Aminotransferase Class I and II Gene Family in the Jinjiang Oyster (Crassostrea ariakensis): Genomewide Identification, Phylogenetic Analysis and Expression Profiles after Salinity Stress
Abstract
:1. Introduction
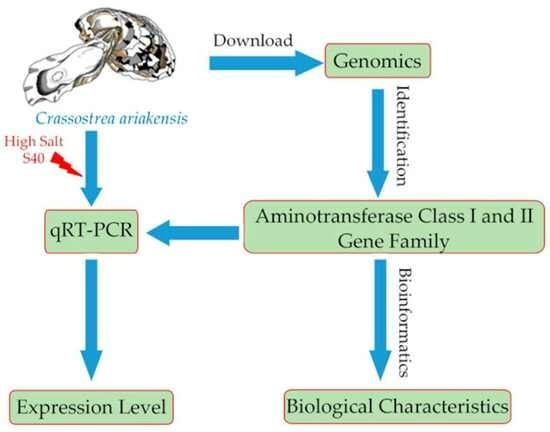
2. Materials and Methods
2.1. Identification of Aminotransferase Class I and II Gene Sequences
2.2. Bioinformatics Analysis of Aminotransferase Class I and II Gene Sequences in C. ariakensis
2.3. Multiple Comparisons and Structural Analysis of Aminotransferase Class I and II Gene Sequences
2.4. Salinity Challenge and Sample Preparation
2.5. Expression Analysis of Aminotransferase Class I and II under High-Salt Stress
3. Results
3.1. Gene Identification and Protein Characteristic Analysis
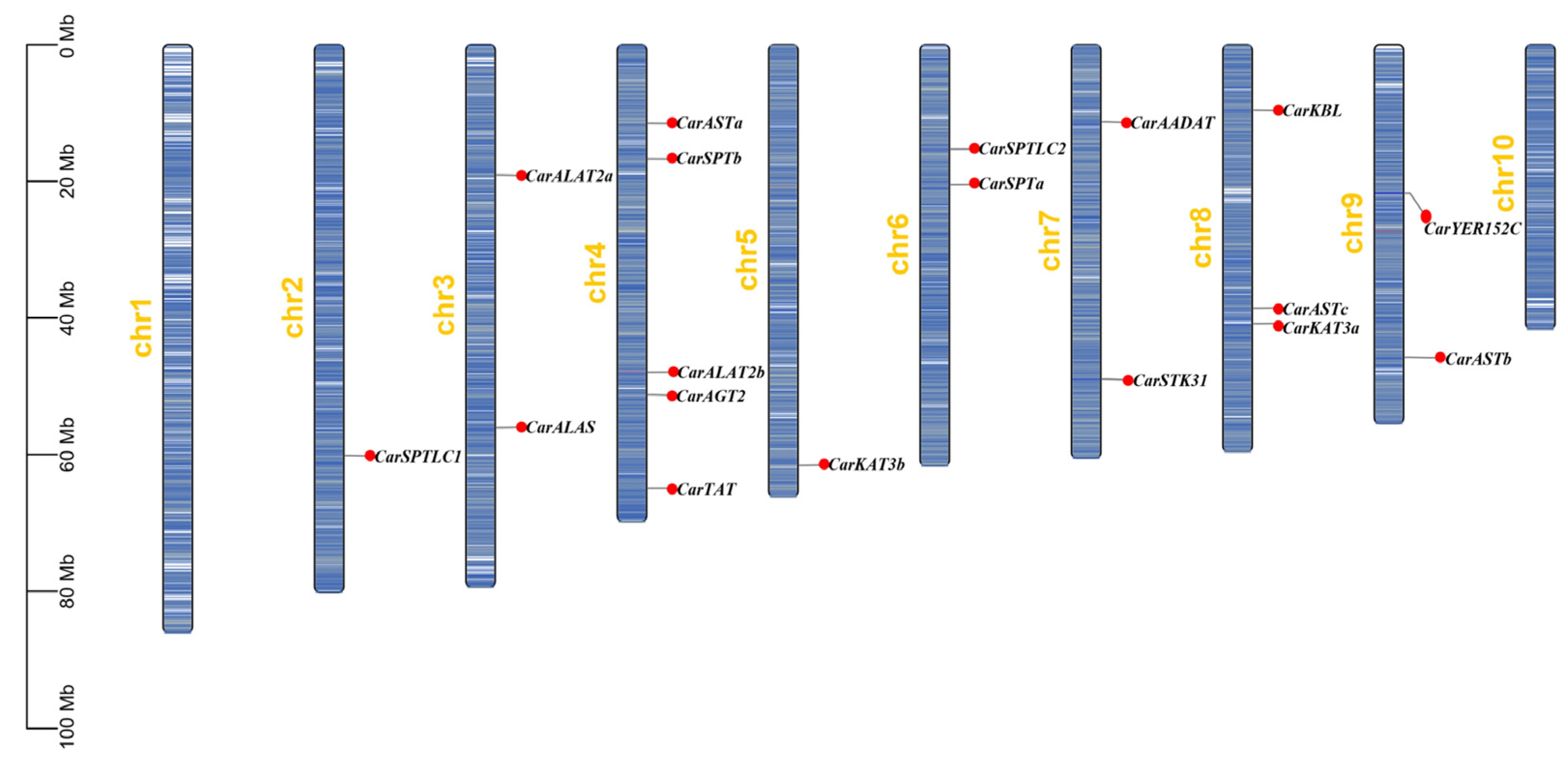
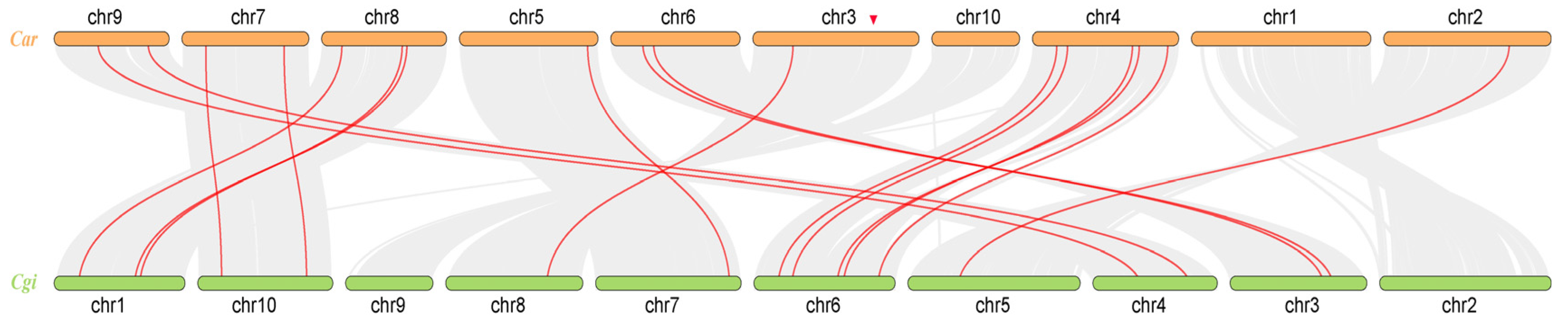
3.2. Chromosome Localization Analysis and Gene Covariance
3.3. Gene Structural Characterization
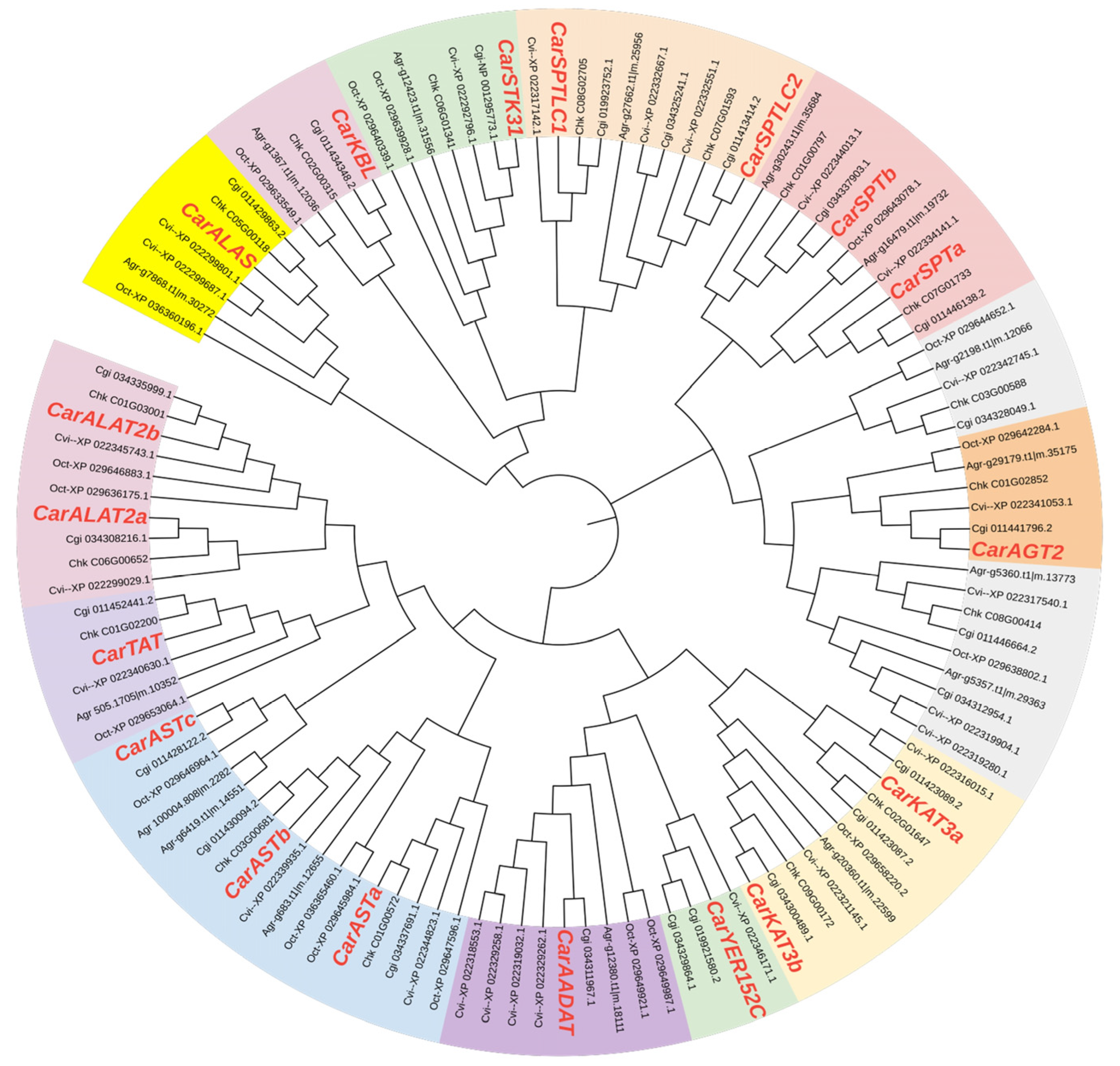
3.4. Evolutionary Analysis of Aminotransferase Class I and II Gene Family Members
3.5. Analysis of Gene Expression Patterns under Acute High-Salt Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Z.; Wang, T. Responses of estuarine circulation and salinity to the loss of intertidal flats—A modeling study. Cont. Shelf Res. Companion J. Deep-Sea Res. Prog. Oceanogr. 2015, 111 Pt B, 159–173. [Google Scholar] [CrossRef]
- Munari, M.; Matozzo, V.; Marin, M.G. Combined effects of temperature and salinity on functional responses of haemocytes and survival in air of the clam Ruditapes philippinarum. Fish Shellfish. Immunol. 2011, 30, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Carregosa, V.; Figueira, E.; Gil, A.M.; Pereira, S.; Pinto, J.; Soares, A.M.; Freitas, R. Tolerance of Venerupis philippinarum to salinity: Osmotic and metabolic aspects. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 171, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, B.; Niu, D.; Li, J. Changes in metabolism and immunity in response to acute salinity stress in chinese razor clams from different regions. Aquac. Rep. 2021, 19, 100624. [Google Scholar] [CrossRef]
- Cao, W.; Bi, S.; Chi, C. Effects of High Salinity Stress on the Survival, Gill Tissue, Enzyme Activity and Free Amino Acid Content in Razor Clam Sinonovacula constricta. Front. Mar. Sci. 2022, 9, 839614. [Google Scholar] [CrossRef]
- She, Z.C.; Jia, Z.; Peng, Y.H. Effects of salinity stress on some biochemical indexes of Hong Kong oyster. Ocean Sci. 2019, 43, 40–45. [Google Scholar]
- Li, N.; Wang, R.J.; Zhao, Y.C.; Shen, M.; Su, W.; Zhao, C.; Li, Y.Q. Effects of high salinity on growth index, plasma osmotic pressure and Na+-K+-ATPase activities of Litopenaeus vannamei. J. Zhejiang Ocean Univ. (Nat. Sci.) 2017, 36, 196–201. [Google Scholar] [CrossRef]
- Zhang, L.; Zang, W.; Deng, P.; Zou, W.; Ding, F. Effect of Ca2+, Mg2+ and salinity on survival, growth and shrimp taste of Litopenaeus vannamei and flavor. J. Fish. 2012, 36, 914–921. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, Q.; Du, M.; Zhang, Z.; Wang, W.; Zhang, G.; Li, A.; Li, L. Genome-wide mapping of regulatory variants for temperature- and salinity-adaptive genes reveals genetic basis of genotype-by-environment interaction in Crassostrea ariakensis. Environ. Res. 2013, 236 Pt 1, 116614. [Google Scholar] [CrossRef]
- Pourmozaffar, S.; Tamadoni Jahromi, S.; Rameshi, H.; Sadeghi, A.; Bagheri, T.; Behzadi, S.; Gozari, M.; Zahedi, M.R.; Abrari Lazarjani, S. The role of salinity in physiological responses of bivalves. Rev. Aquac. 2020, 12, 1548–1566. [Google Scholar] [CrossRef]
- Hosoi, M.; Kubota, S.; Toyohara, M.; Toyohara, H.; Hayashi, I. Effect of salinity change on free amino acid content in pacific oyster. Fish. Sci. 2003, 69, 395–400. [Google Scholar] [CrossRef]
- Laverty, G.; Skadhauge, E. Adaptation of teleosts to very high salinity. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2012, 163, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.Y.; Lee, H.J.; Yang, Y.H.; Joo, H.S.; Kim, B.G. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli k12. Iran. J. Microbiol. 2004, 11, 915–925. [Google Scholar] [CrossRef] [PubMed]
- McCampbell, A.; Truong, D.; Broom, D.C.; Allchorne, A.; Gable, K.; Cutler, R.G.; Mattson, M.P.; Woolf, C.J.; Frosch, M.P.; Harmon, J.M.; et al. Mutant sptlc1 dominantly inhibits serine palmitoyltransferase activity in vivo and confers an age-dependent neuropathy. Hum. Mol. Genet. 2005, 14, 3507–3521. [Google Scholar] [CrossRef]
- Samsonova, M.V.; Min’kova, N.O.; Lapteva, T.I.; Mikodina, E.V.; Filippovich, Y.B. Aspartate and alanine aminotransferases in early development of the keta. Russ. J. Dev. Biol. 2003, 34, 14–18. [Google Scholar] [CrossRef]
- Huang, Q.; Lin, H.; Wang, R.; Huang, Z.; Wang, J. Effect of dietary vitamin B6 supplementation on growth and intestinal microflora of juvenile golden pompano (Trachinotus ovatus). Aquac. Res. Acta Sci. Nat. Univ. Sunyatseni 2019, 50, 2359–2370. [Google Scholar] [CrossRef]
- Zhang, S.M.; Li, J.F.; Wen, H.S.; Lv, L.K.; Li, L.M.; Zhao, J. The impact and physiological mechanism of acute temperature coercion on the metabolic function and blood indicators of Xu’s liver. J. Ocean Univ. China Nat. Sci. Ed. 2018, 48, 32–38. [Google Scholar] [CrossRef]
- Goh, D.L.M.; Patel, A.; Thomas, G.H.; Salomons, G.S.; Schor, D.S.; Jakobs, C.; Geraghty, M.T. Characterization of the human gene encoding α-aminoadipate aminotransferase (AADAT). Mol. Genet. Metab. 2002, 76, 172–180. [Google Scholar] [CrossRef]
- Wu, B.; Chen, X.; Yu, M.; Ren, J.; Hu, J.; Shao, C.; Zhou, L.; Sun, X.; Yu, T.; Zheng, Y.; et al. Chromosome-level genome and population genomic analysis provide insights into the evolution and environmental adaptation of Jinjiang oyster Crassostrea ariakensis. Mol. Ecol. Resour. 2021, 22, 1529–1544. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Ajamieh, H.H.; Berlanga, J.; Merino, N.; Sanchez, G.M.; Carmona, A.M.; Cepero, S.M.; Giuliani, A.; Re, L.; Leon, O.S. Role of protein synthesis in the protection conferred by ozone-oxidative-preconditioning in hepatic ischaemia/reperfusion. Transpl. Int. 2010, 18, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Pogorelova, T.N.; Gunko, V.O.; Mikhelson, A.A.; Nikashina, A.A.; Mikhelson, A.F.; Lebedenko, E.Y. The activity of enzymes of amino acid metabolism of the placenta in different terms of the physiological and complicated pregnancy. Klin. Lab. Diagn. 2019, 64, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Kitade, H.; Satoi, S.; Zhang, Z.T.; Kaibori, M.; Kwon, A.H.; Kamiyama, Y.; Okumura, T. Increased nitric oxide production in hepatocytes is involved in liver dysfunction following obstructive jaundice. J. Surg. Res. 2002, 106, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Wejksza, K.; Rzeski, W.; Okuno, E.; Kandefer-Szerszen, M.; Albrecht, J.; Turski, W.A. Demonstration of kynurenine aminotransferases I and II and characterization of kynurenic acid synthesis in oligodendrocyte cell line (OLN-93). Neurochem. Res. 2005, 30, 963–968. [Google Scholar] [CrossRef]
- Wang, Y.P.; Sharda, A.; Xu, S.N.; Van Gastel, N.; Man, C.H.; Choi, U.; Leong, W.Z.; Li, X.; Scadden, D.T. Malic enzyme 2 connects the Krebs cycle intermediate fumarate to mitochondrial biogenesis. Cell Metab. 2021, 33, 1027–1041.e8. [Google Scholar] [CrossRef]
- Daniel, R.M.; Dines, M.; Petach, H.H. The denaturation and degradation of stable enzymes at high temperatures. Biochem. J. 1996, 317, 1–11. [Google Scholar] [CrossRef]
- Yao, Y.J.; Khoo, K.S.; Chung, M.C.; Li, S.F. Determination of isoelectric points of acidic and basic proteins by capillary electrophoresis. J. Chromatogr. A 1994, 680, 431–435. [Google Scholar] [CrossRef]
- Hoefgen, S.; Bissell, A.U.; Huang, Y.; Gherlone, F.; Raguž, L.; Beemelmanns, C.; Valiante, V. Desaturation of the Sphingofungin Polyketide Tail Results in Increased Serine Palmitoyltransferase Inhibition. Microbiol. Spectr. 2022, 10, e01331-22. [Google Scholar] [CrossRef]
- Ortega, A.; Becker, V.M.; Alvarez, R.; Lepock, J.R.; Gonzalez-Serratos, H. Interaction of D-600 with the transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Am. J. Physiol. Cell Physiol. 2000, 279, C166–C172. [Google Scholar] [CrossRef]
- Smith, E.C.; Smith, S.E.; Carter, J.R.; Webb, S.R.; Gibson, K.M.; Hellman, L.M.; Fried, M.G.; Dutch, R.E. Trimeric transmembrane domain interactions in paramyxovirus fusion proteins: Roles in protein folding, stability, and function. J. Biol. Chem. 2013, 288, 35726–35735. [Google Scholar] [CrossRef]
- Ceppi, P.; Colombo, S.; Francolini, M.; Raimondo, F.; Borgese, N.; Masserini, M. Two tail-anchored protein variants, differing in transmembrane domain length and intracellular sorting, interact differently with lipids. Proc. Natl. Acad. Sci. USA 2005, 102, 16269–16274. [Google Scholar] [CrossRef]
- Li, A.; Dai, H.; Guo, X.; Zhang, Z.; Zhang, K.; Wang, C.; Wang, X.; Wang, W.; Chen, H.; Li, X.; et al. Genome of the estuarine oyster provides insights into climate impact and adaptive plasticity. Commun. Biol. 2021, 4, 1287. [Google Scholar] [CrossRef]
- Liu, X.; Hu, B.; Chu, C. Nitrogen assimilation in plants: Current status and future prospects. J. Genet. Genom. 2021, 49, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Chong-Robles, J.; Charmantier, G.; Boulo, V.; Lizárraga-Valdéz, J.; Enríquez-Paredes, L.M.; Giffard-Mena, I. Osmoregulation pattern and salinity tolerance of the white shrimp litopenaeus vannamei (boone, 1931) during post-embryonic development. Aquaculture 2014, 422–423, 261–267. [Google Scholar] [CrossRef]
- Wang, H.; Teng, M.X.; Liu, P.P.; Zhao, M.Y. Selection Signatures of Pacific White Shrimp Litopenaeus vannamei Revealed by Whole-Genome Resequencing Analysis. Front. Mar. Sci. 2022, 9, 844597. [Google Scholar] [CrossRef]
- Yao, L.J.; Yang, P.; Luo, W.Q.; Li, S.M. Macrophage-stimulating activity of European eel (Anguilla anguilla) peptides in RAW264.7 cells mediated via NF-κB and MAPK signaling pathways. Food Funct. 2020, 11, 10968–10978. [Google Scholar] [CrossRef]
- Zhang, X.L.; Lu, Y.S.; Jian, J.C.; Wu, Z.H. Cloning and expression analysis of recombination activating genes (rag1/2) in red snapper (lutjanus sanguineus). Fish Shellfish. Immunol. 2012, 32, 534–543. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Abbreviation | Sequence (5′–3′) | Product Size (bp) | Efficiency (%) |
|---|---|---|---|---|
| Serine palmitoyltransferase 1 | CarSPTLC1 | GTGCTATCCCAGCTTACTCGAAACG | 172 | 95.99 |
| GCTCCAGATCCTCCATGTCATTGTG | ||||
| Serine palmitoyltransferase 2 | CarSPTLC2 | TGGTGCTGTTGGAGGTTACATTGG | 140 | 95.09 |
| GGAGTAGATAGCCGAGTGGGAGTAC | ||||
| Serine—pyruvate aminotransferase | CarSPTa | GAGGTGCTGATGGATGAC | 245 | 96.38 |
| GCTATGGCGTAGGAGAAC | ||||
| Serine—pyruvate aminotransferase | CarSPTb | AACAAGGTGCTGAATCGTA | 335 | 97.99 |
| GCGTAATCGGACACATCT | ||||
| 5-aminolevulinate synthase | CarALAS | GGCTTCATCTTCACCACCAGTCTC | 244 | 97.21 |
| CTATCAGGTTCTGTCGCAGGTAACG | ||||
| Alanine aminotransferase 2 | CarALAT2a | CTCCCGATGCTTTCTACTGCTTCC | 213 | 96.23 |
| CCTCCTTCTCTCCGAACCCACTC | ||||
| Alanine aminotransferase 2 | CarALAT2b | TGCCTTCCCTCAAATCAAGATGCC | 397 | 98.11 |
| GTTCCTGGTTGCTGTCCGAATCC | ||||
| Aspartate aminotransferase | CarASTa | TGGTTGGGAGAAGTGAAAGGAATGG | 165 | 97.20 |
| GGTCAATGATGTGTTGCCAGTTACG | ||||
| Aspartate aminotransferase | CarASTb | GTCATAACCCAACTGGAGTGAACCC | 185 | 95.01 |
| GTTACCAGAAGCGAAGCCCTGATAG | ||||
| Aspartate aminotransferase, cytoplasmic | CarASTc | GCTCTAGGATTCTGGATGCTGTTGG | 121 | 98.86 |
| TGTCTGGATTCCTCGCCTCTTGAG | ||||
| Kynurenine—oxoglutarate transaminase 3 | CarKAT3a | TTCGCCCCACCAAAGCACATTC | 89 | 99.24 |
| GCTGCTGAAGTTCTCGGCTGTAG | ||||
| Kynurenine—oxoglutarate transaminase 3 | CarKAT3b | GCTCCAGATGATGGCAGACTTGTG | 118 | 97.35 |
| AACATCCCAGGCAGAGAGGCTATC | ||||
| Kynurenine/alpha-aminoadipate aminotransferase | CarAADAT | TCAAAGCCGTATGTTCCCAGTCTTC | 312 | 97.68 |
| CAGTCACAAACCCCACCCTCATTC | ||||
| Tyrosine aminotransferase | CarTAT | CAACGGCTACAACCCATCCATAGG | 164 | 95.29 |
| CAGCCACCAGTAAGAACCACATCC | ||||
| Alanine—glyoxylate aminotransferase 2 | CarAGT2 | AATGACTTGGCGGTTCTG | 381 | 97.82 |
| CTGTGTTGTTCCTCCTACTC | ||||
| Serine/threonine-protein kinase 31 | CarSTK31 | GCGAGCCGTCAGGAATTAGGTTATC | 169 | 98.00 |
| GCTGGCAGGTTCATAGAGTTGGTAG | ||||
| 2-amino-3-ketobutyrate coenzyme A ligase | CarKBL | CCAGTTGTTGCTTGTGCCAGTAAAG | 135 | 98.90 |
| GCGGAATCGTTTGGTGTTGTTGAG | ||||
| Uncharacterized protein | CarYER152C | TGTGATCTGTTGTTTGGTGGACCTG | 402 | 99.68 |
| AGAAGAGCCTGCGTTGCCTTTAAG | ||||
| β-actin | CTGTGCTACGTTGCCCTGGACTT | 122 | 94.77 | |
| TGGGCACCTGAATCGCTCGTT |
| Gene ID | Gene Name | Protein Length (aa) | Molecular Weight (kDa) | pI | Instability Index | Grand Average of Hydropathicity | Localization |
|---|---|---|---|---|---|---|---|
| Ori.tig00000232.98 | CarSPTLC1 | 1982 | 221.36 | 5.91 | 40.44 | −0.429 | NC a, CP b |
| Ori.tig01225668.108 | CarSPTLC2 | 1107 | 123.53 | 8.82 | 34.46 | −0.215 | NC a, CP a, MB b |
| Ori.tig00000276.148 | CarSPTa | 415 | 45.95 | 8.49 | 40.33 | −0.05 | Mit a,b, CP a |
| Ori.tig00000013.86 | CarSPTb | 431 | 47.13 | 8.82 | 35.18 | −0.144 | Mit a,b |
| Ori.tig00014637.33 | CarALAS | 615 | 57.73 | 7.12 | 44.74 | −0.295 | Mit a,b, CP a |
| Ori.tig01225679.19 | CarALAT2a | 529 | 58.74 | 7.02 | 32.77 | −0.208 | CP a,b |
| Ori.tig00057033.282 | CarALAT2b | 825 | 92.59 | 9.03 | 41.08 | −0.35 | Mit a,b, CP a |
| Ori.tig00001151.290 | CarASTa | 428 | 47.04 | 8.95 | 32.22 | −0.25 | Mit a,b, CP a |
| Ori.tig01227070.450 | CarASTb | 407 | 45.71 | 7.13 | 30.46 | −0.237 | CP a,b |
| Ori.tig00000007.252 | CarASTc | 550 | 62.22 | 5.80 | 41.40 | −0.095 | PM a, CP b |
| Ori.tig00000007.312 | CarKAT3a | 468 | 52.51 | 8.61 | 39.30 | −0.31 | Mit a,b |
| Ori.tig00057021.276 | CarKAT3b | 680 | 76.18 | 5.57 | 35.95 | −0.146 | CP a,b |
| Ori.tig00000052.211 | CarAADAT | 459 | 51.1 | 5.87 | 39.9 | −0.218 | NC a, CP a,b |
| Ori.tig00057030.149 | CarTAT | 434 | 48.51 | 7.22 | 44.37 | −0.249 | CP a,b |
| Ori.tig00057033.171 | CarAGT2 | 488 | 53.73 | 8.56 | 29.36 | −0.097 | Mit a,b |
| Ori.tig00000599.319 | CarSTK31 | 478 | 53.52 | 5.49 | 36.65 | −0.093 | CP a, IM b |
| Ori.tig01225420.248 | CarKBL | 425 | 46.34 | 8.68 | 29.02 | −0.167 | Mit a,b |
| Ori.tig00002832.215 | CarYER152C | 461 | 52.18 | 6.68 | 48.08 | −0.268 | NC a, CP a,b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, G.; Liu, Z.; Yu, T.; Zhou, L.; Sun, X.; Li, Z.; Zheng, Y. Aminotransferase Class I and II Gene Family in the Jinjiang Oyster (Crassostrea ariakensis): Genomewide Identification, Phylogenetic Analysis and Expression Profiles after Salinity Stress. Fishes 2023, 8, 459. https://doi.org/10.3390/fishes8090459
Ge G, Liu Z, Yu T, Zhou L, Sun X, Li Z, Zheng Y. Aminotransferase Class I and II Gene Family in the Jinjiang Oyster (Crassostrea ariakensis): Genomewide Identification, Phylogenetic Analysis and Expression Profiles after Salinity Stress. Fishes. 2023; 8(9):459. https://doi.org/10.3390/fishes8090459
Chicago/Turabian StyleGe, Guangyu, Zhihong Liu, Tao Yu, Liqing Zhou, Xiujun Sun, Zhuanzhuan Li, and Yanxin Zheng. 2023. "Aminotransferase Class I and II Gene Family in the Jinjiang Oyster (Crassostrea ariakensis): Genomewide Identification, Phylogenetic Analysis and Expression Profiles after Salinity Stress" Fishes 8, no. 9: 459. https://doi.org/10.3390/fishes8090459
APA StyleGe, G., Liu, Z., Yu, T., Zhou, L., Sun, X., Li, Z., & Zheng, Y. (2023). Aminotransferase Class I and II Gene Family in the Jinjiang Oyster (Crassostrea ariakensis): Genomewide Identification, Phylogenetic Analysis and Expression Profiles after Salinity Stress. Fishes, 8(9), 459. https://doi.org/10.3390/fishes8090459