilizing the TRIzol reagent kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. RNase-free agarose gel electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) were used to detect RNA quality. Then, oligo (dT) beads (Epicenter, Madison, WI, USA) were utilized for mRNA enrichment. Subsequently, the small segments of the fragmented enriched mRNA were reversibly transcribed into cDNA. Then, a short segment buffer of mRNA was obtained by fragmentation and reverse transcribed into cDNA (primers were randomly designed). The cDNA was subsequently degraded by RNaseH to synthesize second-strand cDNA (DNA pol I, dNTPs, and buffer). AMPure XP beads were used to screen the purified double-stranded cDNA (Beckman Coulter, Beverly, MA, USA). The purified cDNA was amplified by PCR, purified by AMPure XP beads again, and sequenced via Illumina HiSeqTM 6000. The products were sequenced to construct a cDNA library via Illumina HiSeqTM 6000 by Gene De Novo Biotechnology Co. (Guangzhou, China).
Raw reads were screened by excluding reads connecting adapters; fastp was used to remove reads that contained over 10% unknown nucleotides (N) or over 50% low-quality (Q-value ≤ 20) bases. Bowtie2 (v2.2.8) was used to compare the sequenced reads to the rRNA database of the species. In the comparison, ribosomal reads were eliminated without permitting mismatch, and the unmapped reads that remained were utilized for posterior transcriptomics analysis. The ribosomal reads in the comparison, without allowing mismatch, were removed, and the remaining unmapped reads were used for the subsequent transcriptomics analysis. Mapping of the clean data to the P. vannamei genome (ncbi_GCA_003789085.1) was carried out. According to the comparison results of HISAT2 (v2.1.0), String Tie (v2.0.4) was used to reconstruct the transcripts, and RSEM (v1.2.19) was used to identify the expression of all genes [17]. The levels of gene expression in each sample were identified by applying the FPKM (number of reads from a gene per kilobase length per million reads) method [16]. DEseq2 software (v1.42.0) was used to detect significant differentially expressed genes (DEGs) between the control and treatment groups with the screening threshold of |log2-fold change (FC)| > 1 and false discovery rate (FDR) < 0.05. Heatmap clustering analysis of DEG expression profiles was performed. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with DEGs by pairwise comparison were identified using the KEGG database. GO functional categories associated with DEGs were identified using the GO database.
Trend analysis of the expression patterns of DEGs of the cold stress treatment group was conducted utilizing Short Time-series Expression Miner (STEM) software, version 1.3.13 [18]. DEGs were clustered into preset profiles according to their expression patterns, in which profiles with p-values ≤ 0.05 were recognized as significant profiles. Ultimately, GO and KEGG databases were employed to annotate the potential functions of DEGs from profiles.
2.5. Metabolome Analysis
After thawing the samples at 4 °C, 100 μL aliquots were combined with 400 μL of cold methanol/acetonitrile (1:1, v/v) to eliminate the protein, and the mixture was centrifuged for 15 min (14,000× g, 4 °C). The resulting supernatant was dried using a vacuum centrifuge, and the samples were redissolved in 100 μL of acetonitrile/water (1:1, v/v) solvent for LC-MS/MS analysis by ultra-high-pressure liquid chromatography (1290 Infinity LC, Agilent Technologies) combined with quadrupole time-of-flight mass spectrometry (AB Sciex TripleTOF 6600).
The obtained original data were transformed to mzXML format through Proteowizard software (v3.0.8789). Then, the alignments, including the mass-to-charge ratio, retention time, and intensity, were obtained after peak identification, filtration, and alignment. The metabolites were mapped to the Human Metabolome Database (http://www.hmdb.ca, accessed on 13 October 2022), METLIN (http://metlin.scripps.edu, accessed on 13 October 2022), Massbank (http://www.massbank.jp/, accessed on 13 October 2022), LipidMaps (http://www.lipidmaps.org, accessed on 13 October 2022), and McCloud (https://www.mzcloud.org, accessed on 13 October 2022) according to the MS/MS clastotype. The unsupervised dimensionality reduction method of principal component analysis (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA) were applied to all samples. In the OPLS-DA, the variable importance in the projection (VIP) was sorted according to the overall contribution of each variable to the OPLS-DA model, and those variables with VIP ≥ 1 and p < 0.05 were considered significant. Then, the differentially expressed metabolites (DEMs) between different comparison groups were analyzed for pathway enrichment using the KEGG database.
2.6. Combined Analysis
2.6.1. Pathway Model
KEGG pathway maps link transcriptomic information for genes to chemical structures of endogenous molecules, thus enabling integration analysis of genes and metabolites. All identified DEGs and DEMs were analyzed with the KEGG pathway database to obtain their connections in metabolism-related pathways.
2.6.2. Pearson Model
Pearson correlation coefficients were computed to integrate metabolome and transcriptome data. In accordance with the absolute correlation coefficients (p < 0.05), the gene and metabolite pairs were arranged in decreasing order. Correlation network analysis was carried out using Cytoscape (v3.7.1) to depict the precise relationships visually.
2.7. Quantitative Real-Time PCR Analysis
To confirm the RNA-Seq results, ten genes that had a large differential fold between groups, high gene expression, and a relatively high read count were randomly selected for qRT-PCR analysis, and β-actin rRNA of P. vannamei (GenBank: AF300705.2) was selected as the reference gene for internal standardization. cDNA was synthesized from qualified total RNA with the Prime Script™ RT reagent kit with gDNA Eraser (Perfect Real Time, Takara, Beijing, China). qRT-PCR procedure was conducted following the instructions provided with TB Green® Premix Ex Taq™ (Tli RNaseH Plus, Takara, Beijing, China) on a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). Primers were designed using Primer software (v5.0) and AlleleID software (v6.0); all primers were designed as shown in Table 1. The calculation of relative gene expression levels was performed utilizing the 2−△△Ct method.

Table 1.
Primer sequences of unigenes verified by qRT-PCR.
3. Results
3.1. Physiological Parameters
In general, the levels of energy reserves, antioxidant capacity, and immune activities in the hemolymph’s serum of P. vannamei were significantly different under cold stress (Figure 2). When the temperature dropped, the levels of TG decreased, reaching a minimum at 12 °C, which was significantly lower than 24 °C (p < 0.05) (Figure 2A). The changes in MDA, CAT, GPX, and SOD in the serum of P. vannamei are shown in Figure 2B–E. Although the MDA content did not significantly increase after initial cooling compared to 24 °C (p > 0.05), it was significantly higher when dropped to 14 °C and 12 °C compared to 24 °C (p < 0.05). The activity of the CAT enzyme sharply decreased after the temperature declined, and when the temperature dropped to 18 °C, the CAT activity was significantly lower than that at 24 °C (p < 0.05). The activity of GPX and SOD showed a tendency of initial increase, followed by a decrease, and peaked when the temperature dropped to 18 °C; this peak of activity was significantly higher than that at 24 °C (p < 0.05). The activities of two immune-related enzymes, ACP and AKP [20,21], showed a decreasing trend with decreasing temperature and were both highest at 24 °C, significantly higher than those of the shrimp domesticated at 12 °C (p < 0.05) (Figure 2F,G).
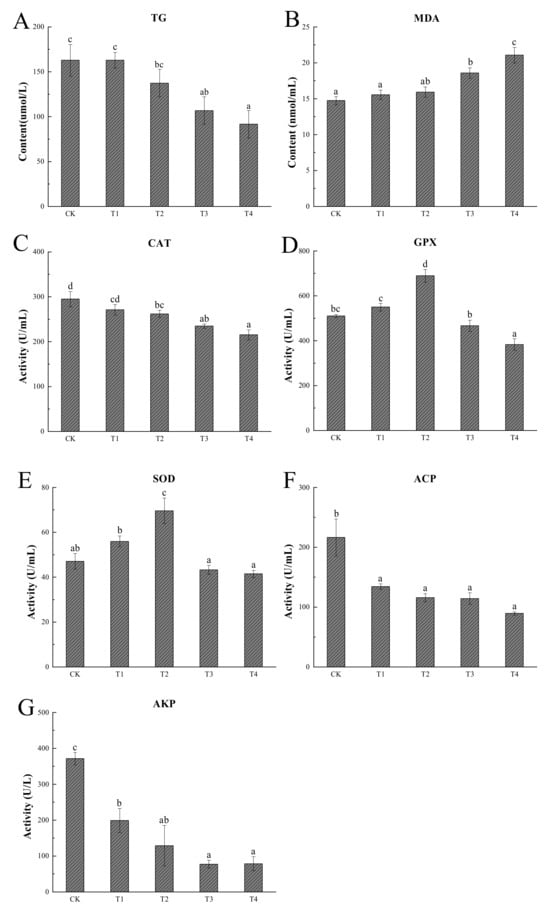
Figure 2.
Results of physiological analysis. Different letters in superscript indicate significant difference (p < 0.05). n=3; CK (24 °C), T1 (20 °C), T2 (18 °C), T3 (14 °C), and T4 (12 °C). Triglyceride (A), malondialdehyde (B), catalase (C), glutathione peroxidase (D), superoxide dismutase (E), acid phosphatase (F), and alkaline phosphatase (G).
3.2. Transcriptome Responses under Cold Stress
The sequencing statistics for the nine libraries generated with the Illumina HiSeqTM6000 platform are presented in Table 2. The raw data per sample ranged from 39.34 to 48.14 Mb. After filtering, the clean data per library were between 39.12 and 47.88 Mb. The Q20 and Q30 values exceeded 97.80% and 93.80%, respectively, while low-quality reads accounted for less than 0.56%. The GC content surpassed 42.55% for all samples. All 1687 DEGs were identified as belonging to three groups, which indicates that these genes might be sensitive to low temperatures. In addition, the gene expression difference seemed to be a gradual process during cold treatment. Specifically, volcano map analysis revealed 140 (109 up and 31 down) DEGs between CK and T2, increasing to 466 (223 up and 243 down) DEGs between T2 and T4, and peaking at 1546 (878 up and 668 down) DEGs between CK and T4 (Figure 3A–C). In addition, the results of the heatmap also proved that the expression of DEGs within groups was highly similar, and the three replicates of each group were clustered together, compared with those of other groups (Figure 3D–F).

Table 2.
Quality control of the RNA-Seq data obtained from different samples.
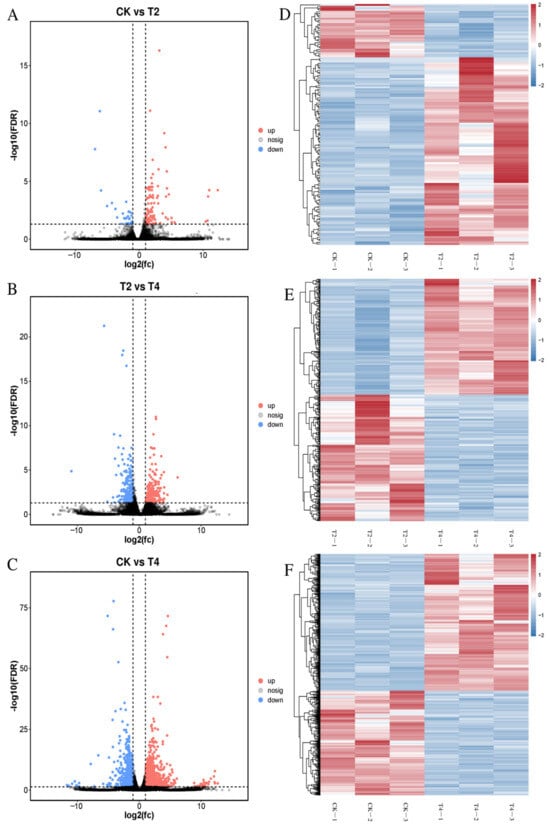
Figure 3.
Differentially expressed genes (DEGs) between treatments. Volcano maps of DEGs in CK vs. T2 (A), T2 vs. T4 (B), and CK vs. T4 (C) and heat maps of DEGs in CK vs. T2 (D), T2 vs. T4 (E), CK vs. T4 (F). The red dots and lines represent significantly upregulated DEGs, the blue dots and lines represent significantly downregulated DEGs (|log2 fold change (FC)| > 1, −log10 (FDR < 0.05)), and the black dots represent signify nonregulated genes in volcano maps. CK (24 °C), T2 (18 °C), and T4 (12 °C).
The DEG dynamic expression modes were clustered with STEM software and generated eight profiles, four of which were significantly enriched (Figure 4). The 298 DEGs in profile 7 exhibited significant upregulation consistently from CK to T4. The 490 DEGs in profile 4 showed no difference between CK and T2 but were significantly upregulated from T2 to T4. Profile 0 contained 316 DEGs with continuous significant downregulation from CK to T4. The DEGs of profile 3 (274 DEGs) showed no significant difference between CK and T2 but were significantly downregulated from T2 to T4.
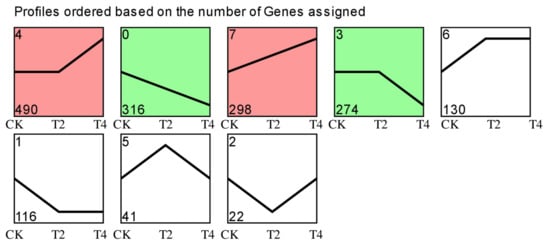
Figure 4.
Trend analysis of DEGs in three different comparisons. Colored blocks indicate significant enrichment trends (p < 0.05). The top-left number represents the trend ID where the gene is located. The bottom-left number represents the number of genes in this trend. CK (24 °C), T2 (18 °C), and T4 (12 °C).
GO enrichment analysis was performed on DEGs from the significantly enriched profiles to elucidate their biological roles (Figure 5A–D); most terms of the profiles were biological processes. The biological processes mainly included cellular processes, single-organism processes, and metabolic processes. The cellular components mainly comprise cells, cell parts, and organelles. The molecular functions are mostly related to binding and catalytic activity.
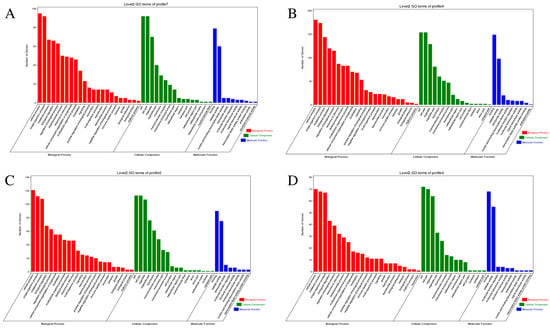
Figure 5.
GO enrichment analysis results of the significantly enriched profiles (profile 7 (A), profile 4 (B), profile 0 (C), and profile 3 (D)) in trend analysis. The X-axis displays different functions of biological processes (red), cellular components (green), and molecular functions (blue). The Y-axis represents the number of genes.
DEGs of significantly enriched profiles were aligned with the KEGG database to discriminate the significantly changed pathways, and the top 20 enrichment pathways of each profile are presented in Figure 6. In profile 7 (Figure 6A), spliceosome, progesterone-mediated oocyte maturation, ferroptosis, and other pathways were enriched when the temperature dropped from CK to T4. The upregulation of DEGs in ferroptosis may indicate that the antioxidant defense of the organism was gradually inactivated, which could lead to the accumulation of toxic lipid ROS. In profile 4 (Figure 6B), spliceosome, antifolate resistance, ABC transporters, MAPK signaling pathway, and several signaling pathways between T2 and T4 were significantly enriched, indicating that these pathways were induced to activate in response to 12 °C. In profile 0 (Figure 6C), protein processing in the endoplasmic reticulum, cysteine and methionine metabolism, antigen processing and presentation, and other pathways were significantly enriched from CK to T4, indicating that immunity may be suppressed or damaged under long-term cold stress, while the downregulation of DEGs in certain metabolic processes, such as protein processing and synthesis, may demonstrate the organism’s attempt to save energy and more effectively allocate resources to cope with environmental stress; in profile 3 (Figure 6D), several pathways were significantly enriched from T2 to T4, such as pantothenate and CoA biosynthesis, metabolic pathways, beta-alanine metabolism, and glycolysis/gluconeogenesis. The downregulation of DEGs in glycolysis/gluconeogenesis may indicate that sustained low temperature inhibited carbohydrate metabolism.
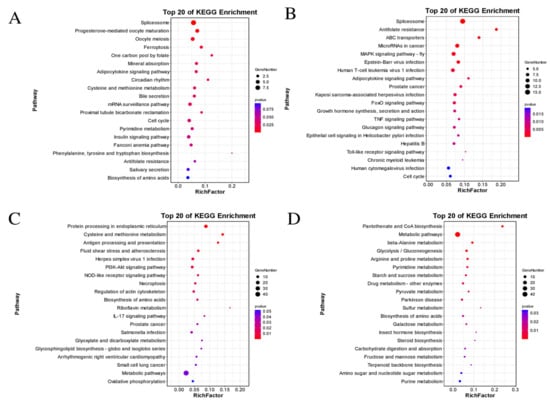
Figure 6.
KEGG enrichment analysis results of the significantly enriched profiles (profile 7 (A), profile 4 (B), profile 0 (C), and profile 3 (D)) in trend analysis. The size and color of each circle are based on the gene number and p-value.
3.3. Metabolome Responses under Cold Stress
Metabolome analysis was conducted to explore the metabolic alterations in the plasma of P. vannamei. To maximize the metabolite coverage, the POS and NEG modes were both used in LC-MS detection. Qualitative and quantitative metabolome analysis detected 12,772 and 11,903 metabolites in positive and negative modes, respectively. Multivariate statistical approaches, including PCA and OPLS-DA, were utilized to evaluate the metabolic data. PCA showed significant differences in the metabolites of P. vannamei treated with different temperatures (Figure 7A,B). The score plot of OPLS-DA revealed a clear separation between different treatment groups (Figure 7C,D), indicating a significant change in the metabolite profiles under cold stress. Additionally, multivariate statistical analysis was carried out using OPLS-TDA and permutation testing to screen the differentially abundant metabolites (Figure S1A–L).
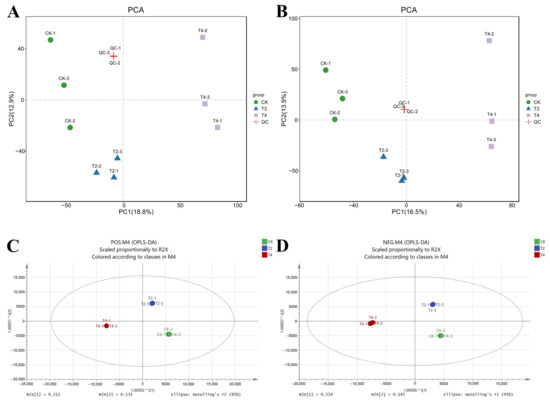
Figure 7.
PCA and OPLS-DA score diagram of control (CK) and treatment groups (T2, T4). PCA score diagram of samples in positive (A) or negative (B) ion mode; QC represents the equal mixed samples from all samples used for quality control. OPLS-DA score diagram of positive (C) and negative (D) ion modes. CK (24 °C), T2 (18 °C), and T4 (12 °C).
Based on VIP values of OPLS-DA, DEMs between experimental groups were identified. The results revealed 76 DEMs between CK and T2 (48 positive modes, 28 negative modes), 118 DEMs between CK and T4 (74 positive modes, 44 negative modes), and 72 DEMs between T2 and T4 (40 positive modes, 32 negative modes) (Figure 8).
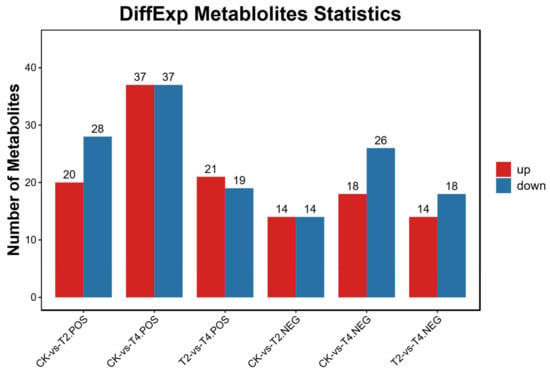
Figure 8.
Number of differentially expressed metabolites (DEMs) in three different comparisons. POS: positive ion mode; NEG: negative ion mode. CK (24 °C), T2 (18 °C), and T4 (12 °C).
To further explore the potential metabolic pathway changes in P. vannamei at different temperature points after cold stress, we performed further analysis of DEMs using the KEGG database (Figure 9). KEGG enrichment analysis results showed that when temperature decreased from CK to T2 (Figure 9A), the metabolic pathways related to mineral absorption, porphyrin metabolism, aminoacyl-tRNA biosynthesis and glycine, serine, and threonine metabolism were significantly enriched. However, when the temperature further decreased from T2 to T4, only the purine metabolism pathway was enriched (Figure 9B). The significant decrease in enriched pathways from T2 to T4 implies metabolic suppression and loss of homeostasis in P. vannamei due to excessive cold stress beyond the tolerance limit of the organism. The enrichment results also showed that the purine metabolism, steroid hormone biosynthesis, and porphyrin metabolism pathways were significantly enriched in CK vs. T4 (Figure 9C).

Figure 9.
DEMs’ KEGG enrichment of CK vs. T2 (A), T2 vs. T4 (B), and CK vs. T4 (C). The size and color of each circle are based on the metabolite number and p-value. CK (24 °C), T2 (18 °C), and T4 (12 °C).
3.4. Identification of Key Genes and Metabolites by Omics Analysis
High-throughput sequencing and mass spectrometry were used to jointly analyze metabolome and transcriptome data of P. vannamei under cold stress and to determine the correlation between DEGs and DEMs. Transcriptome and metabolome KEGG annotation results revealed that shrimp responded to stress during temperature drops through many different biological processes, and that these responses mainly involved in carbohydrate, lipid, amino acid and nucleotide metabolism (Figure 10). Most genes related to the biosynthesis of carbohydrate, lipid, amino acid, and nucleotide metabolites were upregulated, while some genes were downregulated. Cold stress altered glycine, glycerate (glyceric acid), arginine, dihydrouracil, xanthosine, inosine, hypoxanthine, and stearic acid, and the expression levels of genes involved in their synthesis or degradation (AMT, SHMT1, Aldh16a1, ASL, DPYS, DPYD, NT5E, HPRT1, and ELOVL7) were also significantly altered. The Pearson correlation coefficient, the values of which range from -1 to +1, is a statistical measure used to assess the correlation between two variables, indicating the strength of covariance between them. The correlation between genes and metabolites implicated in amino acid, lipid, and nucleotide metabolism was evaluated by calculating the Pearson coefficient of gene expression and metabolite abundance. A network graph was constructed of DEGs and DEMs (|correlation coefficients| > 0.6) screened from the dataset (Figure 11). Then, we screened some key DEGs and DEMs of P. vannamei associated with cold stress in some important pathways (Table S1).
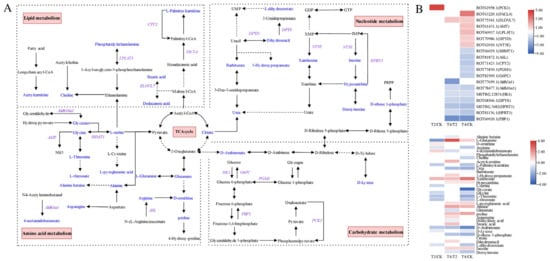
Figure 10.
Carbohydrate metabolism, lipid metabolism, amino acid metabolism, and nucleotide metabolism pathways and annotated differential expression metabolites and genes of P. vannamei under cold stress. (A) Network diagram of four metabolism pathways. Blue represents annotated DEMs, and purple represents annotated DEGs. Dotted lines represent multiple steps between metabolite conversion pathways. (B) Expressions of related differential metabolites and genes’ horizontal heatmap. Red represents significant upregulation, blue represents significant downregulation, and no color (white) represents no significant difference.
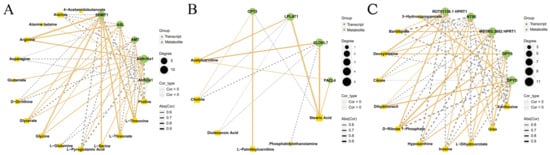
Figure 11.
Correlation network of amino acid metabolism (A), lipid metabolism (B), and nucleotide metabolism (C) pathways related metabolites and genes.
3.5. Quantitative Real-Time PCR (qRT-PCR) Validation
The expression of 10 candidate DEGs, which had a large differential fold between groups, high gene expression, and a relatively high read count, was analyzed by means of qRT-PCR to validate the RNA-Seq results. The trends of these DEGs showed high congruence between qRT-PCR and RNA-Seq data (Figure 12), supporting the reliability of the transcriptomic dataset.
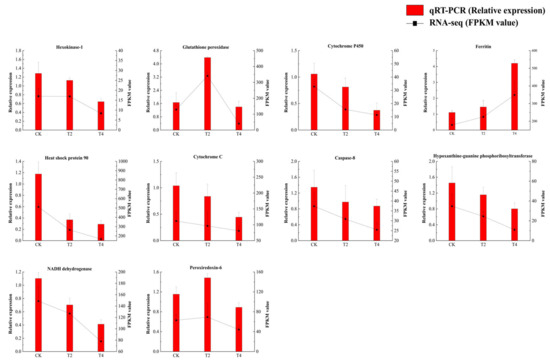
Figure 12.
Expression of DEGs in three different stages. Data are presented as mean ± SD of three replicates (n = 3). CK (24 °C), T2 (18 °C), and T4 (12 °C).
4. Discussion
In crustaceans, hemolymph serves important functions, including transporting nutrients, metabolic waste, hormones, and neuropeptides [22,23,24], while also acting as an important mediator of the immune response; the regulation of hemolymph homeostasis is critical to the health of animals [25,26,27]. Changes in metabolites under low-temperature conditions can be influenced by changes in genes and vice versa. Upregulation or downregulation of certain genes under low-temperature conditions may lead to the activation or inhibition of related metabolic pathways, resulting in an increase or decrease in metabolite levels. Furthermore, changes in metabolites can regulate gene expression through feedback mechanisms. For example, the upregulation of certain metabolites under low-temperature conditions induced the expression of related genes by activating signaling pathways [28].
4.1. Cold Stress-Induced Alterations to Carbohydrate Metabolism
Under long-term cold stress, the metabolic response of the organism may shift toward conserving energy and prioritizing survival. Based on the transcriptome results, we observed that the gluconeogenesis metabolism pathway was significantly downregulated in hemocytes under cold stress. PCK1, FBP1, and G6PC are three key enzymes that limit the steps of gluconeogenesis and control glucose synthesis [29,30]. Phosphoenolpyruvate carboxykinase is considered to be the first committed step in gluconeogenesis, which catalyzes the reversible decarboxylation of oxaloacetate to produce phosphoenolpyruvate and carbon dioxide [31]. The expression of PCK1 was upregulated when the temperature dropped from CK to T2, and we speculated that the gluconeogenesis pathway was initially activated under cold stress. FBP1 acts as the rate-limiting enzyme that catalyzes the hydrolysis of fructose 1,6-bisphosphate to inorganic phosphate and fructose-6-phosphate [32], and G6PC is involved in the last link in gluconeogenesis by catalyzing the hydrolysis of glucose-6-phosphate to glucose [33]. In our experiments, the expression of FBP1 and G6PC was downregulated under cold stress, indicating that their activity was reduced, leading to reduced glucose synthesis. PGM1 plays a central role in cellular glucose homeostasis, mediating the switch between glycolysis and gluconeogenesis through the conversion of glucose 1-phosphate (G-1-P) and glucose 6-phosphate (G-6-P) [34]. PGM1 can maintain energy supply and balance glucose metabolism within cells [35]. Studies have shown that deletion or deficiency of PGM1 leads to abnormal glycogen accumulation and metabolism [36]. Therefore, we speculated that the downregulation of PGM1 expression under cold stress may limit glycogen breakdown, reduce the rate of glucose release in glycogen, inhibit glucose production, lead to glucose homeostasis imbalance, and hinder cellular energy metabolism and ATP production. Notably, the results of PGM1 expression in broilers under low-temperature stress were similar to ours, with the same significant downregulation [37]. Although the upregulation of PCK1 meant the organism initially attempted to activate gluconeogenesis, the downregulation of FBP1 and G6PC might indicate that the gluconeogenesis process was hindered at extremely low temperatures. In the case of prolonged reduced activity and a low metabolic rate, gluconeogenesis, as an energy consumption pathway, may be downregulated to reduce energy consumption. When cobia (Rachycentron canadum) is under hypoxic stress, it also inhibits gluconeogenesis [38], which is consistent with our findings.
4.2. Alteration of Amino Acid Metabolism under Cold Stress
The reduction in some amino acids may be consumed as an energy source, while the increase in some amino acids may perform a specific function. Metabolomic data revealed significant changes in the levels of many metabolites involved in amino acid metabolic pathways under cold stress, such as arginine, glycine, glycerate, L-serine, L-threonine, L-threonate, alanine betaine, and glutamate (glutamic acid), which decreased significantly. Furthermore, RNA-Seq data indicated significant changes in the expression levels of genes which are involved in amino acid metabolism induced by cold stress.
The reduction in these amino acid contents is affected by several factors under cold stress. Low temperature inhibits protein synthesis [39] and the organism’s demand for and utilization of amino acids is reduced. Furthermore, if carbohydrates are insufficient or cannot be metabolized, amino acids become a more vital energy source [40] and are used to generate energy to cope with the low-temperature environment, resulting in their consumption. Some amino acids, such as glycine and glutamate, can produce energy. Glycine plays a critical role in metabolic regulation and antioxidant response, which can prevent tissue damage and enhance antioxidant capacity and possesses anti-inflammatory, immune regulation, and cellular protective effects [41]. We observed significant downregulation of SHMT1 expression and upregulation of AMT expression upon a temperature drop from CK to T4, accompanied by a significant decrease in glycine content. Glycine can be synthesized from serine via the serine hydroxymethyltransferase SHMT [42,43]. Moreover, with the assistance of the carrier protein GCSH, glycine can be cleaved and decomposed into carbon dioxide molecules, ammonia, and carbon units by AMT [44]. We speculated that SHMT1 downregulation and AMT upregulation under cold stress led to a decrease in glycine levels. This is consistent with previous studies, which showed that the glycine content of P. vannamei significantly decreased when the temperature continued to drop [45]. We found that when the temperature dropped from T2 to T4, the content of glutamate significantly decreased. Glutamate is an alternative energy source used in the TCA cycle under energy depletion conditions to increase the production of α-ketoglutarate [46]. In this study, the decreased content of glycine and glutamate under cold stress may be due to the increased consumption of these amino acids by organisms in cold environments to obtain more energy, leading to a decrease in the concentration of these amino acids. Aldehyde dehydrogenase (ALDH) is the key enzyme that catalyzes the oxidation of glyceraldehyde to glycerate [47]. The downregulation of Aldh16a1 expression in our experiments slowed the conversion rate from glyceraldehyde to glycerate, resulting in a decrease in glycerate content. Arginine plays an important role in anabolic processes and is mainly used for synthesizing proteins to promote growth [48]. We found that ASL expression was significantly downregulated, and the arginine content was significantly reduced, when temperature decreased. ASL catalyzes the cleavage of argininosuccinic acid (arginosuccinate) to arginine [49]. The downregulation of ASL expression in our study may inhibit the conversion process of argininosuccinic acid to arginine, resulting in a decrease in arginine levels. These results indicated that cold stress has a significant impact on the amino acid metabolism of shrimp; when carbohydrate utilization is hindered, it shifts toward using amino acids as an energy source. Additionally, there was a decrease in amino acids associated with growth and other specific functions. This metabolic response may affect the overall health and physiological processes of shrimp, including growth, development, and immune function.
In addition, some other amino acids, such as alanine, proline, D-ornithine, and L-glutamine, were significantly increased when the temperature decreased in our study. The accumulation of proline and alanine under cold stress seems to be a common feature of invertebrates [50,51,52]. Arginine can be broken down into ornithine, which can be used to produce proline [53]. Proline acts as an osmoprotectant, stabilizes macromolecules (nucleic acids, proteins, biofilms) [54], regulates cellular redox homeostasis [55,56], removes excess reactive oxygen species (ROS) and hydroxyl radicals to reduce oxidative damage [57,58], drives the oxidative pentose phosphate pathway to generate energy [59], and increases PO activity to improve immune defense [60]. Proline has a protective effect against cold stress in abiotic stress, and the accumulation of proline contributes to the cold tolerance and low-temperature adaptation of organisms [55,61]. Proline can provide important energy reserves for cold tolerance and cold acclimation, and the oxidation of proline to alanine can theoretically provide metabolic energy for 14 ATP molecules [62]. Previous studies have found that cold adaptation as well as cold tolerance in crustaceans are associated with a significant increase in proline levels [28,45]. In our experiment, the decrease in arginine levels was accompanied by a significant increase in D-ornithine and proline concentrations, indicating that some amino acids were broken down to produce more proline under cold stress, which may improve the tolerance of the organism to low temperature through mechanisms such as osmotic protection, antioxidant defense, and energy metabolism. This may be one of the important mechanisms by which organisms improve cold resistance. Glutamine, as a defense mechanism against ROS, can prevent the harmful effects of ROS [63]. Chen et al. [64] demonstrated that glutamine protects against H2O2-induced oxidative stress in cells of Jian carp (Cyprinus carpio var. Jian) due to its antioxidant potential. In addition, it has been reported that glutamine is crucial for the immune system due to its enhancement of many functional parameters of immune cells, such as T lymphocyte proliferation, B lymphocyte differentiation, cytokine production, macrophage phagocytosis, and neutrophil bactericidal effects [65,66,67,68]. Chien et al. [69] found that the higher concentration of glutamine in the hepatopancreas of shrimp that consumed feed containing Bacillus subtilis E20 may be related to improvements in immunity and antioxidant status. In this study, glutamine levels were significantly lower at the beginning of the cooling period, while glutamine levels increased significantly after the temperature dropped to T2, probably in response to ROS generation and immune defense by cold stress, and the organism increased glutamine content to play important physiological regulatory roles, including antioxidant and immune regulation. The accumulation of amino acids such as proline, alanine, D-ornithine, and L-glutamine under cold stress contributed to the enhanced low-temperature tolerance of P. vannamei through mechanisms including osmotic protection, antioxidant defense, energy metabolism, and reduction in oxidative stress damage. These results further revealed that amino acids are the main energy resource in the response of P. vannamei to cold stress.
4.3. Effect of Cold Stress on Lipid Metabolism
Lipids are one of the energy sources that are most easy to obtain, with TG decomposing into glycerol and fatty acids to provide energy [70]. The TG levels of Cherax quadricarinatus and European seabass (Dicentrarchus labrax) decreased significantly at lower temperatures [71,72]. Likewise, we observed that TG levels in the serum of P. vannamei decreased significantly during low-temperature stress, indicating that TG was consumed to produce ATP in order to provide energy. Long-chain free fatty acids (LCFFAs) serve as sources of metabolic energy, substrates for cell membrane biosynthesis (glycolipids and phospholipids), and precursors for many intracellular signaling molecules [73,74,75]. Stearic acid is an important long-chain fatty acid that plays an important role in cellular biological functions. ELOVL7 is an enzyme involved in fatty acid elongation, which promotes the biosynthesis of stearic acid by catalyzing the extension of 16-carbon palmitoyl coenzyme A by adding two-carbon units to the carboxyl terminus of the fatty acid chain using malonyl coenzyme A as a substrate [76,77]. In this study, stearic acid levels increased significantly when the temperature dropped from T2 to T4, probably because the organism increased the synthesis of longer-chain fatty acyl coenzyme A by upregulating the expression level of ELOVL7 to promote stearic acid biosynthesis. Acetylcarnitine can indirectly help the oxidative metabolism of fatty acids in mitochondria by facilitating the entry of fatty acids into mitochondria and ultimately producing ATP to maintain cellular activities. Acetyl-CoA is an important intermediate metabolite that enters the TCA cycle and is oxidized to produce energy [78]. When the production of short-chain acyl CoA exceeds the TCA cycle flux, the excess acetyl-CoA is converted to acetylcarnitine [79]. In this study, the increase in acetylcarnitine under cold stress might be explained by the catabolic metabolism of substrates during β-oxidation that exceeds the utilization capacity of acetyl coenzyme A in the TCA cycle. This indicated that P. vannamei adjusted lipid metabolism in response to cold stress, increasing energy production to sustain cellular energy homeostasis under low-temperature stress.
4.4. Cold Stress-Induced Alterations to Nucleotide Metabolism
Purines are key components of cellular energy systems (e.g., ATP, NAD), signaling (e.g., GTP, cAMP, cGMP), and pyrimidines, RNA, and DNA production [80]. Hypoxanthine and xanthosine are metabolites of purine metabolism. Hypoxanthine is a natural purine base that is produced during purine metabolism and then converted into xanthine (X) and uric acid (UA) under the action of xanthine oxidase (XO) while producing reactive oxygen species (ROS), causing oxidative stress and damage to cells and tissues [81]. HPRT1 can catalyze the formation of IMP from hypoxanthine [82]. In our experiment, HPRT1 expression was downregulated, and the hypoxanthine content increased when the temperature decreased. We speculated that low temperatures may disrupt the nucleotide metabolism of shrimp and inhibit the conversion of hypoxanthine to IMP, leading to the accumulation of hypoxanthine, which could impact the energy system of shrimp. NT5E can catalyze the generation of xanthosine from xanthosine monophosphate (XMP) [83] and can also be used to degrade inosinic acid (IMP) to inosine [84]. The expression of NT5E was upregulated in response to cold, promoting an increase in the xanthosine and inosine contents. DPYD can catalyze the reduction of uracil to dihydrouracil, while DPYS catalyzes the hydrolysis of dihydrouracil to N-amino-β-alanine [85]. In this experiment, the expression of DPYD was upregulated by the organism under cold stress to promote the conversion of uracil to form dihydrouracil, while the expression of DPYS was downregulated to inhibit the catabolic process of dihydrouracil, resulting in the accumulation of dihydrouracil. This alteration in pyrimidine metabolism might affect nucleotide synthesis and cellular processes related to nucleic acids. D-Ribose 1-phosphate can be synthesized via D-ribose 5-phosphate and subsequently converted to 5-phospho-β-D-ribose 1-pyrophosphoric acid (PRPP) [86]. In this experiment, we speculated that the decrease in D-ribose 1-phosphate under cold stress may limit the biosynthesis of nucleotides, thereby affecting the synthesis and repair of DNA and RNA, resulting in the inhibition of cell growth and reproduction. Overall, these changes indicated that cold stress could lead to disordered nucleotide metabolism, which may have negative effects on energy metabolism, signal transduction, and nucleotide synthesis ability. The imbalance between purines and pyrimidines can cause stress on shrimp metabolism and physiology.
4.5. Effects of Cold Stress on Antioxidants, Immune Defense, and Osmoregulation
In healthy organisms, reactive oxygen species (ROS) production and elimination remain in dynamic equilibrium. However, environmental stress can spur excessive ROS, peroxides, and oxygen radical generation, inducing oxidative stress [87,88,89]. Excess ROS may damage DNA and proteins and react specifically with lipids to induce the production of MDA [90,91]. It has been shown that the MDA content of C. quadricarinatus significantly increased when the temperature dropped from 25 °C to 9 °C [92]. Similarly, our study found that the MDA content in the serum of P. vannamei significantly increased under cold stress, suggesting that cold stress caused lipid peroxidation. To reduce or repair oxidative damage, crustaceans utilize endogenous antioxidant defense systems, including SOD, CAT, and GSH-Px, and other antioxidant enzymes to eliminate reactive oxygen species (ROS) and combat oxidative stress, thereby maintaining internal balance [93,94,95]. SOD serves as the first line of defense to clear ROS and can be quickly induced when exposed to oxidative stress, participating in the detoxification of superoxide free radicals [96]. GPX plays an important role in protecting cellular membranes from damage due to lipoperoxidation, owing to its function in terminating radical chain propagation by rapid reduction to yield further radicals [97]. Our study found that GPX and SOD activities initially increased as the temperature decreased, reaching a peak at 18 °C, indicating enhancement of the organism’s antioxidant defense system to protect cells from the adverse effects of oxidative stress at low temperatures. However, as the temperature continued to decrease, GPX and SOD activities significantly decreased, possibly because 12 °C is close to the lowest temperature P. vannamei can tolerate, with oxidative damage becoming severe and beyond the repair capacity of the organism. Thus, the amount of GPX and SOD in the body was insufficient to eliminate excess superoxide. CAT is considered to be the most direct enzyme for scavenging reactive oxygen species (ROS) in the antioxidant defense system of organisms and plays a core role in protecting cells from oxidative stress [98,99]. CAT activity levels in the serum showed a significant decrease under cold stress from CK to T4, indicating a decreased capacity to eliminate ROS, which can potentially elevate the risk of cell components to oxidative damage. In addition, transcriptome analysis showed that genes related to antioxidant activity (GPX, Trx-2, Txndc12) were downregulated after extreme low-temperature stress. Trx-2 is a mitochondrion-specific member of the thioredoxin (Trx) superfamily, and previous studies have found that abalone Trx-2 regulates mitochondrial oxidative stress by catalyzing a reduction in protein disulfide bonds, clearing ROS, and minimizing DNA damage [100]. Txndc12 plays an important role in redox regulation and oxidative stress defense by clearing ROS that control and maintain cellular signaling rates to reduce oxidative stress and homeostasis [101]. Txndc12 has been proven to function as an antioxidant enzyme and is involved in immune defense mechanisms in various fish species such as Sebastes schlegelii [102], Epinephelus akaara [103], Danio rerio [104], and Oplegnathus fasciatus [105]. Therefore, when the temperature dropped to T4, these antioxidant genes were downregulated at the mRNA level, and related antioxidant enzyme activity also showed a decreasing trend in the phenotype, indicating that low temperature destroyed the redox balance in shrimp, leading to oxidative damage.
In low-temperature environments, the metabolic rate of P. vannamei will decrease, which means that their immune system may not function as efficiently as at normal temperatures. The immune defense system can be activated by oxidative stress in aquatic animals [106,107]. AKP and ACP play important roles in the immune system, and AKP and ACP in crustaceans participate in the first line of nonspecific immunity [108,109,110]. ACP, as a key compound of lysosomal enzymes, which is one of the important marker enzymes of lysozyme’s ability to break down invading organisms, plays a crucial role in the immune system and has been used as a marker of macrophage activation in animal models [111,112,113]. ACP has been shown to participate in the response of red claw crayfish to cold stress, and its immune enzyme activity is inhibited at low temperatures [6]. AKP is a hydrolase which can enhance the abilities of recognition and phagocytosis in the pathogen by changing the surface structure of the pathogen [108,114]. Our results showed that AKP and ACP activities decreased significantly after low-temperature stress, indicating that cold stress damaged the nonspecific immune barrier of the organism and compromised the immune system of P. vannamei. Transcriptome analysis showed that the expression of LYZ and CTL2 was downregulated in P. vannamei under low-temperature stress. Lysozymes are important defense molecules in the innate immune system of shrimp such as Litopenaeus stylirostris [115], Fenneropenaeus chinensis [116], and P. vannamei [117], participating in various types of innate immune responses. C-type lectin plays an immunomodulatory role in the innate immunity of crustaceans, including cell adhesion [118], bacterial clearance [119], and proPO activation [120]. It has been found that the CTL involved in the immune response increases in shrimp [121,122,123]. Under cold stress, the downregulation of these genes in P. vannamei weakened its immune recognition and defense system. In summary, cold stress caused stress responses and metabolic disorders in P. vannamei, disrupting the antioxidant system and leading to immune suppression.
In this study, the levels of alanine and proline, which have osmoregulatory effects, increased significantly in the metabolome results, indicating that the organism may respond to low-temperature stress through osmotic regulation. During the process of cold adaptation, shrimp and crabs regulate the quantity and distribution of various ion channels on the cell membrane to modify the composition and concentration of intracellular ions, thereby maintaining normal physiological activities [124]. In cold environments, shrimp and crabs reduce the temperature difference between their body and the surrounding water by adjusting the ion concentration and osmotic pressure of body fluids, thereby reducing heat loss [3]. Calcium ions play a crucial role in signal transduction and osmotic regulation within cells; hence, their concentration must be tightly controlled within a strict range. Calcium transporting ATPase (Ca2+ pump) is a major participant in maintaining intracellular Ca2+ homeostasis [125] and is responsible for transporting calcium ions across the plasma membrane or organelle membrane, thereby maintaining intracellular and extracellular calcium concentration gradients. Arbuscular mycorrhizal (AM) symbiosis has been shown to improve plant tolerance to temperature stress, and increasing osmotic agent accumulation is one of the important mechanisms for improving low-temperature tolerance [126]. Liu et al. [127] observed that inoculation with AM significantly upregulated the expression of Ca2+-ATPase in cucumber roots under low-temperature conditions. Similarly, other studies have also observed that Ca2+-ATPase activity in mud crab (Scylla serrata [128] and Scylla paramamosain [129]) was significantly increased at low temperatures. The expression of ATP2C1 was significantly upregulated when the temperature dropped to T4, and we believe this is the result of compensatory mechanisms for low-temperature effects on Ca2+-ATPase at the mRNA level. It is worth noting that cold acclimation also significantly increased the mRNA levels of calcium-transporting ATPase in goldfish (Carassius auratus) [130], which is consistent with our research findings. Therefore, ectothermic animals exhibit high levels of Ca2+-ATPase in their cells under low-temperature conditions, which could be a crucial osmoregulatory mechanism in response to cold stress. In addition, our research also observed that, in comparison to CK, ATP2B3 and SLO were significantly upregulated under the T4 temperature conditions. Plasma membrane calcium ATPase (PMCA) is a type of Ca2+ pump, also known as the plasma membrane Ca2+ pump, located in the plasma membrane, which actively expels Ca2+ out of the cell to regulate intracellular Ca2+ levels [131,132]. We speculated that under low-temperature stress, shrimp upregulate ATP2B3 to maintain a lower intracellular Ca2+ concentration. This is consistent with previous research; in freshwater crayfish (Procambarus clarkii), from room temperature (23 °C) to 4 °C for 28 days, PMCA mRNA was significantly upregulated [133]. Calcium-activated potassium channels (SLOs) are a class of membrane protein channels whose activity is regulated by intracellular calcium ion concentration [134]. In this experiment, the significant downregulation of SLOs under cold conditions may be attributed to the reduced intracellular Ca2+ concentration. Furthermore, the downregulation of SLOs can prevent excessive K+ outflow by reducing the rate of K+ efflux, contributing to maintaining intracellular and extracellular ion balance and membrane potential stability in cells and avoiding unnecessary ion loss. Therefore, we speculated that Ca2+ regulation is an important regulatory mechanism in response to low-temperature stress.
5. Conclusions
This study employed an integrated multi-omics approach, combining physiology, transcriptome, and metabolome data, to explore the regulatory mechanisms involved in the response of P. vannamei to cold stress. The results indicated that cold stress can cause significant changes in enzyme activity, gene expression, and metabolic product levels in P. vannamei (Figure 13). Due to the inhibition of carbohydrate metabolism, organisms accelerate the consumption of certain amino acids, such as glycine and glutamate, to obtain more energy to maintain their energy metabolism. In addition, the accumulation of some amino acids, such as proline, alanine, and L-glutamine, may improve cold tolerance through mechanisms such as osmotic protection, antioxidant defense, and energy metabolism. Meanwhile, organisms regulate lipid metabolism, e.g., by enhancing fatty acid oxidation to increase energy production and synthesizing long-chain fatty acids such as stearic acid to maintain cell membrane stability. This also included regulating Ca2+ pumps and ion channels to maintain intracellular ion homeostasis and osmotic balance. These mechanisms enable shrimp to better cope with cold-induced physiological stresses and metabolic disorders. However, when the temperature dropped to the tolerance limit of P. vannamei, metabolic homeostasis and physiological balance were disrupted. The antioxidant defense was initially activated but became overwhelmed at extremely low temperatures, ultimately leading to oxidative damage. Additionally, immune levels were found to be severely suppressed under cold conditions. These findings could contribute to a better comprehension of the molecular mechanism underlying hemolymph in the P. vannamei response to cold stress and provide some important references to study the response to cold stress in shrimp.
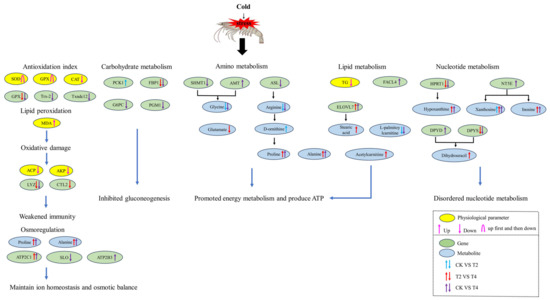
Figure 13.
The potential regulatory mechanisms of P. vannamei under cold stress. CK (24 °C), T2 (18 °C), and T4 (12 °C).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fishes9010036/s1, Figure S1: OPLS-DA score plots and OPLS-DA permutation test. Table S1: The key DEGs and DEMs of P. vannamei under cold stress.
Author Contributions
J.Z., W.S. and X.W. designed the study. J.Z., R.Z., H.L. and L.W. performed the experiment. W.S., J.Z., X.W., R.Z. and C.G. analyzed the data. W.S. and J.Z. wrote the manuscript. X.W. review the manuscript. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS[2021]122), Jiangsu Agricultural Industry Technology System (JATS [2023]154,380).
Institutional Review Board Statement
The sample collection and experimental protocols were approved by the Ethics Committee for Animal Experiments of the Jiangsu Institute of Marine Fisheries (Approval Code: 2022-3-1). All shrimps handling and methods were performed according to the relevant guidelines.
Data Availability Statement
Transcriptome data involved in this present study have been deposited in Sequences Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra, accessed on 30 October 2023), the accession number of our submission is SUB13936776. Metabolome data involved in this present study have been deposited in Metabolights (MTBLS) (https://www.ebi.ac.uk/metabolights/, accessed on 29 December 2023), the accession number of our submission is MTBLS8925.The data that support the study findings are available from the corresponding author upon request.
Acknowledgments
The authors thank the editors and reviewers for their valuable comments and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liao, I.C.; Chien, Y.-H. The Pacific White Shrimp, Litopenaeus vannamei, in Asia: The World’s Most Widely Cultured Alien Crustacean. In The Wrong Place—Alien Marine Crustaceans: Distribution, Biology and Impacts; Springer: Dordrecht, The Netherlands, 2011; pp. 489–519. [Google Scholar] [CrossRef]
- Xu, D.; Wu, J.; Sun, L.; Qin, X.; Fan, X.; Zheng, X. Combined stress of acute cold exposure and waterless duration at low temperature induces mortality of shrimp Litopenaeus vannamei through injuring antioxidative and immunological response in hepatopancreas tissue. J. Therm. Biol. 2021, 100, 103080. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wang, Q.; Shao, H.; Xu, Y.; Liu, P.; Li, J. Effects of low temperature on shrimp and crab physiology, behavior, and growth: A review. Front. Mar. Sci. 2021, 8, 746177. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, W.; Jiang, Q.; Ye, Y.; Tian, J.; Huang, Y.; Du, X.; Li, Y.; Zhao, Y.; Liu, Z. Effects of Low Temperature on Antioxidant and Heat Shock Protein Expression Profiles and Transcriptomic Responses in Crayfish (Cherax destructor). Antioxidants 2022, 11, 1779. [Google Scholar] [CrossRef]
- Qiu, J.; Wang, W.-N.; Wang, L.-j.; Liu, Y.-F.; Wang, A.-L. Oxidative stress, DNA damage and osmolality in the Pacific white shrimp, Litopenaeus vannamei exposed to acute low temperature stress. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 154, 36–41. [Google Scholar] [CrossRef]
- Wu, D.; Huang, Y.; Chen, Q.; Jiang, Q.; Li, Y.; Zhao, Y. Effects and transcriptional responses in the hepatopancreas of red claw crayfish Cherax quadricarinatus under cold stress. J. Therm. Biol. 2019, 85, 102404. [Google Scholar] [CrossRef]
- Mengal, K.; Kor, G.; Kozák, P.; Niksirat, H. Effects of environmental factors on the cellular and molecular parameters of the immune system in decapods. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2023, 276, 111332. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Palafox, J.; Martinez-Palacios, C.A.; Ross, L.G. The effects of salinity and temperature on the growth and survival rates of juvenile white shrimp, Penaeus vannamei, Boone, 1931. Aquaculture 1997, 157, 107–115. [Google Scholar] [CrossRef]
- Li, W.; Luan, S.; Luo, K.; Sui, J.; Xu, X.; Tan, J.; Kong, J. Genetic parameters and genotype by environment interaction for cold tolerance, body weight and survival of the Pacific white shrimp Penaeus vannamei at different temperatures. Aquaculture 2015, 441, 8–15. [Google Scholar] [CrossRef]
- Kumlu, M.; Kumlu, M.; Turkmen, S. Combined effects of temperature and salinity on critical thermal minima of pacific white shrimp Litopenaeus vannamei (Crustacea: Penaeidae). J. Therm. Biol. 2010, 35, 302–304. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, C.; Zhu, K.; Liu, Y.; Zhao, C.; Jiang, M.; Zhu, C.; Li, G. Effects of Natural and Synthetic Astaxanthin on Growth, Body Color, and Transcriptome and Metabolome Profiles in the Leopard Coralgrouper (Plectropomus leopardus). Animals 2023, 13, 1252. [Google Scholar] [CrossRef]
- Xiang, Q.-Q.; Yan, H.; Luo, X.-W.; Kang, Y.-H.; Hu, J.-M.; Chen, L.-Q. Integration of transcriptomics and metabolomics reveals damage and recovery mechanisms of fish gills in response to nanosilver exposure. Aquat. Toxicol. 2021, 237, 105895. [Google Scholar] [CrossRef]
- Yue, H.; Wu, J.; Fu, P.; Ruan, R.; Ye, H.; Hu, B.; Chen, X.; Li, C. Effect of glutamine supplementation against soybean meal-induced growth retardation, hepatic metabolomics and transcriptome alterations in hybrid sturgeon Acipenser baerii♀× A. schrenckii ♂. Aquac. Rep. 2022, 24, 101158. [Google Scholar] [CrossRef]
- Sun, X.; Tu, K.; Li, L.; Wu, B.; Wu, L.; Liu, Z.; Zhou, L.; Tian, J.; Yang, A. Integrated transcriptome and metabolome analysis reveals molecular responses of the clams to acute hypoxia. Mar. Environ. Res. 2021, 168, 105317. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.F.; Dai, T.M.; Zhong, S.Q.; Jin, M.; Sun, P.; Zhou, Q.C. Vibrio parahaemolyticus infection impaired intestinal barrier function and nutrient absorption in Litopenaeus vannamei. Fish Shellfish Immunol. 2020, 99, 184–189. [Google Scholar] [CrossRef]
- Liu, F.; Li, S.; Yu, Y.; Sun, M.; Xiang, J.; Li, F. Effects of ammonia stress on the hemocytes of the Pacific white shrimp Litopenaeus vannamei. Chemosphere 2020, 239, 124759. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Fan, L.; Liao, G.; Wang, Z.; Liu, H.; Cheng, K.; Hu, J.; Yang, Y.; Zhou, Z. Insight into three water additives: Revealing the protective effects on survival and stress response under cold stress for Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2023, 139, 108845. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Zhao, Z.; Luo, L.; Wang, S.; Zhang, R.; Xu, W.; Qiao, G. Immune and intestinal microbiota responses to aerial exposure stress in Chinese mitten crab (Eriocheir sinensis). Aquaculture 2021, 541, 736833. [Google Scholar] [CrossRef]
- Amorim, V.E.; Gonçalves, O.; Capela, R.; Fernández-Boo, S.; Oliveira, M.; Dolbeth, M.; Arenas, F.; Cardoso, P.G. Immunological and oxidative stress responses of the bivalve Scrobicularia plana to distinct patterns of heatwaves. Fish Shellfish Immunol. 2020, 106, 1067–1077. [Google Scholar] [CrossRef]
- Jemec Kokalj, A.; Leonardi, A.; Perc, V.; Dolar, A.; Drobne, D.; Križaj, I. Proteomics of the haemolymph of the terrestrial crustacean Porcellio scaber reveals components of its innate immunity under baseline conditions. Biochimie 2023, 213, 12–21. [Google Scholar] [CrossRef]
- Fredrick, W.S.; Ravichandran, S. Hemolymph proteins in marine crustaceans. Asian Pac. J. Trop. Biomed. 2012, 2, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Ma, X.; Huang, L.; Zhong, S.; Xing, Y.; Chen, X. Interaction analysis of hemolymph extracellular vesicles miRNA and hemocytes mRNA reveals genes and pathways associated with molting in Scylla paramamosain. Front. Mar. Sci. 2022, 9, 971648. [Google Scholar] [CrossRef]
- Söderhäll, I. Crustacean hematopoiesis. Dev. Comp. Immunol. 2016, 58, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gianazza, E.; Eberini, I.; Palazzolo, L.; Miller, I. Hemolymph proteins: An overview across marine arthropods and molluscs. J Proteom. 2021, 245, 104294. [Google Scholar] [CrossRef]
- Qyli, M.; Aliko, V.; Faggio, C. Physiological and biochemical responses of Mediterranean green crab, Carcinus aestuarii, to different environmental stressors: Evaluation of hemocyte toxicity and its possible effects on immune response. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 231, 108739. [Google Scholar] [CrossRef]
- Ren, X.; Yu, Z.; Xu, Y.; Zhang, Y.; Mu, C.; Liu, P.; Li, J. Integrated transcriptomic and metabolomic responses in the hepatopancreas of kuruma shrimp (Marsupenaeus japonicus) under cold stress. Ecotoxicol. Environ. Saf. 2020, 206, 111360. [Google Scholar] [CrossRef] [PubMed]
- Van den Berghe, G. Disorders of gluconeogenesis. J. Inherit. Metab. Dis. 1996, 19, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Yip, J.; Geng, X.; Shen, J.; Ding, Y. Cerebral Gluconeogenesis and Diseases. Front. Pharmacol. 2017, 7, 521. [Google Scholar] [CrossRef]
- Lea, P.J.; Chen, Z.H.; Leegood, R.C.; Walker, R.P. Does phosphoenolpyruvate carboxykinase have a role in both amino acid and carbohydrate metabolism? Amino Acids 2001, 20, 225–241. [Google Scholar] [CrossRef]
- Jurica, M.S.; Mesecar, A.; Heath, P.J.; Shi, W.; Nowak, T.; Stoddard, B.L. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 1998, 6, 195–210. [Google Scholar] [CrossRef] [PubMed]
- González-Santiago, A.E.; Vargas-Guerrero, B.; García-López, P.M.; Martínez-Ayala, A.L.; Domínguez-Rosales, J.A.; Gurrola-Díaz, C.M. Lupinus albus Conglutin Gamma Modifies the Gene Expressions of Enzymes Involved in Glucose Hepatic Production In Vivo. Plant Foods Hum. Nutr. 2017, 72, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Stiers, K.M.; Kain, B.N.; Graham, A.C.; Beamer, L.J. Induced Structural Disorder as a Molecular Mechanism for Enzyme Dysfunction in Phosphoglucomutase 1 Deficiency. J. Mol. Biol. 2016, 428, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Morava, E. Galactose supplementation in phosphoglucomutase-1 deficiency; review and outlook for a novel treatable CDG. Mol. Genet. Metab. 2014, 112, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Nolting, K.; Park, J.H.; Tegtmeyer, L.C.; Zühlsdorf, A.; Grüneberg, M.; Rust, S.; Reunert, J.; Du Chesne, I.; Debus, V.; Schulze-Bahr, E.; et al. Limitations of galactose therapy in phosphoglucomutase 1 deficiency. Mol. Genet. Metab. Rep. 2017, 13, 33–40. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Xing, L.; Liu, H.; Li, X.; Bao, J. Gene Expression Profiling of Broiler Liver under Cold Stress by High-Throughput Sequencing Technology. J. Poult. Sci. 2017, 54, 185–196. [Google Scholar] [CrossRef][Green Version]
- Yang, E.-J.; Amenyogbe, E.; Zhang, J.-D.; Wang, W.-Z.; Huang, J.-S.; Chen, G. Integrated transcriptomics and metabolomics analysis of the intestine of cobia (Rachycentron canadum) under hypoxia stress. Aquac. Rep. 2022, 25, 101261. [Google Scholar] [CrossRef]
- Jones, P.G.; Inouye, M. The cold-shock response—A hot topic. Mol. Microbiol. 1994, 11, 811–818. [Google Scholar] [CrossRef]
- Ji, H.; Bachmanov, A.A. Differences in postingestive metabolism of glutamate and glycine between C57BL/6ByJ and 129P3/J mice. Physiol. Genom. 2007, 31, 475–482. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Hamano, M.; Tomonaga, S.; Osaki, Y.; Oda, H.; Kato, H.; Furuya, S. Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells. Nutrients 2020, 12, 3018. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, K.; Shen, J.; Wang, Y.; Jeyaraj, A.; Hu, S.; Chen, X.; Ding, Z.; Li, X. Glycine-Induced Phosphorylation Plays a Pivotal Role in Energy Metabolism in Roots and Amino Acid Metabolism in Leaves of Tea Plant. Foods 2023, 12, 334. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, A.-L.; Xian, J.-A. Variation of free amino acid and carbohydrate concentrations in white shrimp, Litopenaeus vannamei: Effects of continuous cold stress. Aquaculture 2011, 317, 182–186. [Google Scholar] [CrossRef]
- Gul, Z.; Buyukuysal, R.L. Glutamate-induced modulation in energy metabolism contributes to protection of rat cortical slices against ischemia-induced damage. Neuroreport 2021, 32, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Feng, C.; Lip, H.; Bruce, W.R.; O’Brien, P.J. Cytotoxic molecular mechanisms and cytoprotection by enzymic metabolism or autoxidation for glyceraldehyde, hydroxypyruvate and glycolaldehyde. Chem. Biol. Interact. 2011, 191, 315–321. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef]
- Nagamani, S.C.S.; Erez, A.; Lee, B. Argininosuccinate lyase deficiency. Genet. Med. 2012, 14, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.G.; Fleurat-Lessard, F.; Lavenseau, L.; Gérard, F.; Peypelut, L.; Bonnot, G. The effect of cold acclimation and deacclimation on cold tolerance, trehalose and free amino acid levels in Sitophilus granarius and Cryptolestes ferrugineus (Coleoptera). J. Insect Physiol. 1998, 44, 955–965. [Google Scholar] [CrossRef]
- Li, Y.P.; Goto, M.; Ito, S.; Sato, Y.; Sasaki, K.; Goto, N. Physiology of diapause and cold hardiness in the overwintering pupae of the fall webworm Hyphantria cunea (Lepidoptera: Arctiidae) in Japan. J. Insect Physiol. 2001, 47, 1181–1187. [Google Scholar] [CrossRef]
- Yi, S.-X.; Adams, T.S. Effect of pyriproxyfen and photoperiod on free amino acid concentrations and proteins in the hemolymph of the Colorado potato beetle, Leptinotarsa decemlineata (Say). J. Insect Physiol. 2000, 46, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-C.; Zeng, W.-P.; Wang, H.-L.; Wang, T.; Wang, Y.-L.; Xie, F.-J. Dietary arginine requirement of juvenile Pacific white shrimp, Litopenaeus vannamei. Aquaculture 2012, 364, 252–258. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Savouré, A.; Szabados, L. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef]
- Dou, M.; Li, Y.; Sun, Z.; Li, L.; Rao, W. L-proline feeding for augmented freeze tolerance of Camponotus japonicus Mayr. Sci. Bull. 2019, 64, 1795–1804. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline Mechanisms of Stress Survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.A.; Banu, M.N.A.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Proline and glycinebetaine enhance antioxidant defense and methylglyoxal detoxification systems and reduce NaCl-induced damage in cultured tobacco cells. J. Plant Physiol. 2008, 165, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Misener, S.R.; Chen, C.-P.; Walker, V.K. Cold tolerance and proline metabolic gene expression in Drosophila melanogaster. J. Insect Physiol. 2001, 47, 393–400. [Google Scholar] [CrossRef]
- Xie, S.-W.; Tian, L.-X.; Li, Y.-M.; Zhou, W.; Zeng, S.-L.; Yang, H.-J.; Liu, Y.-J. Effect of proline supplementation on anti-oxidative capacity, immune response and stress tolerance of juvenile Pacific white shrimp, Litopenaeus vannamei. Aquaculture 2015, 448, 105–111. [Google Scholar] [CrossRef]
- Luo, P.; Chen, L.; Chen, Y.; Shen, Y.; Cui, Y. RmZAT10, a novel Cys2/His2 zinc finger transcription factor of Rosa multiflora, functions in cold tolerance through modulation of proline biosynthesis and ROS homeostasis. Environ. Exp. Bot. 2022, 198, 104845. [Google Scholar] [CrossRef]
- Shang, Q.; Pan, Y.; Peng, T.; Yang, S.; Lu, X.; Wang, Z.; Xi, J. Proteomics analysis of overexpressed plasma proteins in response to cold acclimation in Ostrinia furnacalis. Arch. Insect Biochem. Physiol. 2015, 90, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Jin, L.; Zheng, W.; Zhang, C.; Zhang, L.; Chen, Y.; Guan, J.; Fei, H. NMR-based serum metabolomics study reveals a innovative diagnostic model for missed abortion. Biochem. Biophys. Res. Commun. 2018, 496, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, X.-Q.; Feng, L.; Liu, Y.; Jiang, J. Effects of glutamine on hydrogen peroxide-induced oxidative damage in intestinal epithelial cells of Jian carp (Cyprinus carpio var. Jian). Aquaculture 2009, 288, 285–289. [Google Scholar] [CrossRef]
- Newsholme, P. Why is L-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J. Nutr. 2001, 131, 2515S–2522S. [Google Scholar] [CrossRef] [PubMed]
- Wells, S.M.; Kew, S.; Yaqoob, P.; Wallace, F.A.; Calder, P.C. Dietary glutamine enhances cytokine production by murine macrophages. Nutrition 1999, 15, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Coëffier, M.s.; Miralles-Barrachina, O.; Le Pessot, F.; Lalaude, O.; Daveau, M.; Lavoinne, A.; Lerebours, E.; Déchelotte, P. Influence of glutamine on cytokine production by human gut in vitro. Cytokine 2001, 13, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Kew, S.; Wells, S.M.; Yaqoob, P.; Wallace, F.A.; Miles, E.A.; Calder, P.C. Dietary glutamine enhances murine T-lymphocyte responsiveness. J. Nutr. 1999, 129, 1524–1531. [Google Scholar] [CrossRef]
- Chien, C.-C.; Lin, T.-Y.; Chi, C.-C.; Liu, C.-H. Probiotic, Bacillus subtilis E20 alters the immunity of white shrimp, Litopenaeus vannamei via glutamine metabolism and hexosamine biosynthetic pathway. Fish Shellfish Immunol. 2020, 98, 176–185. [Google Scholar] [CrossRef]
- Ferreira, N.G.C.; Morgado, R.; Santos, M.J.G.; Soares, A.M.V.M.; Loureiro, S. Biomarkers and energy reserves in the isopod Porcellionides pruinosus: The effects of long-term exposure to dimethoate. Sci. Total Environ. 2015, 502, 91–102. [Google Scholar] [CrossRef]
- Islam, M.J.; Kunzmann, A.; Slater, M.J. Extreme winter cold-induced osmoregulatory, metabolic, and physiological responses in European seabass (Dicentrarchus labrax) acclimatized at different salinities. Sci. Total Environ. 2021, 771, 145202. [Google Scholar] [CrossRef]
- Wu, D.; Liu, Z.; Yu, P.; Huang, Y.; Cai, M.; Zhang, M.; Zhao, Y. Cold stress regulates lipid metabolism via AMPK signalling in Cherax quadricarinatus. J. Therm. Biol. 2020, 92, 102693. [Google Scholar] [CrossRef]
- Bergström, S. Prostaglandins: Members of a New Hormonal System. Science 1967, 157, 382–391. [Google Scholar] [CrossRef]
- Grimminger, F.; Mayer, K.; Kiss, L.; Wahn, H.; Walmrath, D.; Seeger, W. Synthesis of 4-and 5-series leukotrienes in the lung microvasculature challenged with Escherichia coli hemolysin: Critical dependence on exogenous free fatty acid supply. Am. J. Respir. Cell Mol. Biol. 1997, 16, 317–324. [Google Scholar] [CrossRef]
- McArthur, M.J.; Atshaves, B.P.; Frolov, A.; Foxworth, W.D.; Kier, A.B.; Schroeder, F. Cellular uptake and intracellular trafficking of long chain fatty acids. J. Lipid Res. 1999, 40, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Simińska, D.; Jeżewski, D.; Kojder, K.; Tomasiak, P.; Tarnowski, M.; Chlubek, D.; Baranowska-Bosiacka, I. Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men. Brain Sci. 2022, 12, 1356. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Sato, Y.; Sassa, T.; Ohno, Y.; Kihara, A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 2011, 585, 3337–3341. [Google Scholar] [CrossRef] [PubMed]
- Ling, R.; Chen, G.; Tang, X.; Liu, N.; Zhou, Y.; Chen, D. Acetyl-CoA synthetase 2(ACSS2): A review with a focus on metabolism and tumor development. Discov. Oncol. 2022, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, Y.; Goto-Inoue, N.; Fujii, N.L. Role of carnitine acetylation in skeletal muscle. J. Phys. Fit. Sports Med. 2014, 3, 163–168. [Google Scholar] [CrossRef]
- Law, K.P.; Han, T.-L.; Mao, X.; Zhang, H. Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: A longitudinal metabolomics study of Chinese pregnant women part 2. Clin. Chim. Acta 2017, 468, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Lawal, A.T.; Adeloju, S.B. Progress and recent advances in fabrication and utilization of hypoxanthine biosensors for meat and fish quality assessment: A review. Talanta 2012, 100, 217–228. [Google Scholar] [CrossRef]
- Vasiliou, V.; Sandoval, M.; Backos, D.S.; Jackson, B.C.; Chen, Y.; Reigan, P.; Lanaspa, M.A.; Johnson, R.J.; Koppaka, V.; Thompson, D.C. ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein–protein interactions with HPRT1. Chem. Biol. Interact. 2013, 202, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Itoh, R.; Kimura, K. IMP–GMP 5′-nucleotidase in reptiles: Occurrence and tissue distribution in a crocodile and three species of lizards. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005, 142, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Uemoto, Y.; Ohtake, T.; Sasago, N.; Takeda, M.; Abe, T.; Sakuma, H.; Kojima, T.; Sasaki, S. Effect of two non-synonymous ecto-5′-nucleotidase variants on the genetic architecture of inosine 5′-monophosphate (IMP) and its degradation products in Japanese Black beef. BMC Genom. 2017, 18, 874. [Google Scholar] [CrossRef] [PubMed]
- Van Kuilenburg, A.B.P.; Meijer, J.; Dobritzsch, D.; Meinsma, R.; Duran, M.; Lohkamp, B.; Zoetekouw, L.; Abeling, N.G.G.M.; van Tinteren, H.L.G.; Bosch, A.M. Clinical, biochemical and genetic findings in two siblings with a dihydropyrimidinase deficiency. Mol. Genet. Metab. 2007, 91, 157–164. [Google Scholar] [CrossRef]
- Bozdech, Z.; Ginsburg, H. Data mining of the transcriptome of Plasmodium falciparum: The pentose phosphate pathway and ancillary processes. Malar. J. 2005, 4, 17. [Google Scholar] [CrossRef]
- Liu, B.; Gao, Q.; Liu, B.; Song, C.; Sun, C.; Liu, M.; Liu, X.; Liu, Y.; Li, Z.; Zhou, Q.; et al. Application of Transcriptome Analysis to Understand the Adverse Effects of Hypotonic Stress on Different Development Stages in the Giant Freshwater Prawn Macrobrachium rosenbergii Post-Larvae. Antioxidants 2022, 11, 440. [Google Scholar] [CrossRef]
- Matozzo, V.; Gallo, C.; Marin, M.G. Effects of temperature on cellular and biochemical parameters in the crab Carcinus aestuarii (Crustacea, Decapoda). Mar. Environ. Res. 2011, 71, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-l.; Liu, P.; Li, J.; Gao, B.-Q.; Chen, P. Physiological responses of swimming crab Portunus trituberculatus under cold acclimation: Antioxidant defense and heat shock proteins. Aquaculture 2014, 434, 11–17. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Wang, W.-N.; Zhou, J.; Wang, P.; Tian, T.-T.; Zheng, Y.; Liu, Y.; Mai, W.-J.; Wang, A.-L. Oxidative stress, DNA damage and antioxidant enzyme gene expression in the Pacific white shrimp, Litopenaeus vannamei when exposed to acute pH stress. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 428–435. [Google Scholar] [CrossRef]
- Wu, D.-L.; Liu, Z.-Q.; Huang, Y.-H.; Lv, W.-W.; Chen, M.-H.; Li, Y.-M.; Zhao, Y.-L. Effects of cold acclimation on the survival, feeding rate, and non-specific immune responses of the freshwater red claw crayfish (Cherax quadricarinatus). Aquac. Int. 2018, 26, 557–567. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, Y.; Dong, H.; Zhang, J. Effect of desiccation on oxidative stress and antioxidant response of the black tiger shrimp Penaeus monodon. Fish Shellfish Immunol. 2016, 58, 10–17. [Google Scholar] [CrossRef]
- Tu, H.T.; Silvestre, F.; Meulder, B.D.; Thome, J.-P.; Phuong, N.T.; Kestemont, P. Combined effects of deltamethrin, temperature and salinity on oxidative stress biomarkers and acetylcholinesterase activity in the black tiger shrimp (Penaeus monodon). Chemosphere 2012, 86, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Cao, J.; Mao, Y.; Su, Y.; Wang, J. Effects of thermal stress on oxidative stress and antioxidant response, heat shock proteins expression profiles and histological changes in Marsupenaeus japonicus. Ecol. Indic. 2019, 101, 780–791. [Google Scholar] [CrossRef]
- Gu, Z.; Wei, H.; Cheng, F.; Wang, A.; Liu, C. Effects of air exposure time and temperature on physiological energetics and oxidative stress of winged pearl oyster (Pteria penguin). Aquacult. Rep. 2020, 17, 100384. [Google Scholar] [CrossRef]
- Adams, J.; Lauterburg, B.; Mitchell, J. Plasma glutathione and glutathione disulfide in the rat: Regulation and response to oxidative stress. J. Pharmacol. Exp. Ther. 1983, 227, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Galasso, M.; Gambino, S.; Romanelli, M.G.; Donadelli, M.; Scupoli, M.T. Browsing the oldest antioxidant enzyme: Catalase and its multiple regulation in cancer. Free. Radic. Biol. Med. 2021, 172, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.; Han, G.; Wan, J.; Zhang, S.; Yang, J.; Zong, W.; Niu, Q.; Liu, R. Catalase and superoxide dismutase response and the underlying molecular mechanism for naphthalene. Sci. Total Environ. 2020, 736, 139567. [Google Scholar] [CrossRef]
- De Zoysa, M.; Pushpamali, W.A.; Whang, I.; Kim, S.J.; Lee, J. Mitochondrial thioredoxin-2 from disk abalone (Haliotis discus discus): Molecular characterization, tissue expression and DNA protection activity of its recombinant protein. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 149, 630–639. [Google Scholar] [CrossRef]
- Wei, J.; Ji, H.; Guo, M.; Qin, Q. Isolation and characterization of a thioredoxin domain-containing protein 12 from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 2012, 33, 667–673. [Google Scholar] [CrossRef]
- Thulasitha, W.S.; Umasuthan, N.; Jayasooriya, R.G.P.T.; Noh, J.K.; Park, H.-C.; Lee, J. A thioredoxin domain-containing protein 12 from black rockfish Sebastes schlegelii: Responses to immune challenges and protection from apoptosis against oxidative stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 185, 29–37. [Google Scholar] [CrossRef]
- Hanchapola, H.A.C.R.; Liyanage, D.S.; Omeka, W.K.M.; Lim, C.; Kim, G.; Jeong, T.; Lee, J. Thioredoxin domain-containing protein 12 (TXNDC12) in red spotted grouper (Epinephelus akaara): Molecular characteristics, disulfide reductase activities, and immune responses. Fish Shellfish Immunol. 2023, 132, 108449. [Google Scholar] [CrossRef]
- Kulatunga, D.C.M.; Dananjaya, S.H.S.; Nikapitiya, C.; Godahewa, G.I.; Cho, J.; Kim, C.-H.; Lee, J.; De Zoysa, M. Stress-immune responses and DNA protection function of thioredoxin domain containing 12 in zebrafish (Danio rerio). Fish Shellfish Immunol. 2019, 84, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Thulasitha, W.S.; Kim, Y.; Umasuthan, N.; Jayasooriya, R.G.P.T.; Kim, G.-Y.; Nam, B.-H.; Noh, J.K.; Lee, J. Thioredoxin domain-containing protein 12 from Oplegnathus fasciatus: Molecular characterization, expression against immune stimuli, and biological activities related to oxidative stress. Fish Shellfish Immunol. 2016, 54, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Long, X.; Liu, Z.; Cheng, Y.; Yan, S. Effect of copper nanoparticles and copper sulphate on oxidation stress, cell apoptosis and immune responses in the intestines of juvenile Epinephelus coioides. Fish Shellfish Immunol. 2015, 44, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Pan, X.; Cao, L.; Ma, B.; Fu, Z. Embryonic exposure to cis-bifenthrin enantioselectively induces the transcription of genes related to oxidative stress, apoptosis and immunotoxicity in zebrafish (Danio rerio). Fish Shellfish Immunol. 2013, 34, 717–723. [Google Scholar] [CrossRef]
- Ming, J.-H.; Ye, J.-Y.; Zhang, Y.-X.; Xu, P.; Xie, J. Effects of dietary reduced glutathione on growth performance, non-specific immunity, antioxidant capacity and expression levels of IGF-I and HSP70 mRNA of grass carp (Ctenopharyngodon idella). Aquaculture 2015, 438, 39–46. [Google Scholar] [CrossRef]
- Rajalakshmi, S.; Mohandas, A. Copper-induced changes in tissue enzyme activity in a freshwater mussel. Ecotoxicol. Environ. Saf. 2005, 62, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, J.; Wang, X.; Cai, X.; Lin, Z.; Shi, Q.; Li, E.; Qin, J.G.; Chen, L. N-acetylcysteine provides protection against the toxicity of dietary T-2 toxin in juvenile Chinese mitten crab (Eriocheir sinensis). Aquaculture 2021, 538, 736531. [Google Scholar] [CrossRef]
- Ming, Z. Effect of lipopolysaccharide and Vibrio anguillarum on the activities of phosphatase, superoxide dismutase and the content of hemocyanin in the serum of Fenneropenaeus chinesis. Mar. Sci. 2004, 28, 25–30. [Google Scholar]
- Song, Z.-F.; Wu, T.-X.; Cai, L.-S.; Zhang, L.-J.; Zheng, X.-D. Effects of dietary supplementation with Clostridium butyricum on the growth performance and humoral immune response in Miichthys miiuy. J. Zhejiang Univ. Sci. B 2006, 7, 596–602. [Google Scholar] [CrossRef]
- Li, J.; Wu, Z.-B.; Zhang, Z.; Zha, J.-W.; Qu, S.-Y.; Qi, X.-Z.; Wang, G.-X.; Ling, F. Effects of potential probiotic Bacillus velezensis K2 on growth, immunity and resistance to Vibrio harveyi infection of hybrid grouper (Epinephelus lanceolatus♂ × E. fuscoguttatus♀). Fish Shellfish Immunol. 2019, 93, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Xie, J.; Xu, P.; Ge, X.; Liu, W.; Ye, J. Effects of emodin and vitamin C on growth performance, biochemical parameters and two HSP70s mRNA expression of Wuchang bream (Megalobrama amblycephala Yih) under high temperature stress. Fish Shellfish Immunol. 2012, 32, 651–661. [Google Scholar] [CrossRef]
- Mai, W.; Hu, C. cDNA cloning, expression and antibacterial activity of lysozyme C in the blue shrimp (Litopenaeus stylirostris). Prog. Nat. Sci. 2009, 19, 837–844. [Google Scholar] [CrossRef]
- Yao, C.-L.; Wu, C.-G.; Xiang, J.-H.; Li, F.; Wang, Z.-Y.; Han, X. The lysosome and lysozyme response in Chinese shrimp Fenneropenaeus chinensis to Vibrio anguillarum and laminarin stimulation. J. Exp. Mar. Biol. Ecol. 2008, 363, 124–129. [Google Scholar] [CrossRef]
- De-La-Re-Vega, E.; García-Galaz, A.; Díaz-Cinco, M.E.; Sotelo-Mundo, R.R. White shrimp (Litopenaeus vannamei) recombinant lysozyme has antibacterial activity against Gram negative bacteria: Vibrio alginolyticus, Vibrio parahemolyticus and Vibrio cholerae. Fish Shellfish Immunol. 2006, 20, 405–408. [Google Scholar] [CrossRef]
- Odintsova, N.A.; Belogortseva, N.I.; Khomenko, A.V.; Chikalovets, I.V.; Luk’yanov, P.A. Effect of lectin from the ascidian on the growth and the adhesion of HeLa cells. Mol. Cell. Biochem. 2001, 221, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.-Y.; Shahzad, T.; Yang, P.-J.; Liu, S.; Yu, X.-Q.; Rao, X.-J. A single-CRD C-type lectin is important for bacterial clearance in the silkworm. Dev. Comp. Immunol. 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Yu, X.-Q.; Gan, H.; Kanost, M.R. Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem. Mol. Biol. 1999, 29, 585–597. [Google Scholar] [CrossRef]
- Bi, J.; Ning, M.; Xie, X.; Fan, W.; Huang, Y.; Gu, W.; Wang, W.; Wang, L.; Meng, Q. A typical C-type lectin, perlucin-like protein, is involved in the innate immune defense of whiteleg shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2020, 103, 293–301. [Google Scholar] [CrossRef]
- Thepnarong, S.; Runsaeng, P.; Rattanaporn, O.; Utarabhand, P. Molecular cloning of a C-type lectin with one carbohydrate recognition domain from Fenneropenaeus merguiensis and its expression upon challenging by pathogenic bacterium or virus. J. Invertebr. Pathol. 2015, 125, 1–8. [Google Scholar] [CrossRef]
- Wongpanya, R.; Sengprasert, P.; Amparyup, P.; Tassanakajon, A. A novel C-type lectin in the black tiger shrimp Penaeus monodon functions as a pattern recognition receptor by binding and causing bacterial agglutination. Fish Shellfish Immunol. 2017, 60, 103–113. [Google Scholar] [CrossRef]
- Masroor, W.; Farcy, E.; Gros, R.; Lorin-Nebel, C. Effect of combined stress (salinity and temperature) in European sea bass Dicentrarchus labrax osmoregulatory processes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 215, 45–54. [Google Scholar] [CrossRef]
- Bonza, M.C.; Martin, H.; Kang, M.; Lewis, G.; Greiner, T.; Giacometti, S.; Van Etten, J.L.; De Michelis, M.I.; Thiel, G.; Moroni, A. A functional calcium-transporting ATPase encoded by chlorella viruses. J. Gen. Virol. 2010, 91 Pt 10, 2620–2629. [Google Scholar] [CrossRef]
- Zhu, X.; Song, F.; Liu, F. Arbuscular Mycorrhizal Fungi and Tolerance of Temperature Stress in Plants. In Arbuscular Mycorrhizas and Stress Tolerance of Plants; Wu, Q.-S., Ed.; Springer: Singapore, 2017; pp. 163–194. [Google Scholar] [CrossRef]
- Liu, A.; Chen, S.; Chang, R.; Liu, D.; Chen, H.; Ahammed, G.J.; Lin, X.; He, C. Arbuscular mycorrhizae improve low temperature tolerance in cucumber via alterations in H2O2 accumulation and ATPase activity. J. Plant Res. 2014, 127, 775–785. [Google Scholar] [CrossRef]
- Kong, X.; Wang, G.; Li, S. Antioxidation and ATPase activity in the gill of mud crab Scylla serrata under cold stress. Chin. J. Oceanol. Limnol. 2007, 25, 221–226. [Google Scholar] [CrossRef]
- Kong, X.; Wang, G.; Li, S. Effects of low temperature acclimation on antioxidant defenses and ATPase activities in the muscle of mud crab (Scylla paramamosain). Aquaculture 2012, 370, 144–149. [Google Scholar] [CrossRef]
- Nelson, T.; McEachron, D.; Freedman, W.; Yang, W.-P. Cold acclimation increases gene transcription of two calcium transport molecules, calcium transporting ATPase and parvalbumin beta, in carassius auratus lateral musculature. J. Therm. Biol. 2003, 28, 227–234. [Google Scholar] [CrossRef]
- Kania, E.; Pająk, B.; Orzechowski, A. Calcium Homeostasis and ER Stress in Control of Autophagy in Cancer Cells. Biomed Res. Int. 2015, 2015, 352794. [Google Scholar] [CrossRef]
- Shull, G.E. Gene knockout studies of Ca2+-transporting ATPases. Eur. J. Biochem. 2000, 267, 5284–5290. [Google Scholar] [CrossRef]
- Gao, Y.; Gillen, C.M.; Whalen, D.R.; Vigo, F.M.; Golshani, A.E.; Wheatly, M.G. Expression of genes encoding Ca2+ exporting proteins in freshwater crayfish Procambarus clarkii during cold exposure. J. Therm. Biol. 2009, 34, 144–151. [Google Scholar] [CrossRef]
- Petersen, O.H.; Maruyama, Y. Calcium-activated potassium channels and their role in secretion. Nature 1984, 307, 693–696. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).