Parasitic Copepods as Biochemical Tracers of Foraging Patterns and Dietary Shifts in Whale Sharks (Rhincodon typus Smith, 1828)
Abstract
1. Introduction
- If P. rhincodonicus were good biochemical indicators of whale shark foraging habitats and trophic positions, then values of δ13C and δ15N in P. rhincodonicus would reliably predict values of δ13C and δ15N in whale shark tissue;
- P. rhincodonicus feeding on whale shark blood and regenerating tissue would show trophic discrimination factors similar to the theoretical values of +3.4‰ and +≤1‰ for δ13C and δ15N values associated with the consumption of tissues [25].
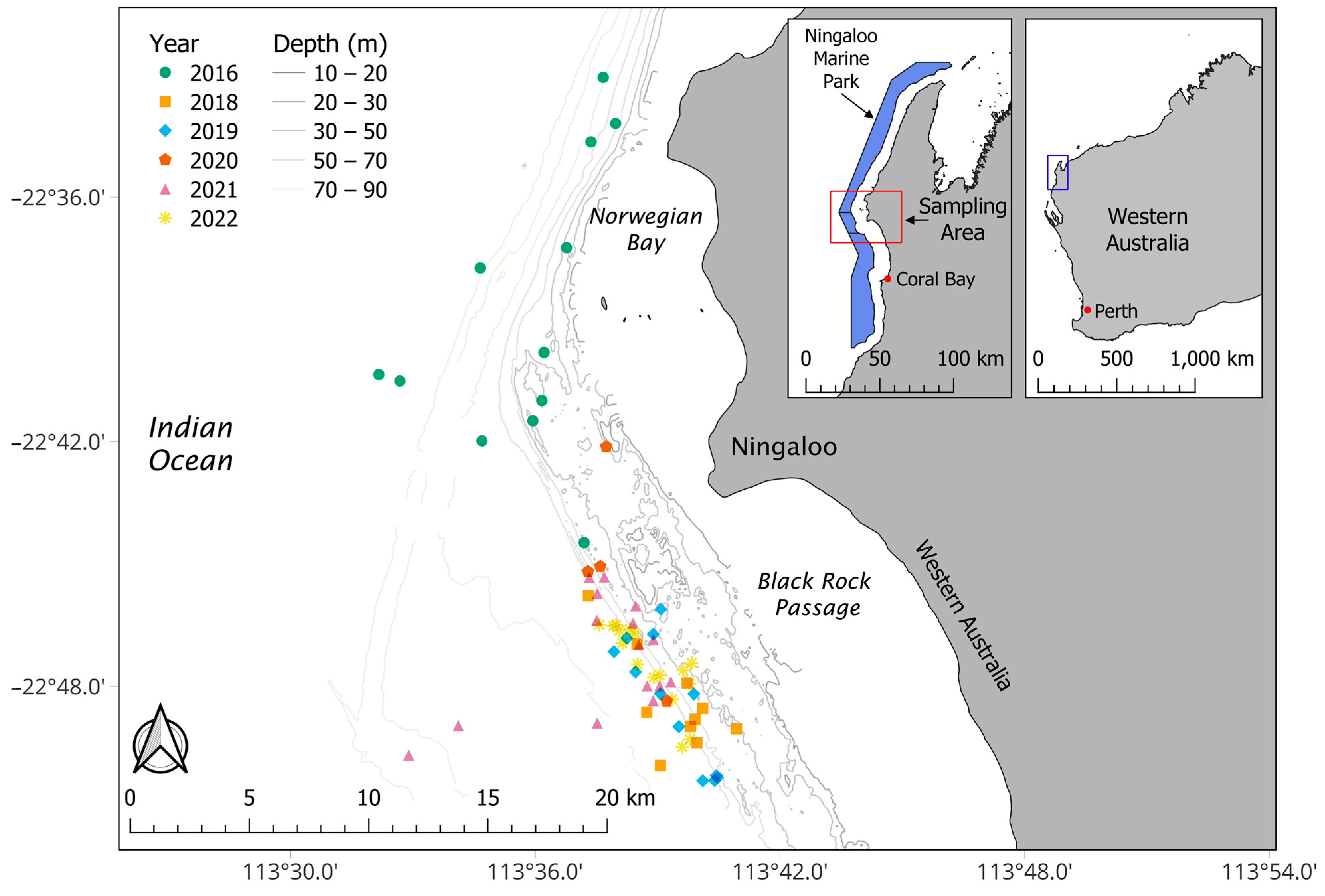
2. Materials and Methods
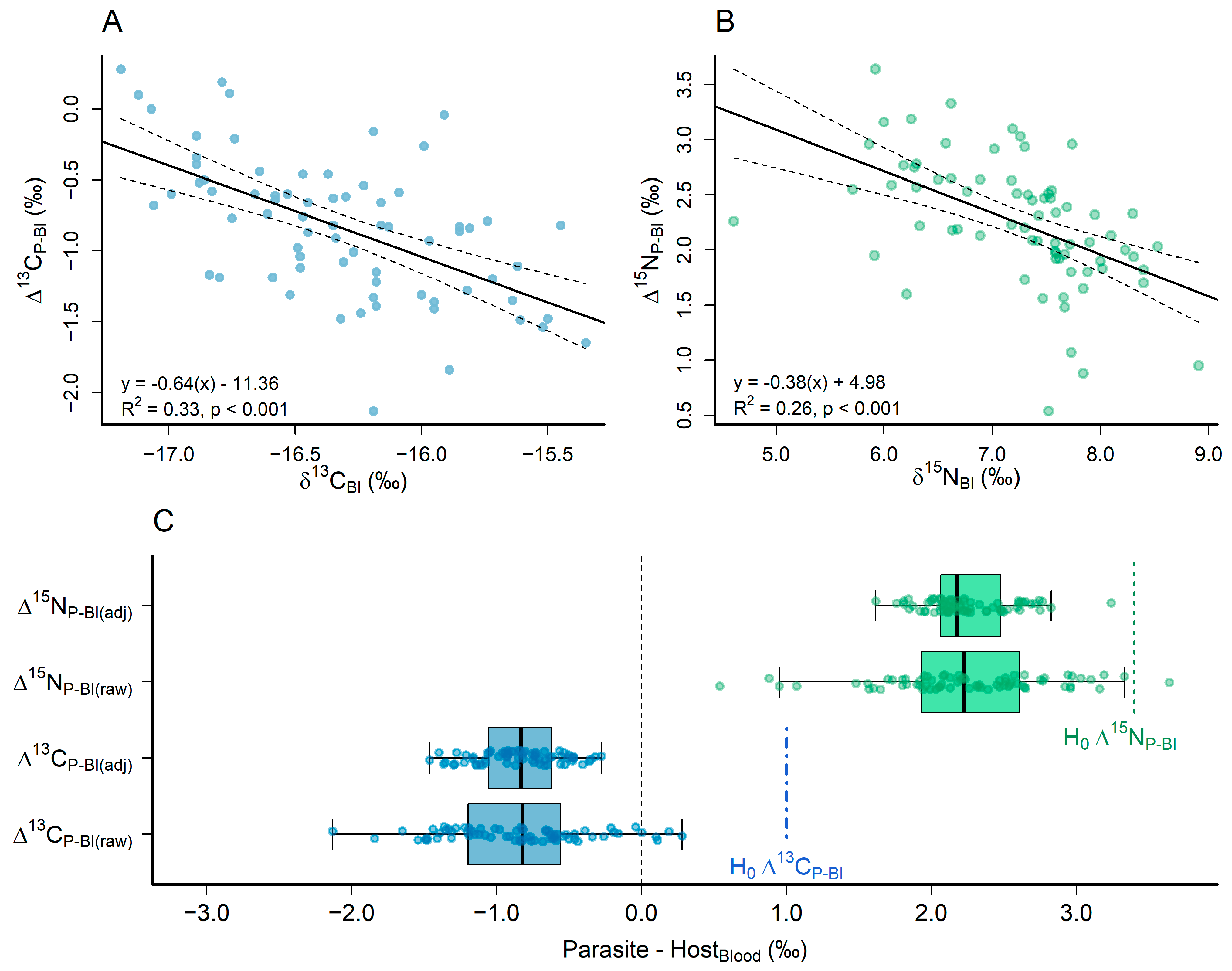
Δ15NP-Bl(adj) = −0.38 × δ13NBl + 4.98
3. Results
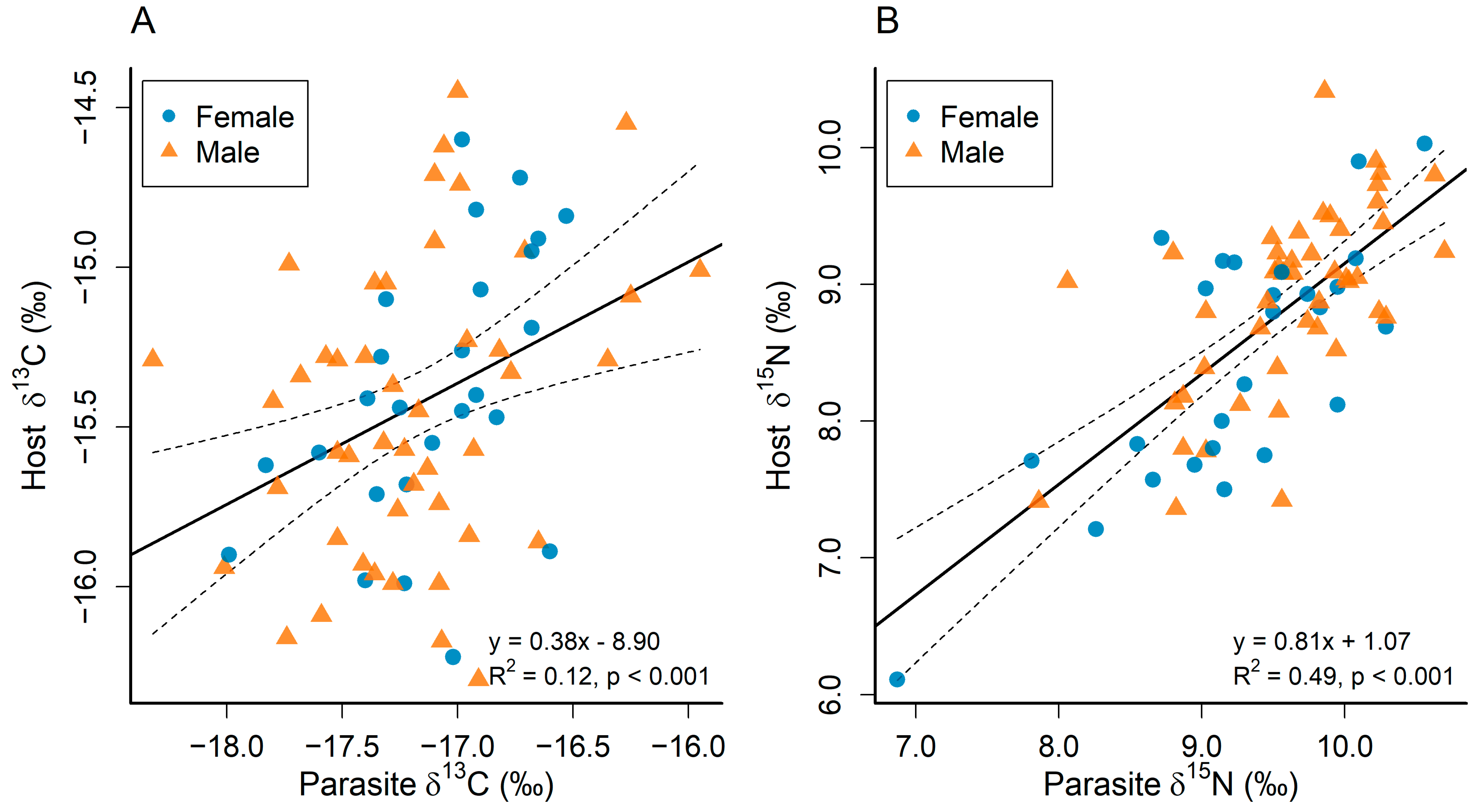
3.1. Comparing the Stable Carbon and Nitrogen Isotope Compositions of the Parasite and the Host
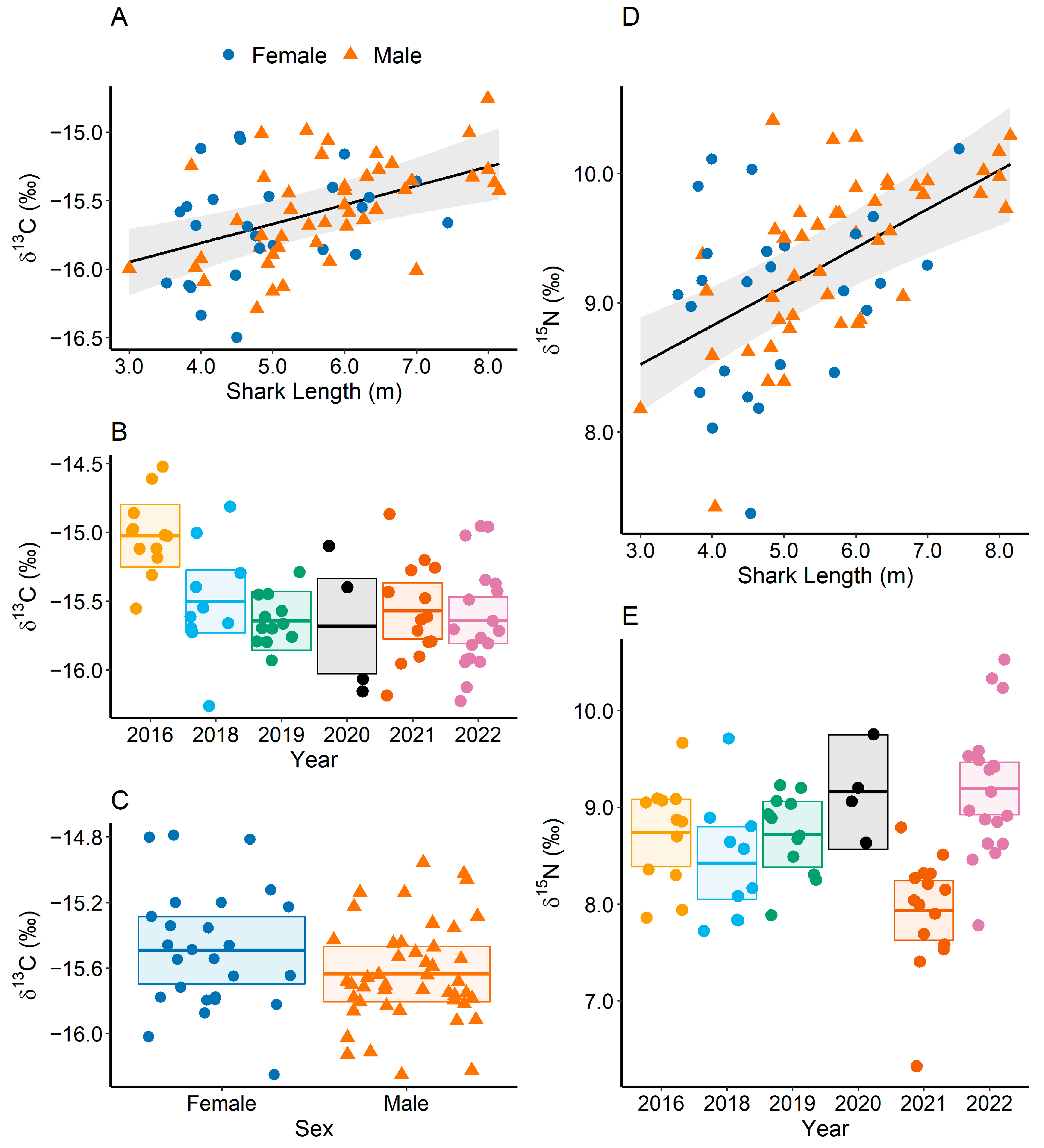
3.2. Drivers of Stable Isotope Composition of Whale Sharks
3.3. Isotopic Niches of Whale Sharks and Parasites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, A. Descriptions of New, or Imperfectly Known Objects of the Animal Kingdom, Found in the South of Africa. Afr. Commer. Advert. 1828, 3, 2. [Google Scholar]
- McClain, C.R.; Balk, M.A.; Benfield, M.C.; Branch, T.A.; Chen, C.; Cosgrove, J.; Dove, A.M.; Gaskins, L.; Helm, R.R.; Hochberg, F.G.; et al. Sizing Ocean Giants: Patterns of Intraspecific Size Variation in Marine Megafauna. PeerJ 2015, 3, e715. [Google Scholar] [CrossRef] [PubMed]
- Compagno, L.J. Sharks of the World: An Annotated and Illustrated Catalogue of Shark Species Known to Date; Food & Agriculture Organization: Rome, Italy, 2001; Volume 2, ISBN 92-5-104543-7. [Google Scholar]
- Pierce, S.J.; Norman, B. Rhincodon typus. The IUCN Red List of Threatened Species 2016; IUCN Red List: London, UK, 2016; p. e.T19488A236529. [Google Scholar] [CrossRef]
- Meekan, M.G.; Taylor, B.M.; Lester, E.; Ferreira, L.C.; Sequeira, A.M.M.; Dove, A.D.M.; Birt, M.J.; Aspinall, A.; Brooks, K.; Thums, M. Asymptotic Growth of Whale Sharks Suggests Sex-Specific Life-History Strategies. Front. Mar. Sci. 2020, 7, 575683. [Google Scholar] [CrossRef]
- Jarman, S.N.; Wilson, S.G. DNA-Based Species Identification of Krill Consumed by Whale Sharks. J. Fish Biol. 2004, 65, 586–591. [Google Scholar] [CrossRef]
- Ketchum, J.T.; Galván-Magaña, F.; Klimley, A.P. Segregation and Foraging Ecology of Whale Sharks, Rhincodon Typus, in the Southwestern Gulf of California. Environ. Biol. Fishes 2013, 96, 779–795. [Google Scholar] [CrossRef]
- Meekan, M.G.; Jarman, S.N.; McLean, C.; Schultz, M.B. DNA Evidence of Whale Sharks (Rhincodon Typus) Feeding on Red Crab (Gecarcoidea Natalis) Larvae at Christmas Island, Australia. Mar. Freshw. Res. 2009, 60, 607–609. [Google Scholar] [CrossRef]
- Motta, P.J.; Maslanka, M.; Hueter, R.E.; Davis, R.L.; de la Parra, R.; Mulvany, S.L.; Habegger, M.L.; Strother, J.A.; Mara, K.R.; Gardiner, J.M.; et al. Feeding Anatomy, Filter-Feeding Rate, and Diet of Whale Sharks Rhincodon Typus during Surface Ram Filter Feeding off the Yucatan Peninsula, Mexico. Zoology 2010, 113, 199–212. [Google Scholar] [CrossRef]
- Sampaio, C.L.S.; Leite, L.; Reis-Filho, J.A.; Loiola, M.; Miranda, R.J.; de Anchieta, C.C.; Nunes, J.; Macena, B.C.L. New Insights into Whale Shark Rhincodon Typus Diet in Brazil: An Observation of Ram Filter-Feeding on Crab Larvae and Analysis of Stomach Contents from the First Stranding in Bahia State. Environ. Biol. Fishes 2018, 101, 1285–1293. [Google Scholar] [CrossRef]
- Boldrocchi, G.; Bettinetti, R. Whale Shark Foraging on Baitfish off Djibouti. Mar. Biodivers. 2019, 49, 2013–2016. [Google Scholar] [CrossRef]
- Montero-Quintana, A.N.; Ocampo-Valdez, C.F.; Vázquez-Haikin, J.A.; Sosa-Nishizaki, O.; Osorio-Beristain, M. Whale Shark (Rhincodon Typus) Predatory Flexible Feeding Behaviors on Schooling Fish. J. Ethol. 2021, 39, 399–410. [Google Scholar] [CrossRef]
- Lester, E.; Cannon, T.; Lawrence, S.; Wilton, J.; Araujo, G.; Chin, A.; Lester, E.; Cannon, T.; Lawrence, S.; Wilton, J.; et al. Whale Sharks (Rhincodon Typus) Feed on Baitfish with Other Predators at Ningaloo Reef. Pac. Conserv. Biol. 2022, 29, 86–87. [Google Scholar] [CrossRef]
- Wyatt, A.S.J.; Matsumoto, R.; Chikaraishi, Y.; Miyairi, Y.; Yokoyama, Y.; Sato, K.; Ohkouchi, N.; Nagata, T. Enhancing Insights into Foraging Specialization in the World’s Largest Fish Using a Multi-Tissue, Multi-Isotope Approach. Ecol. Monogr. 2019, 89, e01339. [Google Scholar] [CrossRef]
- Meekan, M.G.; Virtue, P.; Marcus, L.; Clements, K.D.; Nichols, P.D.; Revill, A.T. The World’s Largest Omnivore Is a Fish. Ecology 2022, 103, e3818. [Google Scholar] [CrossRef]
- Rohner, C.A.; Couturier, L.I.E.; Richardson, A.J.; Pierce, S.J.; Prebble, C.E.M.; Gibbons, M.J.; Nichols, P.D. Diet of Whale Sharks Rhincodon Typus Inferred from Stomach Content and Signature Fatty Acid Analyses. Mar. Ecol. Prog. Ser. 2013, 493, 219–235. [Google Scholar] [CrossRef]
- Marcus, L.; Virtue, P.; Pethybridge, H.R.; Meekan, M.G.; Thums, M.; Nichols, P.D. Intraspecific Variability in Diet and Implied Foraging Ranges of Whale Sharks at Ningaloo Reef, Western Australia, from Signature Fatty Acid Analysis. Mar. Ecol. Prog. Ser. 2016, 554, 115–128. [Google Scholar] [CrossRef]
- Gleiss, A.; Wright, S.; Liebsch, N.; Wilson, R.; Norman, B. Contrasting Diel Patterns in Vertical Movement and Locomotor Activity of Whale Sharks at Ningaloo Reef. Mar. Biol. 2013, 160, 2981–2992. [Google Scholar] [CrossRef]
- Meekan, M.; Fuiman, L.; Davis, R.; Berger, Y.; Thums, M. Swimming Strategy and Body Plan of the World’s Largest Fish: Implications for Foraging Efficiency and Thermoregulation. Front. Mar. Sci. 2015, 2, 64. [Google Scholar] [CrossRef]
- Norman, B.M.; Reynolds, S.; Morgan, D.L. Does the Whale Shark Aggregate along the Western Australian Coastline beyond Ningaloo Reef? Pac. Conserv. Biol. 2016, 22, 72–80. [Google Scholar] [CrossRef]
- Marcus, L.; Virtue, P.; Nichols, P.D.; Ferreira, L.C.; Pethybridge, H.; Meekan, M.G. Stable Isotope Analysis of Dermis and the Foraging Behavior of Whale Sharks at Ningaloo Reef, Western Australia. Front. Mar. Sci. 2019, 6, 546. [Google Scholar] [CrossRef]
- Hussey, N.E.; Brush, J.; McCarthy, I.D.; Fisk, A.T. Delta15N and Delta13C Diet-Tissue Discrimination Factors for Large Sharks under Semi-Controlled Conditions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 155, 445–453. [Google Scholar] [CrossRef]
- Olin, J.A.; Hussey, N.E.; Grgicak-Mannion, A.; Fritts, M.W.; Wintner, S.P.; Fisk, A.T. Variable Δ15N Diet-Tissue Discrimination Factors among Sharks: Implications for Trophic Position, Diet and Food Web Models. PLoS ONE 2013, 8, e77567. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hernández, J.; Nunn, A.D.; Adams, C.E.; Amundsen, P.-A. Causes and Consequences of Ontogenetic Dietary Shifts: A Global Synthesis Using Fish Models. Biol. Rev. 2019, 94, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, M.; Wada, E. Stepwise Enrichment of 15N along Food Chains: Further Evidence and the Relation between Δ15N and Animal Age. Geochim. Cosmochim. Acta 1984, 48, 1135–1140. [Google Scholar] [CrossRef]
- Post, D.M. Using Stable Isotopes to Estimate Trophic Position: Models, Methods, and Assumptions. Ecology 2002, 83, 703–718. [Google Scholar] [CrossRef]
- Newsome, S.D.; Martinez del Rio, C.; Bearhop, S.; Phillips, D.L. A Niche for Isotopic Ecology. Front. Ecol. Environ. 2007, 5, 429–436. [Google Scholar] [CrossRef]
- Kopp, D.; Lefebvre, S.; Cachera, M.; Villanueva, M.C.; Ernande, B. Reorganization of a Marine Trophic Network along an Inshore–Offshore Gradient Due to Stronger Pelagic–Benthic Coupling in Coastal Areas. Prog. Oceanogr. 2015, 130, 157–171. [Google Scholar] [CrossRef]
- Christianen, M.J.A.; Middelburg, J.J.; Holthuijsen, S.J.; Jouta, J.; Compton, T.J.; van der Heide, T.; Piersma, T.; Sinninghe Damsté, J.S.; van der Veer, H.W.; Schouten, S.; et al. Benthic Primary Producers Are Key to Sustain the Wadden Sea Food Web: Stable Carbon Isotope Analysis at Landscape Scale. Ecology 2017, 98, 1498–1512. [Google Scholar] [CrossRef]
- von Biela, V.R.; Newsome, S.D.; Bodkin, J.L.; Kruse, G.H.; Zimmerman, C.E. Widespread Kelp-Derived Carbon in Pelagic and Benthic Nearshore Fishes Suggested by Stable Isotope Analysis. Estuar. Coast. Shelf Sci. 2016, 181, 364–374. [Google Scholar] [CrossRef]
- Duffill Telsnig, J.I.; Jennings, S.; Mill, A.C.; Walker, N.D.; Parnell, A.C.; Polunin, N.V.C. Estimating Contributions of Pelagic and Benthic Pathways to Consumer Production in Coupled Marine Food Webs. J. Anim. Ecol. 2019, 88, 405–415. [Google Scholar] [CrossRef]
- Skinner, C.; Newman, S.P.; Mill, A.C.; Newton, J.; Polunin, N.V.C. Prevalence of Pelagic Dependence among Coral Reef Predators across an Atoll Seascape. J. Anim. Ecol. 2019, 88, 1564–1574. [Google Scholar] [CrossRef]
- Carter, W.A.; Bauchinger, U.; McWilliams, S.R. The Importance of Isotopic Turnover for Understanding Key Aspects of Animal Ecology and Nutrition. Diversity 2019, 11, 84. [Google Scholar] [CrossRef]
- Wilson, S.G.; Polovina, J.J.; Stewart, B.S.; Meekan, M.G. Movements of Whale Sharks (Rhincodon Typus) Tagged at Ningaloo Reef, Western Australia. Mar. Biol. 2006, 148, 1157–1166. [Google Scholar] [CrossRef]
- Hearn, A.; Green, J.R.; Peñaherrera-Palma, C.; Reynolds, S.; Rohner, C.A.; Román, M.; Sequeira, A.M.M. Whale Shark Movements and Migrations. In Whale Sharks: Biology, Ecology and Conservation; Dove, A.D.M., Pierce, S.J., Eds.; CRC Press: Boca Raton, FL, USA, 2021; Volume 1, pp. 105–128. ISBN 978-1-03-204940-3. [Google Scholar]
- Pierce, S.J.; Pardo, S.A.; Rohner, C.A.; Matsumoto, R.; Murakumo, K.; Nozu, R.; Dove, A.D.M.; Perry, C.; Meekan, M.G. Whale Shark Reproduction, Growth, and Demography. In Whale Sharks: Biology, Ecology and Conservation; Dove, A.D.M., Pierce, S.J., Eds.; CRC Press: Boca Raton, FL, USA, 2021; Volume 1, pp. 13–46. ISBN 978-1-03-204940-3. [Google Scholar]
- Ueda, K.; Yanagisawa, M.; Murakumo, K.; Matsumoto, Y.; Sato, K.; Uchida, S. Physical Examination, Blood Sampling and Sedation of Large Elasmobranchs. In The Elasmobranch Husbandry Manual II: Recent Advances in the Care of Sharks, Rays and their Relatives; Smith, M., Warmolts, D., Thoney, D., Hueter, R., Murray, M., Ezcurra, J., Eds.; Ohio Biological Survey, Inc.: Columbus, OH, USA, 2017; pp. 255–262. ISBN 978-0-86727-166-9. [Google Scholar]
- Caira, J.N.; Jensen, K. A Digest of Elasmobranch Tapeworms. J. Parasitol. 2014, 100, 373–391. [Google Scholar] [CrossRef]
- Norman, B.M.; Newbound, D.R.; Knott, B. A New Species of Pandaridae (Copepoda), from the Whale Shark Rhincodon Typus (Smith). J. Nat. Hist. 2000, 34, 355–366. [Google Scholar] [CrossRef]
- Meekan, M.; Austin, C.M.; Tan, M.H.; Wei, N.-W.V.; Miller, A.; Pierce, S.J.; Rowat, D.; Stevens, G.; Davies, T.K.; Ponzo, A.; et al. IDNA at Sea: Recovery of Whale Shark (Rhincodon Typus) Mitochondrial DNA Sequences from the Whale Shark Copepod (Pandarus Rhincodonicus) Confirms Global Population Structure. Front. Mar. Sci. 2017, 4, 420. [Google Scholar] [CrossRef]
- Borucinska, J.D.; Benz, G.W. Lesions Associated with Attachment of the Parasitic Copepod Phyllothyreus Cornutus (Pandaridae: Siphonostomatoida) to Interbranchial Septa of Blue Sharks. J. Aquat. Anim. Health 1999, 11, 290–295. [Google Scholar] [CrossRef]
- Dippenaar, S.M.; Jordaan, A. How Females of Achtheinus Spp. (Pandaridae: Siphonostomatoida) Attach to Their Elasmobranch Hosts with Notes on Their Effects on the Hosts’ Fins. Folia Parasitol. 2015, 62, 005. [Google Scholar] [CrossRef]
- Womersley, F.; Hancock, J.; Perry, C.T.; Rowat, D. Wound-Healing Capabilities of Whale Sharks (Rhincodon Typus) and Implications for Conservation Management. Conserv. Physiol. 2021, 9, coaa120. [Google Scholar] [CrossRef]
- Williams, E.H.; Bunkley-Williams, L. Life Cycle and Life History Strategies of Parasitic Crustacea. In Parasitic Crustacea: State of Knowledge and Future Trends; Smit, N.J., Bruce, N.L., Hadfield, K.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 179–266. ISBN 978-3-030-17385-2. [Google Scholar]
- Sequeira, A.M.M.; Thums, M.; Brooks, K.; Meekan, M.G. Error and Bias in Size Estimates of Whale Sharks: Implications for Understanding Demography. R. Soc. Open Sci. 2016, 3, 150668. [Google Scholar] [CrossRef]
- Bessey, C.; Vanderklift, M.A. Drying Method Has No Substantial Effect on Δ15N or Δ13C Values of Muscle Tissue from Teleost Fishes. Rapid Commun. Mass Spectrom. 2014, 28, 265–273. [Google Scholar] [CrossRef]
- Zalina, B.; Maizah, M.A.; Mohd Uzair, R. Comparisons between Tissues, Preservation, and Desiccation Methods on Stable Isotopes Δ13C and Δ15N of Spot-Tail Sharks (Carcharhinus Sorrah) from the South China Sea. Turk. J. Fish. Aquat. Sci. 2020, 20, 711–716. [Google Scholar] [CrossRef]
- Bennett-Williams, J.; Skinner, C.; Wyatt, A.S.J.; McGill, R.A.R.; Willis, T.J. A Multi-Tissue, Multi-Species Assessment of Lipid and Urea Stable Isotope Biases in Mesopredator Elasmobranchs. Front. Mar. Sci. 2022, 9, 821478. [Google Scholar] [CrossRef]
- Marcus, L.; Virtue, P.; Nichols, P.D.; Meekan, M.G.; Pethybridge, H. Effects of Sample Treatment on the Analysis of Stable Isotopes of Carbon and Nitrogen in Zooplankton, Micronekton and a Filter-Feeding Shark. Mar. Biol. 2017, 164, 124. [Google Scholar] [CrossRef]
- Kolasinski, J.; Rogers, K.M.; Frouin, P. Effects of Acidification on Carbon and Nitrogen Stable Isotopes of Benthic Macrofauna from a Tropical Coral Reef. Rapid Commun. Mass Spectrom. 2008, 22, 2955–2960. [Google Scholar] [CrossRef]
- Skrzypek, G.; Paul, D. Delta13C Analyses of Calcium Carbonate: Comparison between the GasBench and Elemental Analyzer Techniques. Rapid Commun. Mass. Spectrom. 2006, 20, 2915–2920. [Google Scholar] [CrossRef]
- Coplen, T.B. Guidelines and Recommended Terms for Expression of Stable-Isotope-Ratio and Gas-Ratio Measurement Results. Rapid Commun. Mass. Spectrom. 2011, 25, 2538–2560. [Google Scholar] [CrossRef]
- Skrzypek, G. Normalization Procedures and Reference Material Selection in Stable HCNOS Isotope Analyses: An Overview. Anal. Bioanal. Chem. 2013, 405, 2815–2823. [Google Scholar] [CrossRef]
- Riekenberg, P.M.; Briand, M.J.; Moléana, T.; Sasal, P.; van der Meer Marcel, T.J.; Thieltges, D.W.; Letourneur, Y. Isotopic Discrimination in Helminths Infecting Coral Reef Fishes Depends on Parasite Group, Habitat within Host, and Host Stable Isotope Value. Sci. Rep. 2021, 11, 4638. [Google Scholar] [CrossRef]
- Thieltges, D.W.; Goedknegt, M.A.; O’Dwyer, K.; Senior, A.M.; Kamiya, T. Parasites and Stable Isotopes: A Comparative Analysis of Isotopic Discrimination in Parasitic Trophic Interactions. Oikos 2019, 128, 1329–1339. [Google Scholar] [CrossRef]
- Borrell, A.; Aguilar, A.; Gazo, M.; Kumarran, R.P.; Cardona, L. Stable Isotope Profiles in Whale Shark (Rhincodon Typus) Suggest Segregation and Dissimilarities in the Diet Depending on Sex and Size. Environ. Biol. Fishes 2011, 92, 559–567. [Google Scholar] [CrossRef]
- Young, J.N.; Bruggeman, J.; Rickaby, R.E.M.; Erez, J.; Conte, M. Evidence for Changes in Carbon Isotopic Fractionation by Phytoplankton between 1960 and 2010. Glob. Biogeochem. Cycles 2013, 27, 505–515. [Google Scholar] [CrossRef]
- Barton, K. MuMIn: Multi-Model Inference. R Package Version 1.46.0. 2022. Available online: https://CRAN.R-project.org/package=MuMIn (accessed on 18 September 2022).
- Breheny, P.; Burchett, W. Visualization of Regression Models Using Visreg. R J. 2017, 9, 56–71. [Google Scholar] [CrossRef]
- Jackson, A.L.; Parnell, A.C.; Inger, R.; Bearhop, S. Comparing Isotopic Niche Widths among and within Communities: SIBER—Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 2011, 80, 595–602. [Google Scholar] [CrossRef]
- Link, W.A.; Eaton, M.J. On Thinning of Chains in MCMC. Methods Ecol. Evol. 2012, 3, 112–115. [Google Scholar] [CrossRef]
- Makowski, D.; Ben-Shachar, M.S.; Lüdecke, D. BayestestR: Describing Effects and Their Uncertainty, Existance and Significance within the Bayesian Framework. J. Open Source Softw. 2019, 4, 1541. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Pinnegar, J.K.; Campbell, N.; Polunin, N.V.C. REGULAR PAPERS Unusual Stable Isotope Fractionation Patterns Observed for Fish Host—Parasite Trophic Relationships. J. Fish Biol. 2001, 59, 494–503. [Google Scholar] [CrossRef]
- Deudero, S.; Pinnegar, J.K.; Polunin, N.V.C. Insights into Fish Host-Parasite Trophic Relationships Revealed by Stable Isotope Analysis. Dis. Aquat. Org. 2002, 52, 77–86. [Google Scholar] [CrossRef]
- Lafferty, K.D.; Allesina, S.; Arim, M.; Briggs, C.J.; De Leo, G.; Dobson, A.P.; Dunne, J.A.; Johnson, P.T.J.; Kuris, A.M.; Marcogliese, D.J.; et al. Parasites in Food Webs: The Ultimate Missing Links. Ecol. Lett. 2008, 11, 533–546. [Google Scholar] [CrossRef]
- Nachev, M.; Jochmann, M.A.; Walter, F.; Wolbert, J.B.; Schulte, S.M.; Schmidt, T.C.; Sures, B. Understanding Trophic Interactions in Host-Parasite Associations Using Stable Isotopes of Carbon and Nitrogen. Parasites Vectors 2017, 10, 90. [Google Scholar] [CrossRef]
- Hajji, T.; Telahigue, K.; Rabeh, I.; Ben Ammar, R.; Mdaini, Z.; El Cafsi, M.; Ghali, R. Polar and Neutral Lipid Composition of the Copepod Lernaeocera Lusci and Its Host Merluccius Merluccius in Relationship with the Parasite Intensity. Parasitol. Res. 2021, 120, 1979–1991. [Google Scholar] [CrossRef] [PubMed]
- Post, D.M.; Layman, C.A.; Arrington, D.A.; Takimoto, G.; Quattrochi, J.; Montaña, C.G. Getting to the Fat of the Matter: Models, Methods and Assumptions for Dealing with Lipids in Stable Isotope Analyses. Oecologia 2007, 152, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Hussey, N.E.; MacNeil, M.A.; McMeans, B.C.; Olin, J.A.; Dudley, S.F.J.; Cliff, G.; Wintner, S.P.; Fennessy, S.T.; Fisk, A.T. Rescaling the Trophic Structure of Marine Food Webs. Ecol. Lett. 2014, 17, 239–250. [Google Scholar] [CrossRef] [PubMed]
- McMahon, K.W.; McCarthy, M.D. Embracing Variability in Amino Acid Δ15N Fractionation: Mechanisms, Implications, and Applications for Trophic Ecology. Ecosphere 2016, 7, e01511. [Google Scholar] [CrossRef]
- Jenkins, W.G.; Demopoulos, A.W.J.; Nicholson, M.D.; Sikkel, P.C. Stable Isotope Dynamics of Herbivorous Reef Fishes and Their Ectoparasites. Diversity 2020, 12, 429. [Google Scholar] [CrossRef]
- Rohner, C.A.; Prebble, C.E.M. Whale Shark Foraging, Feeding and Diet. In Whale Sharks: Biology, Ecology and Conservation; Dove, A.D.M., Pierce, S.J., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 153–180. ISBN 978-1-03-204940-3. [Google Scholar]
- Demopoulos, A.W.J.; Sikkel, P.C. Enhanced Understanding of Ectoparasite–Host Trophic Linkages on Coral Reefs through Stable Isotope Analysis. Int. J. Parasitol. Parasites Wildl. 2015, 4, 125–134. [Google Scholar] [CrossRef]
- Gilbert, B.M.; Nachev, M.; Jochmann, M.A.; Schmidt, T.C.; Köster, D.; Sures, B.; Avenant-Oldewage, A. Stable Isotope Analysis Spills the Beans about Spatial Variance in Trophic Structure in a Fish Host—Parasite System from the Vaal River System, South Africa. Int. J. Parasitol. Parasites Wildl. 2020, 12, 134–141. [Google Scholar] [CrossRef]
- Pegoraro de Macedo, M.R.; Palomba, M.; Santoro, M. The Current State of Knowledge on Parasitic Copepods (Siphonostomatoida: Pandaridae) of Elasmobranchs. Vet. Clin. N. Am. Exot. Anim. Pract. 2023, 26, 475–509. [Google Scholar] [CrossRef]
- Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis, 10th ed.; Impensis Direct: Stockholm, Sweden, 1758. [Google Scholar] [CrossRef]
- Hewitt, G.C. Eight Species of Parasitic Copepoda on a White Shark. N. Z. J. Mar. Freshw. Res. 1979, 13, 171. [Google Scholar] [CrossRef]
- Benz, G.W.; Mollet, H.F.; Ebert, D.A.; Davis, C.R.; Van Sommeran, S.R. Five Species of Parasitic Copepods (Siphonostomatoida: Pandaridae) from the Body Surface of a White Shark Captured in Morro Bay, California. Pac. Sci. 2003, 57, 39–43. [Google Scholar] [CrossRef]


| Year | Male | Female | Total |
|---|---|---|---|
| 2016 | 7 | 5 | 12 |
| 2018 | 7 | 3 | 10 |
| 2019 | 7 | 5 | 12 |
| 2020 | 4 | 0 | 4 |
| 2021 | 8 | 7 | 15 |
| 2022 | 13 | 6 | 19 |
| Total | 46 | 26 | 72 |
| Linear Models Estimating Host δ13C | |||||||
|---|---|---|---|---|---|---|---|
| Model | Intercept | Parasiteδ13C | Sex (M) | Parasite Sex (M) | ANOVA | R2 | AICc |
| 1 * | −8.90 (<0.001) | 0.38 (<0.001) | - | - | - | 0.12 | 83.21 |
| 2 | −8.93 (<0.001) | 0.38 (<0.001) | −0.02 (0.85) | - | 0.59 | 0.11 | 85.41 |
| 3 | −5.54 (<0.001) | 0.58 (0.01) | −4.64 (0.31) | −0.27 (0.31) | 0.31 | 0.11 | 86.63 |
| Linear Models Estimating Host δ15N | |||||||
| Model | Intercept | Parasiteδ15N | Sex (M) | Parasite Sex (M) | ANOVA | R2 | AICc |
| 1 * | 1.07 (0.25) | 0.81 (<0.001) | - | - | - | 0.49 | 125.95 |
| 2 | 1.24 (0.18) | 0.77 (<0.001) | 0.17 (0.25) | - | 0.40 | 0.49 | 126.79 |
| 3 | 0.59 (0.65) | 0.85 (<0.001) | 1.50 (0.43) | −0.14 (0.48) | 0.48 | 0.49 | 128.57 |
| Model | Y | X-Variables | AICc | ΔAICc | BIC | ΔBIC | wAICc | wBIC | R2 |
|---|---|---|---|---|---|---|---|---|---|
| (1) | δ13C | Length + sex + year | 63.7 | 0.00 | 79.8 | 1.50 | 0.42 | 0.28 | 0.46 |
| 2 * | δ13C | Length + year | 63.8 | 0.15 | 81.3 | 0.00 | 0.39 | 0.59 | 0.44 |
| 3 | δ13C | Length + sex + year + length × sex | 65.7 | 2.01 | 84.8 | 5.09 | 0.15 | 0.05 | 0.47 |
| (1 *) | δ15N | Length + year | 139.2 | 0.00 | 155.1 | 0.00 | 0.41 | 0.83 | 0.49 |
| 2 | δ15N | Length + sex + year + sex × year | 139.8 | 0.66 | 163.1 | 8.05 | 0.29 | 0.02 | 0.57 |
| 3 | δ15N | Length + sex + year | 141.5 | 2.30 | 159.0 | 3.96 | 0.13 | 0.11 | 0.49 |
| 4 | δ15N | Length + sex + year + length × sex | 141.9 | 2.77 | 161.1 | 6.00 | 0.10 | 0.04 | 0.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio, B.J.; Skrzypek, G.; Meekan, M. Parasitic Copepods as Biochemical Tracers of Foraging Patterns and Dietary Shifts in Whale Sharks (Rhincodon typus Smith, 1828). Fishes 2023, 8, 261. https://doi.org/10.3390/fishes8050261
Osorio BJ, Skrzypek G, Meekan M. Parasitic Copepods as Biochemical Tracers of Foraging Patterns and Dietary Shifts in Whale Sharks (Rhincodon typus Smith, 1828). Fishes. 2023; 8(5):261. https://doi.org/10.3390/fishes8050261
Chicago/Turabian StyleOsorio, Brendon James, Grzegorz Skrzypek, and Mark Meekan. 2023. "Parasitic Copepods as Biochemical Tracers of Foraging Patterns and Dietary Shifts in Whale Sharks (Rhincodon typus Smith, 1828)" Fishes 8, no. 5: 261. https://doi.org/10.3390/fishes8050261
APA StyleOsorio, B. J., Skrzypek, G., & Meekan, M. (2023). Parasitic Copepods as Biochemical Tracers of Foraging Patterns and Dietary Shifts in Whale Sharks (Rhincodon typus Smith, 1828). Fishes, 8(5), 261. https://doi.org/10.3390/fishes8050261






