Abstract
Carbon stable isotopes (δ13C) are widely used in ecological studies to understand diet, food web dynamics, and movements of marine fishes. Still, δ13C is influenced by lipid content and often requires chemical extraction or mathematical correction. Here, we developed a species-specific mathematical lipid correction for white muscle tissue of yellowfin tuna (Thunnus albacares), a highly migratory finfish of considerable economic and ecological value. Lipid extraction was conducted on yellowfin tuna white muscle tissue (C:N range: 2.96–6.49), and both linear and non-linear lipid correction models for δ13C were fitted and assessed. Lipid extraction increased δ13C, and to a lesser extent, δ15N values in yellowfin tuna white muscle tissue, but had no effect on δ34S. Both non-linear models provided better fits to the data than the linear model, suggesting an asymptotic relationship between C:N and ∆δ13C. Results support the growing body of evidence that C:N ratios can be used to predict lipid corrected δ13C and highlight the value of mathematical correction approaches. We provide species-specific parameter estimates that can be used for lipid correction of white muscle tissue for δ13C analysis in yellowfin tuna and similar species for which species-specific models have yet to be developed.
Keywords:
lipid extraction; trophic ecology; lipid normalization; Thunnus albacares; Scombridae; nitrogen; sulfur; δ13C; δ15N; δ34S Key Contribution:
Lipid extraction increased δ13C values in yellowfin tuna white muscle tissue, particularly tissues with C:N > 3.5. Species-specific parameters for lipid correction models are provided for use in stable isotope studies.
1. Introduction
Stable isotope analysis is a frequently utilized tool for understanding foraging behavior, food web dynamics, and habitat shifts in marine organisms [1,2,3]. Carbon stable isotopes (δ13C) are among the most widely applied in fish ecology [4,5], and are often used to trace sources of primary production in marine food webs because there is little fractionation between trophic steps [6]. Still, a common concern with δ13C analysis is the fact that lipids tend to have more negative δ13C values than other compounds (e.g., protein) [7]. As a result, interindividual variation in lipid content can affect bulk carbon isotope values (high lipid content leads to more negative values), leading to incorrect interpretation of trophic pathways and/or movements [8,9].
A variety of lipid corrections have been developed to reduce variation in δ13C values caused by interindividual variability in lipid content [9,10]. The most straightforward way to address high lipid content is to directly extract lipids (i.e., lipid extraction) from fish tissue samples prior to stable isotope analysis. However, this approach has other consequences, as lipid extraction can alter isotope values of other elements of interest, such as nitrogen (δ15N) [11,12]. Alternatively, a mathematical lipid correction can be applied [9,13]. Such corrections are typically based on lipid extraction experiments from which the relationship between carbon and nitrogen (C:N) ratios and the difference between bulk carbon and lipid-free carbon (after lipid extraction) can be modeled [10]. The resulting relationship can then be used to estimate lipid-free δ13C based on the C:N ratio while allowing lipids to be incorporated in other isotope analyses (e.g., δ15N) [9]. While mathematical corrections are widely used, lipid relationships vary among species, and species-specific lipid correction equations are unavailable for many marine fishes [9,11,12].
Here we conduct a lipid extraction experiment to develop a mathematical lipid correction equation for yellowfin tuna (Thunnus albacares) white muscle tissue that can be applied to future stable isotope analyses. Yellowfin tuna is a circumtropical, highly migratory predator and is an important component of open ocean ecosystems, supporting economically valuable fisheries around the world. Despite considerable interest in yellowfin tuna dietary studies, a mathematical lipid correction has not yet been developed for the species. The aim of this study was to evaluate linear and non-linear approaches to characterize the relationship between bulk C:N ratios and the change in δ13C values after lipid extraction and develop a mathematical lipid correction equation for yellowfin tuna white muscle tissue.
2. Materials and Methods
Yellowfin tuna (n = 240) were sampled from recreational charter landings in the northern Gulf of Mexico in 2019 and 2020. Epaxial white muscle tissue (5 cm3) anterior to the dorsal fin was removed from each individual and frozen at −20 °C. Individual tissue samples were divided into two aliquots and freeze-dried for 48 h, after which one aliquot was prepared for stable isotope analysis without extraction, while the other aliquot was set aside for the lipid extraction procedure. Samples immediately prepared for stable isotope analysis were homogenized using a mortar and pestle, weighed (1.5 ± 0.025 milligrams), complimented with a combustion catalyst (vanadium pentoxide, 3.0 ± 0.025 milligrams), and loaded into a 5 × 9-mm tin capsule. Stable isotope analysis was performed using an elemental analyzer interfaced with a Thermo Scientific (Waltham, MA, USA) Delta V Advantage continuous flow isotope ratio mass spectrometer (EA-IRMS) at Louisiana State University. We quantified stable isotopes of carbon, nitrogen, and sulfur, given that these isotopes are among the most commonly applied in fish ecology studies [14]. Stable isotope values were reported in delta notation (δ) and per mil units (‰) relative to the international measurement standards Vienna Pee Dee Belemnite (for carbon), atmospheric N2 (for nitrogen), and Vienna Canyon Diablo troilite (for sulfur), using the following equation:
where Rsample and Rstandard represent the ratio of heavy to light isotopes in the sample and the standard, respectively. Sample precision was ±0.1‰ for δ13C, ±0.2‰ for δ15N, and ±0.3‰ for δ34S. Carbon: nitrogen (C:N) ratios were expressed as the % carbon relative to the % nitrogen (by weight) of a sample based on uncorrected percentage element data.
After stable isotope analysis of samples from the first aliquot, C:N ratios were examined as a proxy for the presence of lipid in yellowfin tuna white muscle tissue [10]. Lipid extraction was then performed on 36 samples from the second aliquot that were systematically chosen to represent the range of C:N values (2.96–6.49) observed in samples from the first aliquot [15]. The extraction of lipid from white muscle tissue followed a modified protocol outlined by Kim and Koch [16], using a 2:1 chloroform: methanol solution as the solvent instead of petroleum ether [9]. Samples were weighed to 350 mg, placed in glass scintillation vials with 8 mL of 2:1 chloroform: methanol solution, and sonicated for 15 min in a water bath sonicator. The solution was then decanted, and samples were rinsed by sonicating for 15 min in 8 mL of deionized water. The deionized water was decanted and the entire procedure of sonication in chloroform: methanol solution and rinse in deionized water was then repeated. Finally, the lipid extracted samples were oven dried for 24 h at 50 °C and prepared for stable isotope analysis following the same procedure described earlier for the untreated samples.
Differences in δ13C, δ15N, and δ34S between treated and untreated samples were examined using paired t-tests (α = 0.05). Yellowfin tuna C:N ratios were plotted against the difference in δ13C between treated and untreated samples (∆δ13C), expressed as:
and three models were evaluated for best fit as a lipid correction for yellowfin tuna. First, a linear model was fit to the yellowfin tuna data [10]. However, because relationships between C:N ratios and Δδ13C are not always linear, non-linear least squares was used to solve for two non-linear approaches used by Logan et al. [9] for bluefin tuna (Thunnus thynnus). The first was a three-parameter asymptotic model (non-linear Equation (1)) based on the equation described by Logan et al. [9]:
where a corresponds to the y-asymptote, −b/a corresponds to the x-intercept (C:N ratio when lipid free), and b/c is equal to the y-intercept (C:N = 0). The second non-linear equation was a two-parameter model based on the mass balance equation of Fry [17] expressed as:
where P corresponds to the δ13C discrimination between protein and lipid and F corresponds to the C:N ratio when lipid free [9]. All models were fit to our yellowfin tuna data to develop species-specific parameters that could be used for future studies on yellowfin tuna and similar species. Models were evaluated using Akaike Information Criterion (AIC) and developed and assessed using the R Statistical Programming Environment [18].
3. Results
Yellowfin tuna C:N ratios ranged from 2.96 to 6.49 with a mean (±SE) of 3.93 ± 0.15. In general, higher C:N ratios were observed for larger individuals (Table S1). Lipid extraction of white muscle tissue resulted in a significant increase in δ13C values ranging from 0–3.4‰ with a mean of 1.06 ± 0.18‰ (mean increase ± SE) across all yellowfin tuna samples (paired t-test; p < 0.001). A significant increase in δ15N ± (0.33 ± 0.08‰, mean difference ± SE) was also observed (paired t-test, p < 0.001); however, lipid extraction did not influence δ34S values (0.14 ± 0.08‰) (paired t-test, p > 0.05).
All models indicated that C:N was a good predictor of Δδ13C after lipid extraction. The linear model was described by Δδ13C = 1.16(C:N) − 3.51, with a residual standard error of 0.32 and an adjusted R2 of 0.92 (Figure 1A). Standard errors for parameter estimates were 0.06 for the slope and 0.24 for the y-intercept. Still, AIC indicated the non-linear models provided a better fit to describe the relationship between C:N and Δδ13C for yellowfin tuna (Figure 1). Non-linear equation 1 had the lowest AIC value and residual standard error (AIC = 12.10, RSE = 0.21) compared to non-linear equation 2 (AIC = 13.84, RSE = 0.28) and the linear equation (AIC= 23.21). The difference in AIC between the two non-linear models was considered negligible [19]. Parameter estimates for non-linear Equation (1) were a = 9.36 ± 2.31 (SE), b = −29.36 ± 7.11, and c = 2.18 ± 1.88 (Table 1). In contrast, parameter estimates for non-linear equation 2 were P = 6.64 ± 0.25 (SE) and F = 3.16 ± 0.03 (Table 1). The x-intercept in all models represented the C:N ratio below which δ13C did not increase after lipid extraction (C:N ratio of lipid free sample), and was similar between non-linear Equation (1) (−b/a = 3.14) and non-linear Equation (2) (F = 3.16) but was slightly lower for the linear model (3.03).
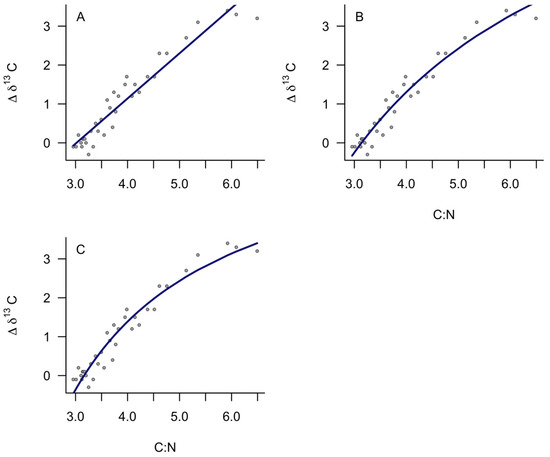
Figure 1.
Model fits showing the relationship between carbon: nitrogen ratios and the change in carbon isotope after lipid extraction for yellowfin tuna (n = 36) across a range of C:N. Models represent (A) Linear, (B) Non-linear Equation (1), and (C) Non-linear Equation (2).

Table 1.
Lipid correction equations for yellowfin tuna with parameter estimates and standard errors (SE). The term δ13CBulk refers to untreated δ13C value, while δ13CLipid-free refers to δ13C value after lipid correction. C:N refers to the carbon: nitrogen ratio of untreated δ13C.
4. Discussion
Lipid extraction of yellowfin tuna white muscle tissue resulted in a substantial increase in δ13C values, suggesting that lipid correction (e.g., either extraction or mathematical) may be necessary for δ13C stable isotope analysis. While it is not uncommon for fish white muscle tissue to have low lipid content, large pelagic predators such as tunas, swordfish, and sharks often have higher lipid content in white muscle and require lipid correction for stable isotope analysis [9,20,21]. Interestingly, the highest C:N ratios were observed in large yellowfin tuna that were captured during winter or early spring. While variance in lipid content can reflect individual or spatio-temporal differences in diet, similar seasonal trends in C:N have been reported for bluefin tuna and albacore (Thunnus alalunga) in the north Atlantic, with increased lipid content during cooler months attributed to reduced allocation to egg production during winter [22]. It is unclear if a similar mechanism applies to yellowfin tuna in the Gulf of Mexico; however, it is worth noting that approximately 75% of the individuals with C:N ratios greater than 4.0 were females. Finally, lipid extraction resulted in small, but significant alteration of δ15N values in yellowfin tuna, which is in agreement with previous studies that have reported more substantial changes in δ15N after lipid extraction [23]. In contrast, our results indicate that extraction has little influence on δ34S. Still, these results suggest that a mathematical correction may be preferred to extraction to correct for lipids without altering interpretation of δ15N or δ34S.
Similar to other marine fishes, a strong predictive relationship was observed between the C:N ratio and Δδ13C for yellowfin tuna white muscle tissue after lipid extraction [24,25]. The finding that both non-linear approaches to lipid correction provided better fits relative to the linear approach is consistent with several recent studies suggesting that the relationship between C:N and Δδ13C is asymptotic in tunas and other large predators [9,12,24]. Linear approaches are also common for lipid correction in marine fishes; however, because the isotopic discrimination in δ13C between lipid and protein is believed to be between 6–7‰ [8], it might be expected that the rate of change in δ13C will approach an asymptote at higher C:N ratios [10,25]. Thus, despite the fact that the linear model provided a good fit in the current study, our results support the notion that a non-linear correction is likely to perform better over a wide range of C:N ratios [9]. The mass balance equation (non-linear Equation (2), [17]) is often recommended for lipid normalization of marine fish muscle tissue [9,24,26] however, non-linear Equation (1) provided the best fit in the current study. Still, differences between both non-linear equations were negligible [19], suggesting that either approach would be appropriate for yellowfin tuna.
Lipid extraction prior to stable isotope analysis is the subject of some debate, and while lipid correction is generally recommended when C:N > 3.5 [10]; other studies have found this reference to be unreliable, as relationships between C:N and lipid content may vary by species, tissue, or trophic grouping [12,27]. Our best fit model indicated that yellowfin tuna white muscle tissue with C:N ratios > 3.14, could be expected to undergo some measure of alteration in δ13C after lipid extraction and could therefore be corrected. Still, differences in δ13C after extraction did not exceed 0.3‰ (mean difference = 0.02) when C:N ratios < 3.3, or 0.5‰ (mean difference = 0.1) when C:N ratios < 3.5. Thus, it appears that in practice, lipid extraction will have little influence on δ13C values for yellowfin tuna white muscle tissue with C:N ratios < 3.5, corroborating the general recommendation of Post et al. [10].
Stable isotope approaches are increasingly incorporated in food web studies to better understand trophic linkages supporting pelagic fish populations [28,29,30]. Our findings suggest lipid extraction or correction is needed for analysis of carbon stable isotopes in yellowfin tuna when lipid content (C:N ratio) is elevated, and highlight the benefits of mathematical correction approaches which reduce time and costs of lipid extraction while avoiding the potential negative impacts of lipid removal on δ15N [9]. While several general (multi-taxa) models have been developed for lipid correction [9,10], species- and tissue-specific lipid correction models generally perform best when available [9,12,26]. Thus, the species-specific parameter estimates for lipid correction in yellowfin tuna white muscle tissue presented here can be applied to other δ13C studies within the general range of C:N ratios included in our models (~2.9–6.5). While species-specific approaches are certainly preferred when available, model parameter estimates presented here may also prove useful for lipid correction of white muscle tissue in other tropical scombrids for which species-specific corrections have yet to be developed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8090446/s1, Table S1: Summary data for yellowfin tuna samples selected for lipid extraction.
Author Contributions
Conceptualization, M.A.D. and M.S.L.; formal analysis, M.A.D. and M.S.L.; investigation, M.S.L.; data curation, M.S.L. and M.A.D.; writing—original draft preparation, M.A.D.; writing—review and editing, M.A.D. and M.S.L.; visualization, M.A.D. and M.S.L.; supervision, M.A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Approval was not required for this research. No live animals were used in the study and no active sampling of fish was conducted by the researchers. All samples were donated by recreational charter anglers from the fishery.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data from this study can be accessed from the authors upon reasonable request.
Acknowledgments
We would like to thank M. Polito at the Stable Isotope Ecology Laboratory at LSU for assistance with lipid extraction experiments and stable isotope analysis. We would also like to acknowledge members of the Fisheries and Movement Ecology Lab at LSU for their help in collecting and processing samples. Finally, we would like to thank the many anglers and charter captains that provided yellowfin tuna samples for this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Layman, C.A.; Arrington, D.A.; Montaña, C.G.; Post, D.M. Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology 2007, 88, 42–48. [Google Scholar] [CrossRef] [PubMed]
- McMahon, K.W.; Hamady, L.L.; Thorrold, S.R. A review of ecogeochemistry approaches to estimating movements of marine animals. Limnol. Oceanogr. 2013, 58, 697–714. [Google Scholar] [CrossRef]
- Matsubayashi, J.; Osada, Y.; Tadokoro, K.; Abe, Y.; Yamaguchi, A.; Shirai, K.; Honda, K.; Yoshikawa, C.; Ogawa, N.O.; Ohkouchi, N.; et al. Tracking long-distance migration of marine fishes using compound-specific stable isotope analysis of amino acids. Ecol. Lett. 2020, 23, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Dance, K.M.; Rooker, J.R.; Shipley, J.B.; Dance, M.A.; Wells, R.J.D. Feeding ecology of fishes associated with artificial reefs in the northwest Gulf of Mexico. PLoS ONE 2018, 13, e0203873. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, A.; Pethybridge, H.; Cassar, N.; Receveur, A.; Allain, V.; Bodin, N.; Bopp, L.; Choy, C.A.; Duffy, L.; Fry, B.; et al. Trends in tuna carbon isotopes suggest global changes in pelagic phytoplankton communities. Glob. Chang. Biol. 2020, 26, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Post, D.M. Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 2002, 83, 703–718. [Google Scholar] [CrossRef]
- DeNiro, M.J.; Epstein, S. Mechanism of Carbon Isotope Fractionation Associated with Lipid Synthesis. Science 1977, 197, 261–263. [Google Scholar] [CrossRef]
- Sweeting, C.J.; Polunin, N.V.C.; Jennings, S. Effects of chemical lipid extraction and arithmetic lipid correction on stable isotope ratios of fish tissues. Rapid Commun. Mass Spectrom. 2006, 20, 595–601. [Google Scholar] [CrossRef]
- Logan, J.M.; Jardine, T.D.; Miller, T.J.; Bunn, S.E.; Cunjak, R.A.; Lutcavage, M.E. Lipid corrections in carbon and nitrogen stable isotope analyses: Comparison of chemical extraction and modelling methods. J. Anim. Ecol. 2008, 77, 838–846. [Google Scholar] [CrossRef]
- Post, D.M.; Layman, C.A.; Arrington, D.A.; Takimoto, G.; Quattrochi, J.; Montaña, C.G. Getting to the fat of the matter: Models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 2007, 152, 179–189. [Google Scholar] [CrossRef]
- Hussey, N.E.; Olin, J.A.; Kinney, M.J.; McMeans, B.C.; Fisk, A.T. Lipid extraction effects on stable isotope values (δ13C and δ15N) of elasmobranch muscle tissue. J. Exp. Mar. Biol. Ecol. 2012, 434, 7–15. [Google Scholar] [CrossRef]
- Cloyed, C.S.; DaCosta, K.P.; Hodanbosi, M.R.; Carmichael, R.H. The effects of lipid extraction on δ13C and δ15N values and use of lipid-correction models across tissues, taxa and trophic groups. Methods Ecol. Evol. 2020, 11, 751–762. [Google Scholar] [CrossRef]
- Skinner, M.M.; Martin, A.A.; Moore, B.C. Is lipid correction necessary in the stable isotope analysis of fish tissues? Rapid Commun. Mass Spectrom. 2016, 30, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Layman, C.A.; Araujo, M.S.; Boucek, R.; Hammerschlag-Peyer, C.M.; Harrison, E.; Jud, Z.R.; Matich, P.; Rosenblatt, A.E.; Vaudo, J.J.; Yeager, L.A.; et al. Applying stable isotopes to examine food-web structure: An overview of analytical tools. Biol. Rev. 2012, 87, 545–562. [Google Scholar] [CrossRef]
- Lovell, M. Seasonal Variation in the Feeding Ecology of Yellowfin Tuna (Thunnus albacares) from the Northern Gulf of Mexico. Master’s Thesis, Louisiana State University, Baton Rouge, LA, USA, 2021. [Google Scholar]
- Kim, S.L.; Koch, P.L. Methods to collect, preserve, and prepare elasmobranch tissues for stable isotope analysis. Environ. Biol. Fishes 2012, 95, 53–63. [Google Scholar] [CrossRef]
- Fry, B. Stable isotopic indicators of habitat use by Mississippi River fish. J. N. Am. Benthol Soc. 2002, 21, 676–685. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 30 June 2023).
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer: New York, NY, USA, 2002; p. 488. [Google Scholar]
- Carlisle, A.B.; Litvin, S.Y.; Madigan, D.J.; Lyons, K.; Bigman, J.S.; Ibarra, M.; Bizzarro, J.J. Interactive effects of urea and lipid content confound stable isotope analysis in elasmobranch fishes. Can. J. Fish Aquat. Sci. 2016, 74, 419–428. [Google Scholar] [CrossRef]
- Logan, J.M.; Golet, W.; Smith, S.C.; Neilson, J.; Van Guelpen, L. Broadbill swordfish (Xiphias gladius) foraging and vertical movements in the north-west Atlantic. J. Fish Biol. 2021, 99, 557–568. [Google Scholar] [CrossRef]
- Goñi, N.; Arrizabalaga, H. Seasonal and interannual variability of fat content of juvenile albacore (Thunnus alalunga) and bluefin (Thunnus thynnus) tunas during their feeding migration to the Bay of Biscay. Prog. Oceanogr. 2010, 86, 115–123. [Google Scholar] [CrossRef]
- Logan, J.M.; Lutcavage, M.E. A comparison of carbon and nitrogen stable isotope ratios of fish tissues following lipid extractions with non-polar and traditional chloroform/methanol solvent systems. Rapid Commun. Mass Spectrom. 2008, 22, 1081–1086. [Google Scholar] [CrossRef]
- Sardenne, F.; Ménard, F.; Degroote, M.; Fouché, E.; Guillou, G.; Lebreton, B.; Hollanda, S.J.; Bodin, N. Methods of lipid-normalization for multi-tissue stable isotope analyses in tropical tuna. Rapid Commun. Mass Spectrom. 2015, 29, 1253–1267. [Google Scholar] [CrossRef]
- Kiljunen, M.; Grey, J.; Sinisalo, T.; Harrod, C.; Immonen, H.; Jones, R.I. A revised model for lipid-normalizing δ13C values from aquatic organisms, with implications for isotope mixing models. J. Appl. Ecol. 2006, 43, 1213–1222. [Google Scholar] [CrossRef]
- Hoffman, J.C.; Sutton, T.T. Lipid correction for carbon stable isotope analysis of deep-sea fishes. Deep Sea Res. Part I 2010, 57, 956–964. [Google Scholar] [CrossRef]
- Patterson, H.K.; Carmichael, R.H. The effect of lipid extraction on carbon and nitrogen stable isotope ratios in oyster tissues: Implications for glycogen-rich species. Rapid Commun. Mass Spectrom. 2016, 30, 2594–2600. [Google Scholar] [CrossRef]
- Le-Alvarado, M.; Romo-Curiel, A.E.; Sosa-Nishizaki, O.; Hernández-Sánchez, O.; Barbero, L.; Herzka, S.Z. Yellowfin tuna (Thunnus albacares) foraging habitat and trophic position in the Gulf of Mexico based on intrinsic isotope tracers. PLoS ONE 2021, 16, e0246082. [Google Scholar] [CrossRef]
- Olson, R.J.; Popp, B.N.; Graham, B.S.; López-Ibarra, G.A.; Galván-Magaña, F.; Lennert-Cody, C.E.; Bocanegra-Castillo, N.; Wallsgrove, N.J.; Gier, E.; Alatorre-Ramírez, V.; et al. Food-web inferences of stable isotope spatial patterns of copepods and yellowfin tuna in the pelagic eastern Pacific Ocean. Prog. Oceanogr. 2010, 86, 124–138. [Google Scholar] [CrossRef]
- Richards, T.M.; Gipson, E.E.; Cook, A.; Sutton, T.T.; Wells, R.J.D. Trophic ecology of meso- and bathypelagic predatory fishes in the Gulf of Mexico. ICES J. Mar. Sci. 2019, 76, 662–672. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).