Molecular Characterization and Expression Analysis of Four Janus Kinases (JAK1, JAK2a, JAK3 and TYK2) from Golden Pompano (Trachinotus ovatus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Molecular Cloning of JAKs cDNA
2.4. DNA Sequence Analysis
2.5. Basal Tissue Expression of trJAK1, trJAK2a, trJAK3 and trTYK2
2.6. Expression of trJAK1, trJAK2a, trJAK3 and trTYK2 post LPS, Poly I:C and V. alginolyticus Stimulation
2.7. qPCR
3. Results
3.1. Sequence Characteristics of trJAK1, trJAK2a, trJAK3 and trTYK2
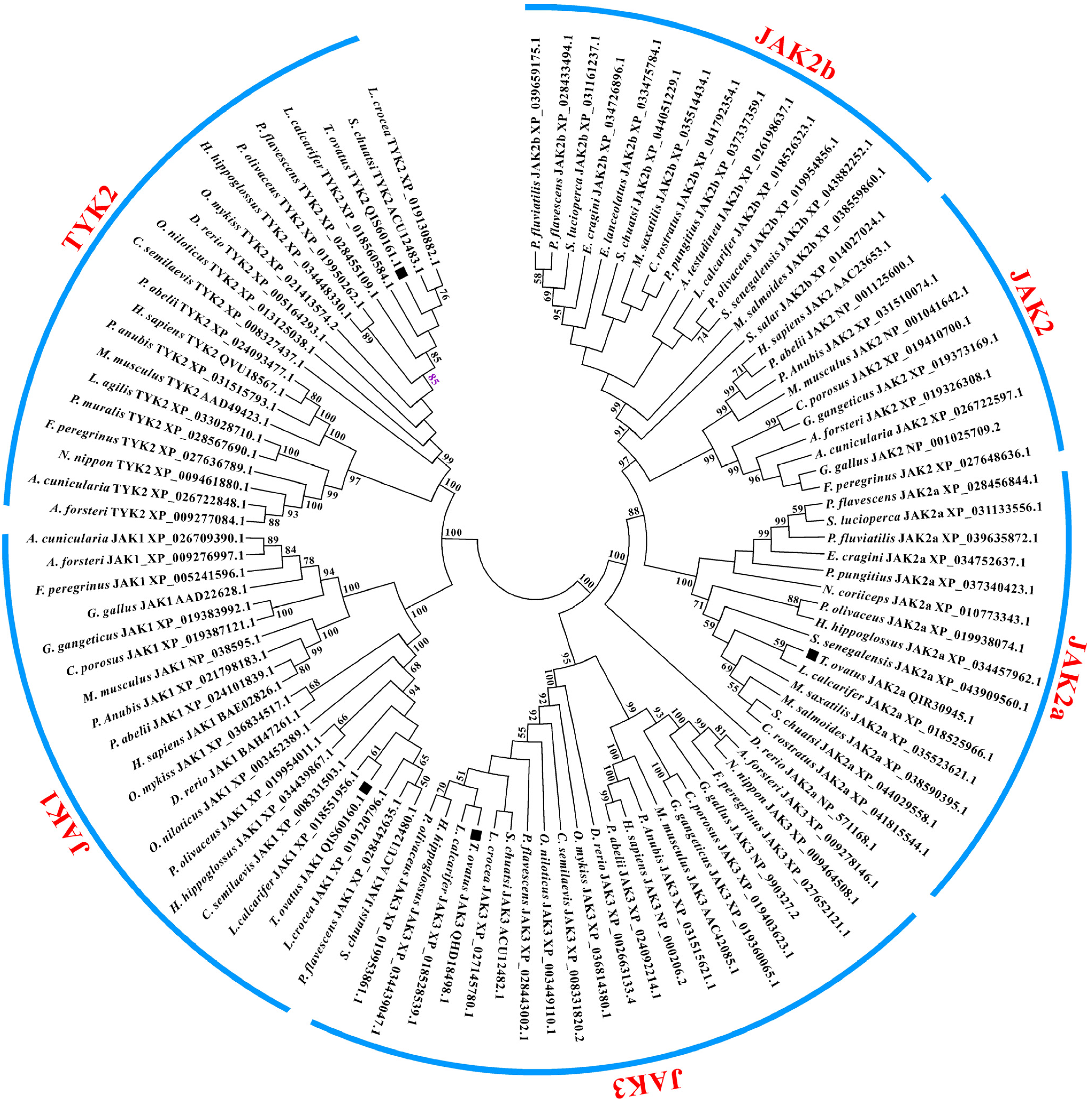
3.2. Homology and Phylogenetic Analysis
3.3. Expression Profiles of trJAK1, trJAK2a, trJAK3 and trTYK2
3.4. Expression of trJAK1, trJAK2a, trJAK3 and trTYK2 after LPS, Poly I:C and V. alginolyticus Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Wu, J.; Ye, B.; Wang, Q.; Xie, X.; Shen, H. Protective effect of curcumin on TNBS-induced intestinal inflammation is mediated through the JAK/STAT pathway. BMC Complement. Altern. Med. 2016, 16, 299. [Google Scholar] [CrossRef] [PubMed]
- Sabaawy, H.E.; Ryan, B.M.; Khiabanian, H.; Pine, S.R. JAK/STAT of all trades: Linking inflammation with cancer development, tumor progression and therapy resistance. Carcinogenesis 2021, 42, 1411–1419. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; Ferdinand, J.R.; O’Shea, J.J. Mechanisms of Jak/STAT Signaling in Immunity and Disease. J. Immunol. 2015, 194, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ezeonwumelu, I.J.; Garcia-Vidal, E.; Ballana, E. JAK-STAT Pathway: A Novel Target to Tackle Viral Infections. Viruses 2021, 13, 2379. [Google Scholar] [CrossRef]
- Raftery, N.; Stevenson, N.J. Advances in anti-viral immune defence: Revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 2017, 74, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, S.; Wang, Y.; Hu, L.; Zhu, L. Inhibition of gp130 alleviates LPS-induced lung injury by attenuating apoptosis and inflammation through JAK1/STAT3 signaling pathway. Inflamm. Res. 2023, 72, 493–507. [Google Scholar] [CrossRef]
- Li, X.; Liu, J. FANCD2 inhibits ferroptosis by regulating the JAK2/STAT3 pathway in osteosarcoma. BMC Cancer 2023, 23, 179. [Google Scholar] [CrossRef]
- Ociepa, K.; Danilewicz, M.; Wągrowska-Danilewicz, M.; Peterson-Jęckowska, R.; Wójcicka-Rubin, A.; Lewkowicz, N.; Zajdel, R.; Żebrowska, A. Expression of the Selected Proteins of JAK/STAT Signaling Pathway in Diseases with Oral Mucosa Involvement. Int. J. Mol. Sci. 2023, 24, 323. [Google Scholar] [CrossRef]
- Wu, J.; Del Duca, E.; Espino, M.; Gontzes, A.; Cueto, I.; Zhang, N.; Estrada, Y.D.; Pavel, A.B.; Krueger, J.G.; Guttman-Yassky, E. RNA Sequencing Keloid Transcriptome Associates Keloids with Th2, Th1, Th17/Th22, and JAK3-Skewing. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Rizi, F.Z.; Ghorbani, A.; Zahtab, P.; Darbaghshahi, N.N.; Ataee, N.; Pourhamzeh, P.; Hamzei, B.; Dolatabadi, N.F.; Zamani, A.; Hooshmand, M. TYK2 single-nucleotide variants associated with the severity of COVID-19 disease. Arch. Virol. 2023, 168, 1–8. [Google Scholar] [CrossRef]
- Villarino, A.V.; Gadina, M.; O’Shea, J.J.; Kanno, Y. Snapshot: JAK-STAT signaling II. Cell 2020, 181, 1696.e1. [Google Scholar] [CrossRef]
- Gadina, M.; Chisolm, D.A.; Philips, R.L.; McInness, I.B.; Changelian, P.S.; O’Shea, J.J. Translating JAKs to Jakinibs. J. Immunol. 2020, 204, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.H.; Yan, S.J.; Lee, T.F.; Chou, C.M.; Chen, S.T.; Hwang, P.P.; Chou, C.K.; Huang, C.J. Complete genomic organization and promoter analysis of the round-spotted pufferfish JAK1, JAK2, JAK3, and TYK2 genes. DNA Cell Biol. 2000, 19, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-J.; Zhang, Y.-F.; Yang, L.-S.; Yang, X.-B.; Wu, Y.-Y.; Liu, D.; Chen, W.-J.; Weng, S.-P.; Yu, X.-Q.; He, J.-G. The JAK and STAT family members of the mandarin fish Siniperca chuatsi: Molecular cloning, tissues distribution and immunobiological activity. Fish Shellfish Immunol. 2009, 27, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Tan, X.-Y.; Xu, Y.-H.; Chen, Q.-L.; Pan, Y.-X. JAK and STAT members of yellow catfish Pelteobagrus fulvidraco and their roles in leptin affecting lipid metabolism. Gen. Comp. Endocrinol. 2016, 226, 14–26. [Google Scholar] [CrossRef]
- Wu, K.; Tan, X.-Y.; Xu, Y.-H.; Shi, X.; Fan, Y.-F.; Li, D.-D.; Liu, X. JAK family members: Molecular cloning, expression profiles and their roles in leptin influencing lipid metabolism in Synechogobius hasta. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 203, 122–131. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, T.; Li, N.; Liu, S.; Xu, X.; Pan, Y.; Tan, S.; Shi, H.; Yang, Y.; Yuan, Z.; et al. JAK and STAT members in channel catfish: Identification, phylogenetic analysis and expression profiling after Edwardsiella ictaluri infection. Dev. Comp. Immunol. 2018, 81, 334–341. [Google Scholar] [CrossRef]
- Ward, A.E.; Rosenthal, B.M. Evolutionary responses of innate immunity to adaptive immunity. Infect. Genet. Evol. 2014, 21, 492–496. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, J.; Wang, J.; Wang, H.; Gao, Z.; Liu, H. Janus kinase 1 in Megalobrama amblycephala: Identification, phylogenetic analysis and expression profiling after Aeromonas hydrophila infection. Fish Shellfish Immunol. 2023, 135. [Google Scholar] [CrossRef]
- Meng, X.Y.; Wang, Z.H.; Yu, X.D.; Zhang, Q.Y.; Ke, F. Development and characterization of a skin cell line from Chinese perch (Siniperca chuatsi ) and its application in aquatic animal viruses. J. Fish Dis. 2022, 45, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guan, Y.; Guan, H.; Mu, Y.; Ding, Y.; Zou, J.; Ouyang, S.; Chen, X. Molecular and Structural Basis of Receptor Binding and Signaling of a Fish Type I IFN with Three Disulfide Bonds. J. Immunol. 2022, 209, 806–819. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Teng, Y.; Wu, H.; Li, X.; Huo, J.; Ao, J.; Chen, X. Large yellow croaker (Lrimichthys crocea) IL-2 modulates humoral immunity via the conserved JAK-STAT5 signal pathway. Fish Shellfish Immunol. 2023, 133. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liao, J.; Zhang, D.; Liu, S.; Zhang, L.; Kang, S.; Xu, L.; Chen, H.; Peng, W.; Zhou, S.; et al. Isolation, Characterization, and Transcriptome Analysis of an ISKNV-Like Virus from Largemouth Bass. Viruses 2023, 15, 398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Shi, H.; Song, N.; Han, F.; Chai, X.; Liu, Q.; Wang, Y.; Gao, T. Comparative transcriptomic analysis of the liver and spleen in marbled rockfish (Sebastiscus marmoratus) challenged with polyriboinosinic polyribocytidylic acid (poly(I:C)). Aquaculture 2022, 554, 738144. [Google Scholar] [CrossRef]
- Zhang, X.; Hao, X.; Ma, W.; Zhu, T.; Zhang, Z.; Wang, Q.; Liu, K.; Shao, C.; Wang, H.-Y. Transcriptome Analysis Indicates Immune Responses against Vibrio harveyi in Chinese Tongue Sole (Cynoglossus semilaevis). Animals 2022, 12, 1144. [Google Scholar] [CrossRef]
- Song, Y.; Dong, X.; Hu, G. Transcriptome analysis of turbot (Scophthalmus maximus) head kidney and liver reveals immune mechanism in response to Vibrio anguillarum infection. J. Fish Dis. 2022, 45, 1045–1057. [Google Scholar] [CrossRef]
- Fu, Q.; Li, Y.; Zhao, S.; Wang, H.; Zhao, C.; Zhang, P.; Cao, M.; Yang, N.; Li, C. Comprehensive identification and expression profiling of immune-related lncRNAs and their target genes in the intestine of turbot (Scophthalmus maximus L.) in response to Vibrio anguillarum infection. Fish Shellfish Immunol. 2022, 130, 233–243. [Google Scholar] [CrossRef]
- Gao, C.; Cai, X.; Ma, L.; Sun, P.; Li, C. Systematic analysis of circRNA-related ceRNA networks of black rockfish (Sebastes schlegelii) in response to Aeromonas salmonicides infection. Fish Shellfish Immunol. 2023, 135, 108648. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, J.; Wu, S.; Li, Y.; Pan, Y. Integrative analysis of miRNA and mRNA expression associated with the immune response in the intestine of rainbow trout (Oncorhynchus mykiss) infected with infectious hematopoietic necrosis virus. Fish Shellfish Immunol. 2022, 131, 54–66. [Google Scholar] [CrossRef]
- Di, G.; Li, H.; Zhao, Y.; Lin, Y.; Lan, D.; Kong, X.; Chen, X. Comprehensive transcriptomic analysis reveals insights into the gill response to hypoxia and Poly I:C in Qihe crucian carp Carassius auratus. Aquac. Rep. 2022, 24, 101154. [Google Scholar] [CrossRef]
- Gao, J.; Guo, H.-Y.; Liu, M.-J.; Zhu, K.-C.; Liu, B.; Liu, B.-S.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Transcriptome Analysis of the Immune Process of Golden Pompano (Trachinotus ovatus) Infected with Streptococcus agalactiae. Fishes 2023, 8, 52. [Google Scholar] [CrossRef]
- Guo, H.-Y.; Li, W.-F.; Zhu, K.-C.; Liu, B.-S.; Zhang, N.; Liu, B.; Yang, J.-W.; Zhang, D.-C. Pathology, Enzyme Activity and Immune Responses after Cryptocaryon irritans Infection of Golden Pompano Trachinotus ovatus (Linnaeus 1758). J. Mar. Sci. Eng. 2023, 11, 262. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, X.; Kong, B.; Sun, Q.; Ji, H.; Liu, S. Effects of magnetic field-assisted immersion freezing at different magnetic field intensities on the muscle quality of golden pompano (Trachinotus ovatus). Food Chem. 2023, 407, 135092. [Google Scholar] [CrossRef]
- Xie, Y.; Gao, S.; Cao, Y.; Ji, Y.; Zhang, Q.; Wei, Y.; Qi, Z. Molecular characterization and functional analysis of DIGIRR from golden pompano (Trachinotus ovatus). Front. Immunol. 2022, 13, 974310. [Google Scholar] [CrossRef]
- Xie, Y.; Lei, K.; He, J.; Wei, Y. Molecular Characterization and Expression Analysis of TAK1, TAB1 and TAB2 of Golden Pompano (Trachinotus ovatus). Fishes 2022, 7, 173. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Geven, E.J.; Klaren, P.H. The teleost head kidney: Integrating thyroid and immune signalling. Dev. Comp. Immunol. 2017, 66, 73–83. [Google Scholar] [CrossRef]
- He, Y.; Wang, E.; Wang, K.; Wang, J.; Fan, W.; Chen, D.; Yang, Q. Morphology of the Spleen in Oreochromis niloticus: Splenic Subregions and the Blood-Spleen Barrier. Animals 2021, 11, 2934. [Google Scholar] [CrossRef]
- Huo, J.; Hu, X.; Bai, J.; Lv, A. Multiomics analysis revealed miRNAs as potential regulators of the immune response in Carassius auratus gills to Aeromonas hydrophila infection. Front. Immunol. 2023, 14, 1098455. [Google Scholar] [CrossRef]
- Akiyoshi, H.; Inoue, A. Comparative Histological Study of Teleost Livers in Relation to Phylogeny. Zoöl. Sci. 2004, 21, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Villarino, A.V.; Kanno, Y.; O’Shea, J.J. Mechanisms and consequences of Jak–STAT signaling in the immune system. Nat. Immunol. 2017, 18, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Wan, Z.; Yang, L.; Li, W.; Wang, Q. JAK/STAT signalling regulates antimicrobial activities in Eriocheir sinensis. Fish Shellfish Immunol. 2019, 84, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.-Q.; Gao, L.; Yan, M.; Kang, C.-J. Peroxiredoxin 4 Interacts with Domeless and Participates in Antibacterial Immune Response Through the JAK/STAT Pathway. Front. Immunol. 2022, 13, 907183. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, B.-R.; Zhang, H.; You, Y.-L.; Li, F.; Wang, X.-W. Interferon functional analog activates antiviral Jak/Stat signaling through integrin in an arthropod. Cell Rep. 2021, 36, 109761. [Google Scholar] [CrossRef]
- Bang, I.S. JAK/STAT signaling in insect innate immunity. Èntomol. Res. 2019, 49, 339–353. [Google Scholar] [CrossRef]
- Takahashi, T.; Shirasawa, T. Molecular cloning of rat JAK3, a novel member of the JAK family of protein tyrosine kinases. FEBS Lett. 1994, 342, 124–128. [Google Scholar] [CrossRef]
- Haan, C.; Kreis, S.; Margue, C.; Behrmann, I. Jaks and cytokine receptors—An intimate relationship. Biochem. Pharmacol. 2006, 72, 1538–1546. [Google Scholar] [CrossRef]
- Shuai, K.; Liu, B. Regulation of JAK–STAT signalling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Haan, S.; Margue, C.; Engrand, A.; Rolvering, C.; de Leur, H.S.-V.; Heinrich, P.C.; Behrmann, I.; Haan, C. Dual Role of the Jak1 FERM and Kinase Domains in Cytokine Receptor Binding and in Stimulation-Dependent Jak Activation. J. Immunol. 2008, 180, 998–1007. [Google Scholar] [CrossRef]
- Hou, Q.; Gong, R.; Liu, X.; Mao, H.; Xu, X.; Liu, D.; Dai, Z.; Wang, H.; Wang, B.; Hu, C. Poly I:C facilitates the phosphorylation of Ctenopharyngodon idellus type I IFN receptor subunits and JAK kinase. Fish Shellfish Immunol. 2017, 60, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Sobhkhez, M.; Hansen, T.; Iliev, D.B.; Skjesol, A.; Jørgensen, J.B. The Atlantic salmon protein tyrosine kinase Tyk2: Molecular cloning, modulation of expression and function. Dev. Comp. Immunol. 2013, 41, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Gao, Q.; Liu, D.; Fu, Y.; Yuan, Y.; Wen, C. Molecular identification of tyrosine kinase 2 (TYK2) gene from common carp Cyprinus carpio and its expression pattern. Acta Hydrobiol. Sin. 2015, 39, 229–233. [Google Scholar]
- Wu, X.; Li, X.; Leng, X.; Guan, L.; Guo, T. Cloning and tissue expression of JAK2 gene in grass carp (Ctenopharyngodon idella). J. Shanghai Ocean Univ. 2012, 21, 21–27. [Google Scholar]
- Bjørgen, H.; Koppang, E.O. Anatomy of teleost fish immune structures and organs. Immunogenetics 2021, 73, 53–63. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Wu, X.; Liu, H. Molecular cloning and expression of TYK2 in Megalobrama amblycephala. J. Huazhong Agric. Univ. 2021, 40, 197–205. [Google Scholar]
- Li, Y.; Xia, P.; Wu, J.; Huang, A.; Bu, G.; Meng, F.; Kong, F.; Cao, X.; Han, X.; Yu, G.; et al. The potential sensing molecules and signal cascades for protecting teleost fishes against lipopolysaccharide. Fish Shellfish Immunol. 2020, 97, 235–247. [Google Scholar] [CrossRef]
- Li, D.; Cui, Z.; Zhao, F.; Zhu, X.; Tan, A.; Deng, Y.; Lai, Y.; Huang, Z. Characterization of snakehead (Channa argus) interleukin-21: Involvement in immune defense against two pathogenic bacteria, in leukocyte proliferation, and in activation of JAK–STAT signaling pathway. Fish Shellfish Immunol. 2022, 123, 207–217. [Google Scholar] [CrossRef]
- Kimura, A.; Naka, T.; Muta, T.; Takeuchi, O.; Akira, S.; Kawase, I.; Kishimoto, T. Suppressor of cytokine signaling-1 selectively inhibits LPS-induced IL-6 production by regulating JAK–STAT. Proc. Natl. Acad. Sci. USA 2005, 102, 17089–17094. [Google Scholar] [CrossRef] [PubMed]
- Kamezaki, K.; Shimoda, K.; Numata, A.; Matsuda, T.; Nakayama, K.-I.; Harada, M. The role of Tyk2, Stat1 and Stat4 in LPS-induced endotoxin signals. Int. Immunol. 2004, 16, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, N.; Gao, X.; Zhang, Y.; Li, X.; Zhang, Y.; Bing, X.; Huang, H.; Zhang, X. The infection of red seabream iridovirus in mandarin fish (Siniperca chuatsi) and the host immune related gene expression profiles. Fish Shellfish Immunol. 2018, 74, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Li, M.; Ding, F.; Ding, Y.; Ao, J.; Hu, S.; Chen, X. De Novo Characterization of the Spleen Transcriptome of the Large Yellow Croaker (Pseudosciaena crocea) and Analysis of the Immune Relevant Genes and Pathways Involved in the Antiviral Response. PLoS ONE 2014, 9, e97471. [Google Scholar] [CrossRef]
- Liu, Q.-N.; Tang, Y.-Y.; Zhou, M.-J.; Luo, S.; Li, Y.-T.; Wang, G.; Zhang, D.-Z.; Yang, H.; Tang, B.-P.; He, W.-F. Differentially expressed genes involved in immune pathways from yellowhead catfish (Tachysurus fulvidraco) after poly (I:C) challenge. Int. J. Biol. Macromol. 2021, 183, 340–345. [Google Scholar] [CrossRef]
- Lu, J.; Xu, D.; Shen, Z.; Lu, L. Differential expression of miRNA in Carassius auratus gibelio in response to cyprinid herpesvirus 2 infection. Dev. Comp. Immunol. 2018, 82, 1–6. [Google Scholar] [CrossRef]
- Lu, J.; Xu, D.; Jiang, Y.; Kong, S.; Shen, Z.; Xia, S.; Lu, L. Integrated analysis of mRNA and viral miRNAs in the kidney of Carassius auratus gibelio response to cyprinid herpesvirus 2. Sci. Rep. 2017, 7, 13787. [Google Scholar] [CrossRef]
- Kotob, M.H.; Kumar, G.; Saleh, M.; Gorgoglione, B.; Abdelzaher, M.; El-Matbouli, M. Differential modulation of host immune genes in the kidney and cranium of the rainbow trout (Oncorhynchus mykiss) in response to Tetracapsuloides bryosalmonae and Myxobolus cerebralis co-infections. Parasites Vectors 2018, 11, 326. [Google Scholar] [CrossRef]
- Neves, J.V.; Caldas, C.; Wilson, J.M.; Rodrigues, P.N.S. Molecular mechanisms of hepcidin regulation in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol. 2011, 31, 1154–1161. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Yu, B.; Huang, S.; Li, Y.; Wu, J. Study on separation of hemolytic Aeromonas hydrophila strain XF-6 from Tenualosa reevesii and its effect on iron metabolism. Southwest China. J. Agric. Sci. 2018, 31, 862–868. [Google Scholar]
- Yan, H.; Wang, H.; Yu, B.; Huang, S.; Li, Y.; Wu, J. Isolation of pathogenic Edwardsiella tarda strain CA26 from Silurus asotus and its effect on immune factors. J. Henan Agric. Sci. 2018, 47, 144–149, 160. [Google Scholar]
- Yang, Y.; Fu, X.; Chen, B.; Wang, W.; Liu, H. Effect of Aeromonas hydiophila infection on iron metabolism in Megalobrama amblycephala liver. J. Huazhong Agric. Univ. 2015, 34, 97–101. [Google Scholar]
- Majoros, A.; Platanitis, E.; Kernbauer-Hölzl, E.; Rosebrock, F.; Müller, M.; Decker, T. Canonical and Non-Canonical Aspects of JAK–STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef]
- Skjesol, A.; Liebe, T.; Iliev, D.B.; Thomassen, E.I.S.; Tollersrud, L.G.; Sobhkhez, M.; Joensen, L.L.; Secombes, C.J.; Jørgensen, J.B. Functional conservation of suppressors of cytokine signaling proteins between teleosts and mammals: Atlantic salmon SOCS1 binds to JAK/STAT family members and suppresses type I and II IFN signaling. Dev. Comp. Immunol. 2014, 45, 177–189. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Wang, S.; Lv, Y.; Deng, H.; Chang, K.; Zhou, P.; Hu, C. Grass carp (Ctenopharyngodon idella) TNK1 modulates JAK-STAT signaling through phosphorylating STAT1. Dev. Comp. Immunol. 2021, 116, 103951. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.-C.; Guo, H.-Y.; Zhang, N.; Liu, B.-S.; Guo, L.; Jiang, S.-G.; Zhang, D.-C. Functional characterization of IRF8 regulation of type II IFN in golden pompano (Trachinotus ovatus). Fish Shellfish Immunol. 2019, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, Y.; Cao, Z.; Sun, Y.; Chen, Y.; Xiang, Y.; Wang, L.; Zhang, S.; Guo, W. Comparative analysis of the expression patterns of IL-1β, IL-11, and IL-34 in golden pompano (Trachinotus ovatus) following different pathogens challenge. Fish Shellfish Immunol. 2019, 93, 863–870. [Google Scholar] [CrossRef]






| Primer Name | Sequence (5′ to 3′) | Application |
|---|---|---|
| Nested-PCR | ||
| JAK1-F1 | AACGAAACGGAAGTAACTTGCCA | trJAK1 partial sequence |
| JAK1-F2 | GGCTCAACTGTCCTCTTCTCCTACT | |
| JAK1-R1 | AAATGAAGCCGCACAACAGATG | |
| JAK1-R2 | CCCAGAAAGTCAAACAGTCGGA | |
| JAK2a-F1 | GACGAGGCAGCTACTTTTATTGGCA | trJAK2a partial sequence |
| JAK2a-F2 | CAGCAGAGGACAACAGGCAGAGTG | |
| JAK2a-R1 | TGCCGTCACTCCAACATCTTCATT | |
| JAK2a-R2 | TGCTGCCATAAACCCAGACATCAC | |
| JAK3-F1 | CACAGCATAAACCCAGAAGTCAAAGAA | trJAK3 partial sequence |
| JAK3-F2 | GGTCCCAGTTTACAAGTACACATCT | |
| JAK3-R1 | GCTTGCTTGAGGTCGTGTCTGTTC | |
| JAK3-R2 | GTGCCCAAAACTGCTGTGTGTCTTA | |
| TYK2-F1 | GAGAATCACGGAGAGTAGGCGACC | trTYK2 partial sequence |
| TYK2-F2 | TGTAACACCCTCAGAAGCTCGTCG | |
| TYK2-R1 | ACCCATCAGCCAGGAAATAAAGACA | |
| TYK2-R2 | GCATCGCACAGCACGAGATACATT | |
| RACE-PCR | ||
| JAK1-5′R1 | TCCTCCTCTGAGTTTGCTGATGCA | trJAK1 5′-UTR |
| JAK1-5′R2 | TTCAGTGGTGCCATGCCAATTTC | |
| JAK1-3′F1 | AGAGCATCAAGGACAACGAGGGATA | trJAK1 3′-UTR |
| JAK1-3′F2 | GACTCCTCCAAGAGCCCGATGA | |
| JAK2a-5′R1 | GGGCATCAGCGACCAGTCTGTA | trJAK2a 5′-UTR |
| JAK2a-5′R2 | TATTGGCCTGCTTGATGCTGATG | |
| JAK2a-3′F1 | AGATCCTCAAGTCCCTCCACCATG | trJAK2a 3′-UTR |
| JAK2a-3′F2 | AAGACGGAACCTGCGGCTGAT | |
| JAK3-5′R1 | CATCCACAGAGGAGTTTCCGAGC | trJAK3 5′-UTR |
| JAK3-5′R2 | CGGTTTCGCTTCTGGATGTCAT | |
| JAK3-3′F1 | TTTGGCAGTGTCGAACTTTGTCG | trJAK3 3′-UTR |
| JAK3-3′F2 | CCGGTGAGCTAGTCGCTGTGAA | |
| TYK2-5′R1 | AAGGCGGTCAATTTGTTGGGAGT | trTYK2 5′-UTR |
| TYK2-5′R2 | GAACCTCCATGCAGCGATTGT | |
| TYK2-3′F1 | GGAGTCTGCGAGAGTACCTTCCCA | trTYK2 3′-UTR |
| TYK2-3′F2 | CATCGAGACCTAGCTGCCCGTAA | |
| UPM-Long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | RACE universal primers |
| UPM-Short | CTAATACGACTCACTATAGGGC | |
| NUP | AAGCAGTGGTATCAACGCAGAGT | |
| Primer Name | Sequence (5′ to 3′) | Application |
|---|---|---|
| JAK1-qF | ACGACCCCAAGAAGAGACCT | trJAK1 expression |
| JAK1-qR | CCCGAGGATCATAGCGACAC | |
| JAK2a-qF | GTTTCCACCTGGGGAGTACG | trJAK2a expression |
| JAK2a-qR | CTGGCTGACTGGTCGAGTTT | |
| JAK3-qF | CTGCGACATGAACTGCAACC | trJAK3 expression |
| JAK3-qR | ACTGTACACCTTTGGTGGGC | |
| TYK2-qF | GGAAGAGCGTCTTGAGCGTA | trTYK2 expression |
| TYK2-qR | AGGTAAGCGGCTCTTTTGCT | |
| β-actin-F | GCTACGTCGCCCTGGACTTC | Gene expression |
| β-actin-R | CTCATGGATTCCGCAGGACTC |
| T. ovatus | L. calcarifer | S. chuatsi | P. flavescens | H. hippoglossus | P. olivaceus | D. rerio | H. sapiens | M. musculus |
|---|---|---|---|---|---|---|---|---|
| JAK1 | 92.9 | 90.0 | 89.4 | 86.9 | 86.8 | 73.8 | 60.9 | 61.3 |
| JAK2a | 95.3 | 92.3 | 91.4 | 89.9 | 88.9 | 69.1 | 69.5 | 69.1 |
| JAK3 | 90.5 | 87.6 | 87.1 | 85.3 | 88.5 | 67.0 | 50.6 | 50.2 |
| TYK2 | 85.5 | 86.3 | 84.3 | 82.1 | 83.2 | 66.3 | 51.9 | 51.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Chen, M.; Han, P.; Liang, X.; Yang, M.; Lu, Z.; Wei, Y. Molecular Characterization and Expression Analysis of Four Janus Kinases (JAK1, JAK2a, JAK3 and TYK2) from Golden Pompano (Trachinotus ovatus). Fishes 2023, 8, 245. https://doi.org/10.3390/fishes8050245
Xie Y, Chen M, Han P, Liang X, Yang M, Lu Z, Wei Y. Molecular Characterization and Expression Analysis of Four Janus Kinases (JAK1, JAK2a, JAK3 and TYK2) from Golden Pompano (Trachinotus ovatus). Fishes. 2023; 8(5):245. https://doi.org/10.3390/fishes8050245
Chicago/Turabian StyleXie, Yushuai, Mingqu Chen, Pengfu Han, Xiang Liang, Meng Yang, Zhuanling Lu, and Youchuan Wei. 2023. "Molecular Characterization and Expression Analysis of Four Janus Kinases (JAK1, JAK2a, JAK3 and TYK2) from Golden Pompano (Trachinotus ovatus)" Fishes 8, no. 5: 245. https://doi.org/10.3390/fishes8050245
APA StyleXie, Y., Chen, M., Han, P., Liang, X., Yang, M., Lu, Z., & Wei, Y. (2023). Molecular Characterization and Expression Analysis of Four Janus Kinases (JAK1, JAK2a, JAK3 and TYK2) from Golden Pompano (Trachinotus ovatus). Fishes, 8(5), 245. https://doi.org/10.3390/fishes8050245





