Transcriptome Sequencing Analysis of Sex-Related Genes in the Gonads of Mytilus unguiculatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Conditions
2.2. RNA Extraction and Library Construction
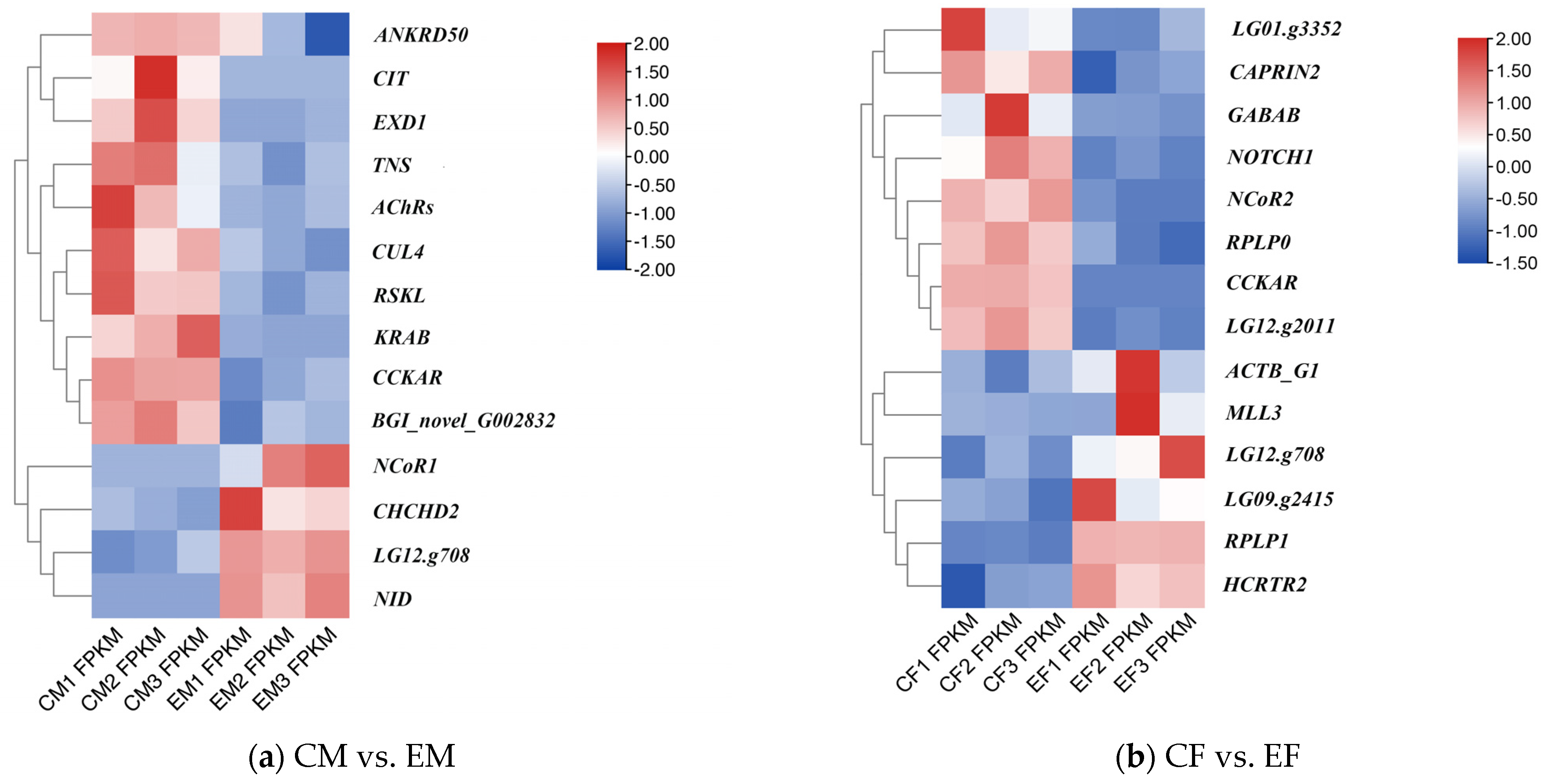
2.3. Data Assembly, Functional Annotation, and Differential Gene Screening and Clustering
2.4. RT-qPCR
3. Results
3.1. Assembly, Annotation, and Bioinformatical Analysis
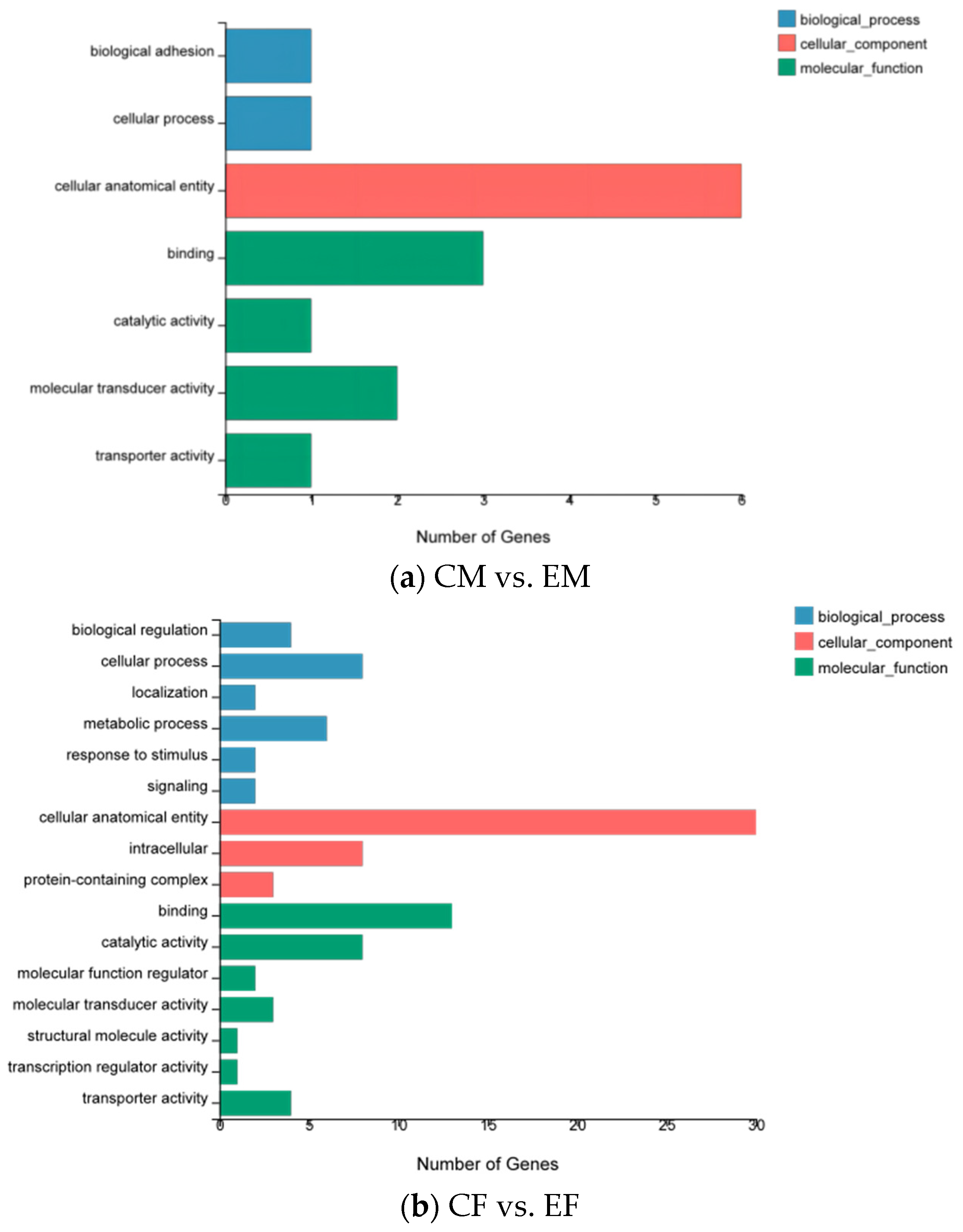
3.2. Functional Annotation and Classification of the Unigenes
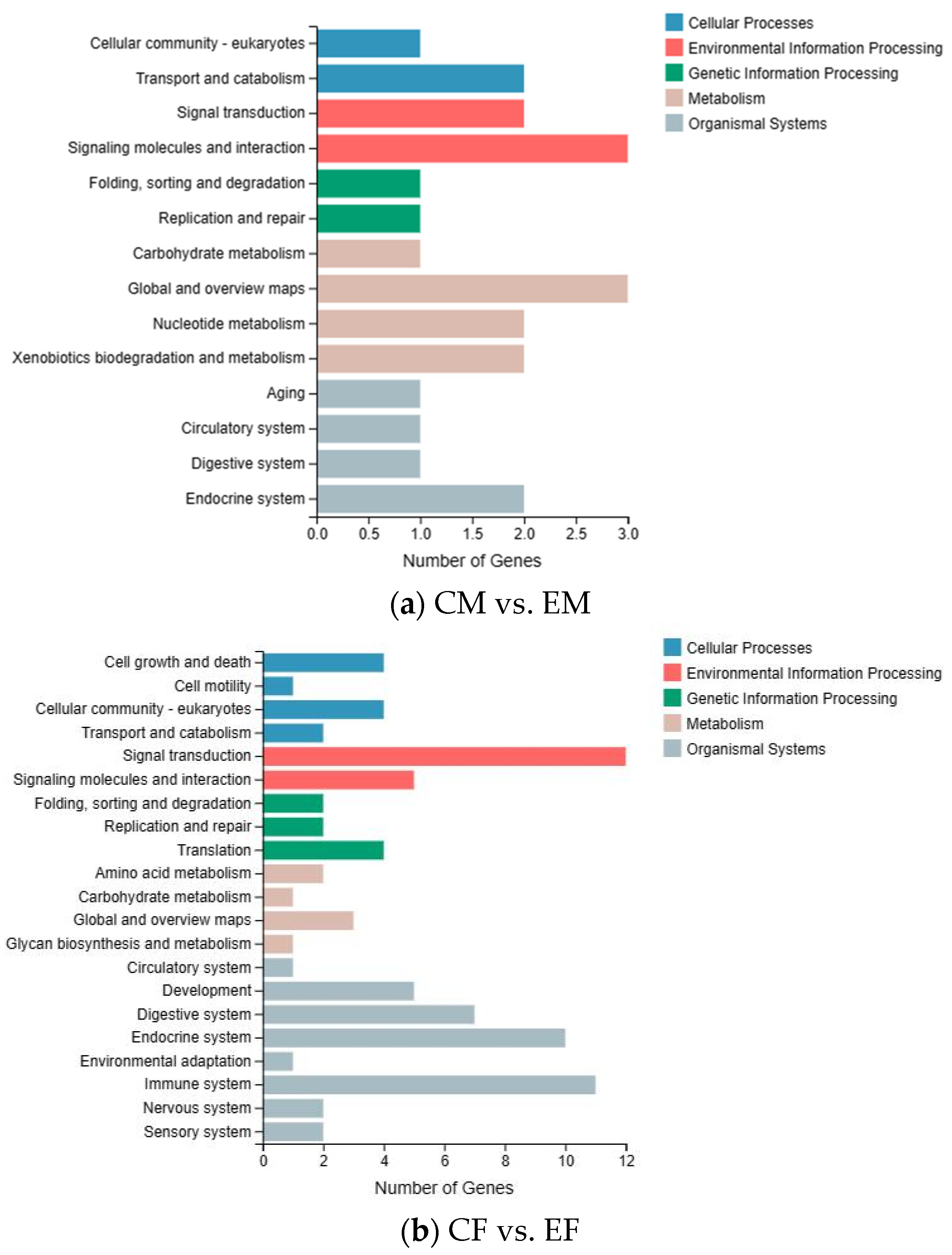
3.3. KEGG Enrichment Analysis
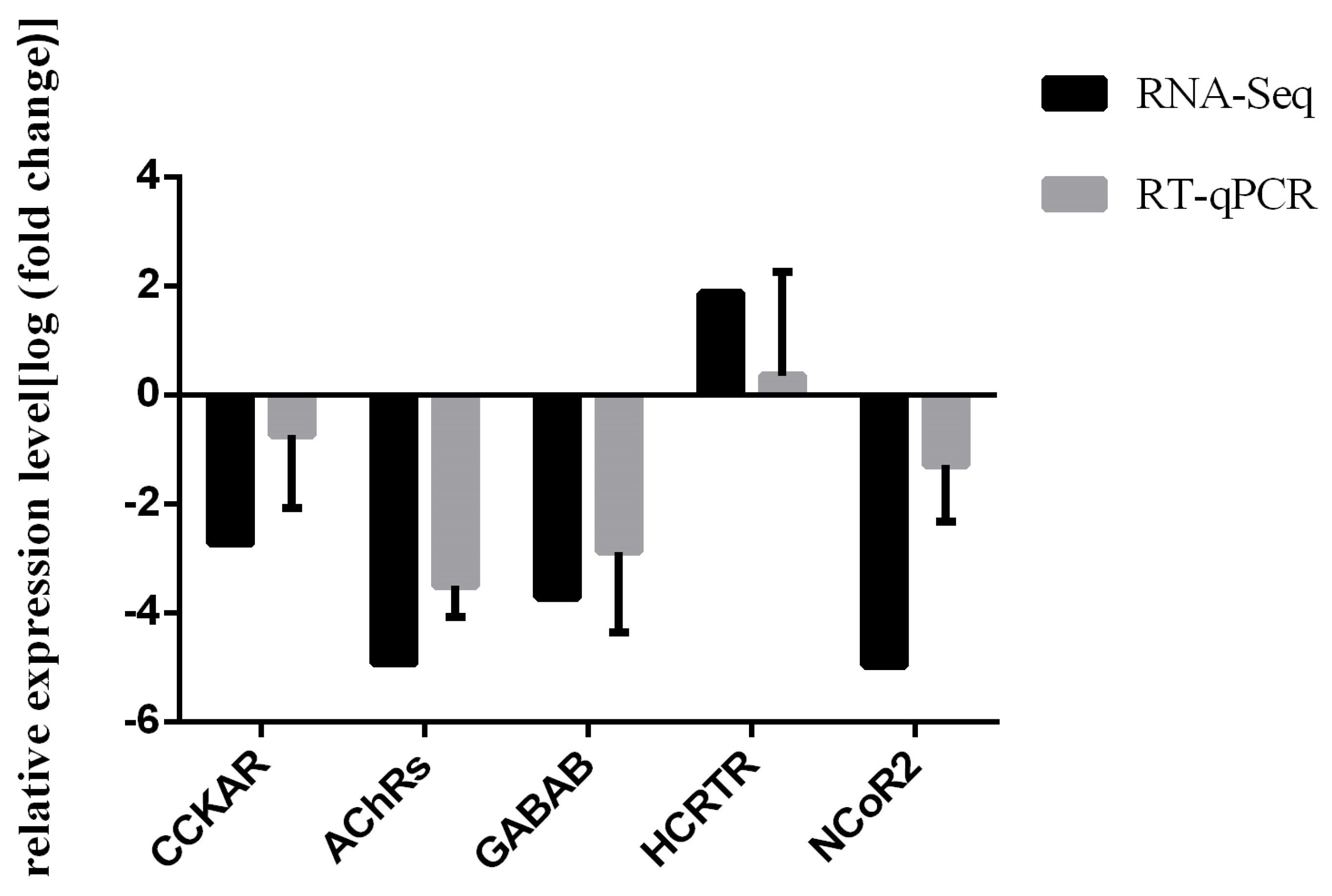
3.4. RT-qPCR Validation
4. Discussion
4.1. Analysis of KEGG Enrichment Pathways
4.2. Key Differentially Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koehn, R.K. The genetics and taxonomy of species in the genus Mytilus. Aquaculture 1991, 94, 125–145. [Google Scholar] [CrossRef]
- Zhang, W.; Li, R.; Chen, X.; Wang, C.; Gu, Z.; Mu, C.; Huang, J. Molecular identification reveals hybrids of Mytilus coruscus × Mytilus galloprovincialis in mussel hatcheries of China. Aquacult. Int. 2020, 28, 85–93. [Google Scholar] [CrossRef]
- Ye, Y.Y.; Li, J.J.; Wu, C.W.; Xu, M.Y.; Guo, B.Y. Genetic analysis of mussel (Mytilus coruscus) populations on the coast of East China Sea revealed by ISSR-PCR markers. Biochem. Syst. Ecol. 2012, 45, 1–6. [Google Scholar] [CrossRef]
- An, H.S.; Lee, J.W. Development of microsatellite markers for the Korean mussel, Mytilus coruscus (Mytilidae) using next-generation sequencing. Int. J. Mol. Sci. 2012, 13, 10583–10593. [Google Scholar] [CrossRef] [PubMed]
- Guinotte, J.M.; Fabry, V.J. Ocean acidification and its potential effects on marine ecosystems. Ann. N. Y. Acad. Sci. 2008, 1134, 320–342. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.D.; Randall, H.A.; Hultgren, K.M.; Miner, B.G.; Sorte, C.J.; Thornber, C.S.; Rodriguez, L.F.; Tomanek, L.; Williams, S.L. The impacts of climate change in coastal marine systems. Ecol. Lett. 2006, 9, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Nigel, W.; Arnell, S.; Gosling, N. The impacts of climate change on river flow regimes at the global scale. J. Hydrol. 2013, 486, 351–364. [Google Scholar]
- Huang, C.M. Studies on the Comparative Anatomy of Four Marine Bivalve Molluscs Gastropods and Byssus; Hainan University: Hainan, China, 2011. [Google Scholar]
- Cheng, L.; Xu, S.L.; Liu, F.; Qi, F. Study on the meat condition index and biochemical compositions of Mytilus unguiculatus in different gonad development stages. ICES J. Mar. Sci. 2013, 31, 68–73. [Google Scholar]
- Wang, M.; Xia, J.; Jawad, M.; Wei, W.; Gui, L.; Liang, X.; Li, M. Transcriptome sequencing analysis of sex-related genes and miRNAs in the gonads of Mytilus coruscus. Front. Mar. Sci. 2022, 9, 1013857. [Google Scholar] [CrossRef]
- Chang, K.M.; Wu, J.F. Study on artificial propagation of mussel Mytilus unguiculatus. South. China Fish. Sci. 2007, 3, 26–30. [Google Scholar]
- Secor, S.M.; Carey, H.V. Integrative physiology of fasting. Compr. Physiol. 2016, 6, 773–825. [Google Scholar] [PubMed]
- Zhao, D. Expression Characteristics and Function of Genes Dmrtl and Foxl2 in the Scallop Patinopecten yessoensis; Shanghai Ocean University: Shanghai, China, 2021. [Google Scholar]
- Wang, H.; Qu, M.; Tang, W.; Liu, S.; Ding, S. Transcriptome Profiling and Expression Localization of Key Sex-Related Genes in a Socially-Controlled Hermaphroditic Clownfish, Amphiprion clarkii. Int. J. Mol. Sci. 2022, 23, 9085. [Google Scholar] [CrossRef] [PubMed]
- Davey, A.J.H.; Jellyman, D.J. Sex Determination in Freshwater Eels and Management Options for Manipulation of Sex. Rev. Fish. Biol. Fish. 2005, 15, 37–52. [Google Scholar] [CrossRef]
- Prevedelli, D.; N’Siala, G.M.; Simonini, R. Gonochorism vs. hermaphroditism: Relationship between life history and fitness in three species of Ophryotrocha (Polychaeta: Dorvilleidae) with different forms of sexuality. J. Anim. Ecol. 2006, 75, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Mei, J.; Ge, C.T.; Liu, X.L.; Gui, J.F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci. 2022, 65, 1091–1122. [Google Scholar] [CrossRef] [PubMed]
- Naimi, A.; Martinez, A.S.; Specq, M.L.; Mrac, A.; Diss, B.; Mathieu, M.; Sourdaine, P. Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 152, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Hong, Y.; Sheng, J.; Peng, K.; Wang, J. De novo transcriptome sequencing to identify the sex-determination genes in Hyriopsis schlegelii. Biosci. Biotechnol. Biochem. 2015, 79, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Sun, Y.; Ma, X.; Wang, J.; Li, R.; Zhang, M.; Wang, S.; Hu, X.; Bao, Z. Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in Scallop Patinopecten yessoensis. Mar. Biotechnol. 2016, 18, 453–465. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, W.; He, M. Proteome and transcriptome analysis of ovary, intersex gonads, and testis reveals potential key sex reversal/differentiation genes and mechanism in Scallop Chlamys nobilis. Mar. Biotechnol. 2018, 20, 220–245. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA -Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2016, 8, 1364. [Google Scholar] [CrossRef]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Lu, Y.; Wang, F.; Liu, L.; Liu, J.; Liu, X. Comparative transcriptome analysis of zebrafish (Danio rerio) brain and spleen infected with spring viremia of carp virus (SVCV). Fish. Shellfish. Immunol. 2017, 69, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, X.; Sun, Z.; Li, F.; Xiang, J. Transcriptome analysis on Chinese shrimp Fenneropenaeus chinensis during WSSV acute infection. PLoS ONE 2013, 8, e58627. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, J.; Guo, H. Transcriptome analysis and discovery of genes involved in immune pathways in Solen strictus (Gould, 1861) under Vibrio anguillarum. Fish Shellfish Immunol. 2019, 88, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhang, Q.; Wang, Z.; Ma, T.; Zhou, J.; Holland, J.W.; Gao, Q. Transcriptome analysis of the endangered Chinese giant salamander (Andrias davidianus): Immune modulation in response to Aeromonas hydrophila infection. Vet. Immunol. Immunopathol. 2016, 169, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.F.; Yu, H.; Li, Q. Examination of the role of CgSox-like in sex determination and gonadal development in the Pacific oyster Crassostrea gigas. Aquaculture 2023, 566, 739234. [Google Scholar] [CrossRef]
- Lian, C.P. Gonadal Development and Reproductive Cycle, Transcriptomic Studies and Analysis of the Effects of Morphological Traits on Live Body Weight in Tapes Conspersus; Guangxi University: Guangxi, China, 2023. [Google Scholar]
- Liao, X.T. Study on the Gonadal Development and Expression of Sex Related Genes Dmrtl and Foxl2 in Clam Cyclina sinensis; Jiangsu Ocean University: Jiangsu, China, 2022. [Google Scholar]
- Sun, D.F.; Yu, H.; Li, Q. Early gonadal differentiation is associated with the antagonistic action of Foxl2 and Dmrt1l in the Pacific oyster. Comp. Biochem. Phys. B 2023, 265, 110831. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kong, H.; Shang, Y.; Dupont, S.; Peng, J.; Wang, X.; Deng, Y.; Hu, M.; Wang, Y. Ocean acidification but not hypoxia alters the gonad performance in the thick shell mussel Mytilus coruscus. Mar. Pollut Bull 2021, 167, 112282. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Regev, A. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Abdi, H. The Bonferonni and Šidák corrections for multiple comparisons. Encycl. Meas. Stat. 2007, 1, 2007. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Wang, J.; Zhou, J.; Yang, Q.; Wang, W.; Liu, Q.; Liu, W.; Liu, S. Effects of 17 α-methyltestosterone on the transcriptome, gonadal histology and sex steroid hormones in Pseudorasbora parva. Theriogenology 2020, 155, 88–97. [Google Scholar] [CrossRef]
- Reardon, J.T.; Sancar, A. Nucleotide excision repair. Prog. Nucleic Acid. Res. Mol. Biol. 2005, 79, 183–235. [Google Scholar]
- Sun, D.F.; Yu, H.; Li, Q. Genome-wide differential DNA methylomes provide insights into the infertility of Triploid Oysters. Mar. Biotechnol. 2022, 24, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Orian, A.; Schwartz, A.L. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays 2000, 22, 442–451. [Google Scholar] [CrossRef]
- Lee, H.J.; Jang, M.; Kim, H.; Kwak, W.; Park, W.; Hwang, J.Y.; Lee, C.K.; Jang, G.W.; Park, M.N.; Kim, H.C.; et al. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLoS ONE 2013, 8, e66267. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Chen, G.; Tu, H.; Yao, X.; Peng, X.; Lan, X.; Tang, Q.; Yi, S.; Xia, Z.; Cai, M.; et al. Transcriptomic analysis and functional gene expression in different stages of gonadal development of Macrobrachium rosenbergii. Fishes 2023, 8, 94. [Google Scholar] [CrossRef]
- Huang, W.; Xu, F.; Qu, T.; Zhang, R.; Li, L.; Que, H.; Zhang, G. Identification of thyroid hormones and functional characterization of thyroid hormone receptor in the Pacific oyster Crassostrea gigas provide insight into evolution of the thyroid hormone system. PLoS ONE 2015, 10, e0144991. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, I.; Pech-Pool, S.M.; Olvera, A.; García-Martínez, I.; Palacios-Pérez, S.; Orozco, A. The importance of thyroid hormone signaling during early development: Lessons from the zebrafish model. Gen. Comp. Endocr. 2023, 334, 114225. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Li, Q.; Yu, H. Gonad transcriptome analysis of the Pacific oyster Crassostrea gigas identifies potential genes regulating the sex determination and differentiation process. Mar. Biotechnol. 2018, 20, 206–219. [Google Scholar] [CrossRef]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Cao, M.; Wang, X.; Wang, G.; Li, J. Identification of wnt2 in the pearl mussel Hyriopsis cumingii and its role in innate immunity and gonadal development. Fish. Shellfish. Immunol. 2021, 118, 85–93. [Google Scholar] [CrossRef]
- Sreenivasan, R.; Jiang, J.; Wang, X.; Bártfai, R.; Kwan, H.Y.; Christoffels, A.; Orbán, L. Gonad differentiation in zebrafish is regulated by the canonical Wnt signaling pathway. Biol. Reprod. 2014, 90, 45. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Duan, S.H.; Wang, G.L.; Li, J.L. Integrated mRNA and miRNA expression profile analysis of female and male gonads in Hyriopsis cumingii. Sci. Rep. 2021, 11, 665. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; He, P.; Zhang, X.; Li, W.; Zhang, L.; Guan, J.; Peng, J. Identification and characterization of microRNAs in the gonads of Crassostrea hongkongensis using high-throughput sequencing. Comp. Biochem. 2019, 31, 100606. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, J. Nicotinic acetylcholine receptors in health and disease. Mol. Neurobiol. 1997, 15, 193–222. [Google Scholar] [CrossRef] [PubMed]
- Corsi, I.; Pastore, A.M.; Lodde, A.; Palmerini, E.; Castagnolo, L.; Focardi, S. Potential role of cholinesterases in the invasive capacity of the freshwater bivalve, Anodonta woodiana (Bivalvia: Unionacea): A comparative study with the indigenous species of the genus, Anodonta sp. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 145, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Bleisch, W.V.; Luine, V.N. Modulation of acetylcholinesterase activity and acetylcholine receptor number in an androgen sensitive neuromuscular system. Soc. Neurosci. Abstr. 1981, 7, 805. [Google Scholar]
- Luine, V.N.; Nottebohm, F.; Harding, C.; McEwen, B.S. Androgen affects cholinergic enzymes in syringeal motor neurons and muscle. Brain Res. 1980, 192, 89–107. [Google Scholar] [CrossRef]
- Bleisch, W.V.; Harrelson, A.L.; Luine, V.N. Testosterone increases acetylcholine receptor number in the “levator ani” muscle of the rat. J. Neurobiol. 1982, 13, 153–161. [Google Scholar] [CrossRef]
- Gutmann, E.; Tucek, S.; Hanzlíková, V. Changes in the choline acetyltransferase and cholinesterase activities in the levator ani muscle of rats following castration. Physiol. Bohemoslov. 1969, 18, 195–203. [Google Scholar]
- Cooper, S.J.; Dourish, C.T. Multiple cholecystokinin (CCK) receptors and CCK-monoamine interactions are instrumental in the control of feeding. Physiol. Behav. 1990, 48, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Moran, T.H. Cholecystokinin and satiety: Current perspectives. Nutrition 2000, 16, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.N.; Corwin, R.L. Biological actions of cholecystokinin. Peptides 1994, 15, 731–755. [Google Scholar] [CrossRef] [PubMed]
- Hökfelt, T.; Skirboll, L.; Rehfeld, J.F.; Goldstein, M.; Markey, K.; Dann, O. A subpopulation of mesencephalic dopamine neurons projecting to limbic areas contains a cholecystokinin-like peptide: Evidence from immunohistochemistry combined with retrograde tracing. Neuroscience 1980, 5, 2093–2124. [Google Scholar] [CrossRef] [PubMed]
- Gulyás, A.I.; Görcs, T.J.; Freund, T.F. Innervation of different peptide-containing neurons in the hippocampus by GABAergic septal afferents. Neuroscience 1990, 37, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Mezey, E.; Reisine, T.D.; Skirboll, L.; Beinfeld, M.; Kiss, J.Z. Cholecystokinin in the medial parvocellular subdivision of the paraventricular nucleus. Co-existence with corticotropin-releasing hormone. Ann. N. Y. Acad. Sci. 1985, 448, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.; Dubos, M.P.; Pasquier, J.; Zatylny, C.; Favrel, P. Emergence of a cholecystokinin/sulfakinin signalling system in Lophotrochozoa. Sci. Rep. 2018, 8, 16424. [Google Scholar] [CrossRef] [PubMed]
- Silveyra, P.; Cataldi, N.I.; Lux-Lantos, V.; Libertun, C. Gonadal steroids modulated hypocretin/orexin type-1 receptor expression in a brain region, sex and daytime specific manner. Regul. Pept. 2009, 158, 121–126. [Google Scholar] [CrossRef]
- Campbell, R.E.; Grove, K.L.; Smith, M.S. Gonadotropin-releasing hormone neurons coexpress orexin 1 receptor immunoreactivity and receive direct contacts by orexin fibers. Endocrinology 2003, 144, 1542–1548. [Google Scholar] [CrossRef]
- Furuta, M.; Funabashi, T.; Kimura, F. Suppressive action of orexin A on pulsatile luteinizing hormone secretion is potentiated by a low dose of estrogen in ovariectomized rats. Neuroendocrinology 2002, 75, 151–157. [Google Scholar] [CrossRef]
- Irahara, M.; Tamura, T.; Matuzaki, T.; Saito, S.; Yasui, T.; Yamano, S.; Kamada, M.; Aono, T. Orexin-A suppresses the pulsatile secretion of luteinizing hormone via beta-endorphin. Biochem. Biophys. Res. Commun. 2001, 281, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Watanobe, H.; Kakizaki, Y.; Suda, T.; Schioth, H.B. A significant participation of orexin-A, a potent orexigenic peptide, in the preovulatory luteinizing hormone and prolactin surges in the rat. Brain Res. 2001, 898, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.; Jain, M.R.; Kalra, P.S.; Kalra, S.P. Orexins, a novel family of hypothalamic neuropeptides, modulate pituitary luteinizing hormone secretion in an ovarian steroid-dependent manner. Regul. Pept. 1998, 78, 133–136. [Google Scholar] [PubMed]
- Russell, S.H.; Small, C.J.; Kennedy, A.R.; Stanley, S.A.; Seth, A.; Murphy, K.G.; Taheri, S.; Ghatei, M.A.; Bloom, S.R. Orexin A interactions in the hypothalamo-pituitary gonadal axis. Endocrinology 2001, 142, 5294–5302. [Google Scholar] [CrossRef]
- Silveyra, P.; Catalano, P.N.; Lux-Lantos, V.; Libertun, C. Impact of proestrous milieu on expression of orexin receptors and prepro-orexin in rat hypothalamus and hypophysis: Actions of Cetrorelix and Nembutal. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E820–E828. [Google Scholar] [CrossRef] [PubMed]
- Small, C.J.; Goubillon, M.L.; Murray, J.F.; Siddiqui, A.; Grimshaw, S.E.; Young, H.; Sivanesan, V.; Kalamatianos, T.; Kennedy, A.R.; Coen, C.W.; et al. Central orexin A has site-specific effects on luteinizing hormone release in female rats. Endocrinology 2003, 144, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Irahara, M.; Tezuka, M.; Kiyokawa, M.; Aono, T. Orexins, orexigenic hypothalamic neuropeptides, suppress the pulsatile secretion of luteinizing hormone in ovariectomized female rats. Biochem. Biophys. Res. Commun. 1999, 264, 759–762. [Google Scholar] [CrossRef]
- Kok, S.W.; Roelfsema, F.; Overeem, S.; Lammers, G.J.; Frolich, M.; Meinders, A.E.; Pijl, H. Pulsatile LH release is diminished, whereas FSH secretion is normal, in hypocretin deficient narcoleptic men. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E630–E636. [Google Scholar] [CrossRef]
- Yin, D.; Lin, D.; Ying, C.; Ma, F.; Yang, Y.; Wang, Y.; Tan, J.; Liu, K. Metabolic mechanisms of Coilia nasus in the natural food intake state during migration. Genomics 2020, 112, 3294–3305. [Google Scholar] [CrossRef]
- Zohar, Y.; Muñoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef]
- Ritter, M.J.; Amano, I.; Imai, N.; Soares, D.; Oliveira, L.; Vella, K.R.; Hollenberg, A.N. Nuclear receptor corepressors, NCOR1 and SMRT, are required for maintaining systemic metabolic homeostasis. Mol. Metab. 2021, 53, 101315. [Google Scholar] [CrossRef] [PubMed]
- Stanya, K.J.; Kao, H.Y. New insights into the functions and regulation of the transcriptional corepressors SMRT and N-CoR. Cell Div. 2009, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Perissi, V.; Jepsen, K.; Glass, C.K.; Rosenfeld, M.G. Deconstructing repression: Evolving models of co-repressor action. Nat. Rev. Genet. 2010, 11, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Oberoi, J.; Fairall, L.; Watson, P.J.; Yang, J.C.; Czimmerer, Z.; Kampmann, T.; Goult, B.T.; Greenwood, J.A.; Gooch, J.T.; Kallenberger, B.C.; et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat. Struct. Mol. Biol. 2011, 18, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.J.; Fairall, L.; Schwabe, J.W. Nuclear hormone receptor co-repressors: Structure and function. Mol. Cell Endocrinol. 2012, 348, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.M.; Watson, P.J.; Fairall, L.; Jamieson, A.G.; Schwabe, J.W.R. Insights into the recruitment of class IIa histone deacetylases (HDACs) to the SMRT/NCoR transcriptional repression complex. J. Biol. Chem. 2015, 290, 18237–18244. [Google Scholar] [CrossRef] [PubMed]
- Desravines, D.C.; Serna, M.I.; Schneider, R.; Mas, P.J.; Aleksandrova, N.; Jensen, M.R.; Blackledge, M.; Hart, D.J. Structural characterization of the SMRT corepressor interacting with histone deacetylase 7. Sci. Rep. 2017, 7, 3678. [Google Scholar] [CrossRef]
- Hörlein, A.J.; Näär, A.M.; Heinzel, T.; Torchia, J.; Gloss, B.; Kurokawa, R.; Ryan, A.; Kamei, Y.; Söderström, M.; Glass, C.K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 1995, 377, 397–404. [Google Scholar] [CrossRef]
- Chen, J.D.; Evans, R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 1995, 377, 454–457. [Google Scholar] [CrossRef]
- Jepsen, K.; Hermanson, O.; Onami, T.M.; Gleiberman, A.S.; Lunyak, V. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 2000, 102, 753–763. [Google Scholar] [CrossRef]
- Mikoláš, P.; Kollárová, J.; Sebková, K.; Saudek, V.; Yilma, P.; Kostrouchová, M.; Krause, M.W.; Kostrouch, Z.; Kostrouchová, M. GEI-8, a homologue of vertebrate nuclear receptor corepressor NCoR/SMRT, regulates gonad development and neuronal functions in Caenorhabditis elegans. PLoS ONE 2013, 8, e58462. [Google Scholar] [CrossRef]

| Gene | Primer Sequence (5′-3′) |
|---|---|
| AChRs | F: TGAGGTTTGGTCAGAGCTGAAATGG |
| R: CCTTGCCTCCCGAACAACACTC | |
| CCKAR | F: TTATCACGAACGCCTTGGTACTGTC |
| R: CGTCGATGCTTCAGGCTCAGTG | |
| HCRTR2 | F: GCAGATAGGGCGTGAGATTAACCG |
| R: TTTCCGTCTTCGCTTTACTGTACCG | |
| GABAB | F: AGTTGAGAAAGGACCCGGTCTATGG |
| R: TTGTCTCCGTCTATGGCCCAGTC | |
| NCoR2 | F: ACCCGATTTTGATGCCGAGAATGG |
| R: AGCCTTGCCTGTCTCAACGAATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Ye, Y.; Yao, R.; Qi, P.; Li, J. Transcriptome Sequencing Analysis of Sex-Related Genes in the Gonads of Mytilus unguiculatus. Fishes 2023, 8, 456. https://doi.org/10.3390/fishes8090456
Ma Y, Ye Y, Yao R, Qi P, Li J. Transcriptome Sequencing Analysis of Sex-Related Genes in the Gonads of Mytilus unguiculatus. Fishes. 2023; 8(9):456. https://doi.org/10.3390/fishes8090456
Chicago/Turabian StyleMa, Yanwen, Yingying Ye, Ronghui Yao, Pengzhi Qi, and Jiji Li. 2023. "Transcriptome Sequencing Analysis of Sex-Related Genes in the Gonads of Mytilus unguiculatus" Fishes 8, no. 9: 456. https://doi.org/10.3390/fishes8090456
APA StyleMa, Y., Ye, Y., Yao, R., Qi, P., & Li, J. (2023). Transcriptome Sequencing Analysis of Sex-Related Genes in the Gonads of Mytilus unguiculatus. Fishes, 8(9), 456. https://doi.org/10.3390/fishes8090456








