Abstract
Beneficial microorganisms can increase nutrient digestion and absorption in farmed fish. This study investigates the effects of supplemental feeding of Bacillus species isolated from the intestines of wild glass eels on the growth, survival, and gene expression of farm-raised eel larvae for 30 days after hatching. Three species of Bacillus (B. velezensis, AJBV; B. subtilis, AJBS; B. licheniformis, AJBL) without hemolytic activity were isolated, and an experiment compared the growth of eel larvae fed an artificial diet supplemented with each Bacillus species. There were no significant differences in the total length and body depth of eel larvae at 30 days after hatching in all groups. During the feeding period, 149 eels survived from the initial 1000 in the control group. On the other hand, 240, 178, and 141 eels survived in the AJBV, AJBS, and AJBL groups, respectively. However, there were no significant differences in survival rates despite the difference in the number of surviving eel larvae among the groups. In the comparison of gene expression of genes involved with growth (growth hormone, growth hormone receptor 1, insulin-like growth factor II-2) and those involved with digestive enzymes (amylase, trypsin, lipase), there were also no significant differences among the groups. Our results confirm that dietary supplementation with each of the three host-associated Bacillus does not affect the growth and survival rates of eel larvae reared on an artificial diet up to the first 30 days after hatching, nor does it significantly affect related gene expression.
Key Contribution:
Dietary supplementation with each of the three host-associated Bacillus does not affect the growth and survival rates of eel larvae reared on an artificial diet up to the first 30 days after hatching, nor does it significantly affect related gene expression.
1. Introduction
The eel has long been esteemed as an important food fish not only in East Asian countries such as Japan, Korea, China, and Taiwan but also in European countries [1]. Among eels, the Japanese eel (Anguilla japonica) is one of the species farmed in East Asian countries [2,3]. To date, all the stocking for eel aquaculture is dependent on wild glass eels, naturally sourced juveniles of eel collected from estuaries [1,4]. However, due to overfishing, environmental degradation, climate change, and other unknown factors, the natural stock of Japanese eels has noticeably declined over the past few decades [5]. This decrease creates an unstable supply of glass eels for seeding, which has led to price increases [1]. Therefore, artificial propagation technology should be developed to protect natural glass eel resources and stabilize the aquaculture industry through a stable supply of glass eels for farming, which in turn can stabilize eel prices [1,3].
Research on the production of artificial eel larvae started in Japan in the 1960s. Currently, active research is conducted not only in Japan but also in Korea and China [6,7]. However, only small-scale production is possible in the laboratory because a suitable rearing system and feed have not yet been found [8]. In addition, artificially produced eel larvae exhibit slower growth and lower survival rates than naturally sourced individuals [2,9]. Therefore, further research progress in the rearing system and feed development are needed to overcome these deficiencies.
Many studies on probiotics have shown consistent results in that they can help improve growth and immunity in various farmed fish species [10]. In addition, recently, it has also been reported that bacteria isolated from the intestines of the fish studied may be the best potential probiotics [11,12,13]. Later these were called host-associated probiotics (HAPs) by Van Doan et al. (2020), who defined them as “bacteria originally isolated from the rearing waters or gastrointestinal tract to the host to improve the growth and health of the host” [14]. HAPs may be highly functional because they can evade the host’s defense system and are better adapted to the host’s intestinal environment [13,14].
This study was conducted to verify the effect of host-associated microorganisms on the growth and survival rate of eel larvae reared on an artificial diet. Three species of isolated host-associated microorganisms were separately supplied to eel larvae for 30 days after hatching, and growth and survival rates and related gene expression were investigated.
2. Materials and Methods
2.1. Bacterial Isolation and Identification
Microorganisms were isolated from wild glass eels captured in the Geumgang River (36°00′14.4″ N 126°43′52.4″ E). The glass eels were ground and suspended in 0.85% NaCl, then spread on MRS (Difco, Detroit, MI, USA), LB (Difco, Detroit, MI, USA), and MB (Difco, Detroit, MI, USA) agar plates and cultured at 37 °C for 24 h. Colonies cultured on agar plates were separated according to size, color, and shape and then identified by performing 16S rRNA sequence analysis using 27F (AGA GTT TGA TCM TGG CTC AG) and 1492R (GGT TAC CTT GTT ACG ACT T) 16S universal primers (Bionics, South Korea). A phylogenetic tree of microorganisms based on 16s rRNA sequences was constructed using the neighbor-joining algorithm of the Molecular Evolutionary Genetics Analysis version 7 (MEGA7) software. Hemolytic activity was determined by incubating the microorganisms on a 5% sheep’s blood agar base plate (Kisan Bio, Seoul, Republic of Korea) and confirming the formation of a clear area around the colony.
2.2. Experimental Artificial Diet Preparation
Table 1 lists the ingredients for preparing the slurry-type feed used in the experiment. Each feed component was mixed and filtered at 90 μm before use [3]. Microorganisms were cultured at 37 °C in LB broth and then washed twice with phosphate-buffered saline and treated to a final concentration of 1 × 1011 colony-forming units (CFU)/mL.

Table 1.
Composition of the basal experimental diet.
2.3. Rearing System and Condition for the Feeding Trial
Eel larvae were provided by the National Institute of Fisheries Science. The rearing system and environment were the same as above [3]. Briefly, a feeding experiment was conducted in a 20 L round acrylic resin tank accommodating 330 eel larvae (3 tank/group, triplicates). Water quality was regularly monitored, and stable environmental parameters were maintained. The water temperature was maintained at 23 ± 0.1 °C, and the flow rate was 0.99–1.13 L/min. Feed was provided five times (09:00, 11:00, 13:00, 15:00, and 17:00) daily at the bottom of each tank after stopping the flow of water.
2.4. Growth and Survival Rate
The growth and survival rates of eel larvae supplied with the experimental feed were calculated 30 days after hatching. Thirty eel larvae were randomly selected per tank, and the total length and body depth were measured under a stereomicroscope (Stereo Discovery V.20, Carl Zeiss, Jena, Germany). Deformed individuals were excluded. The survival rate was calculated using the following equation: Survival rate (%) = the number of surviving eel larvae/number of hatched eel larvae × 100.
2.5. Gene Expression Analysis
Total RNA was isolated by pooling five eel larvae per tank (3tank/group, triplicates). The pooled samples were ground using a disposable tissue grinder pestle (Thermo Fisher Scientific, Waltham, MA, USA), and total RNA was isolated using the Hybrid-R RNA Purification Kit (GeneAll Biotechnology, South Korea) according to the manufacturer’s instructions. Residual DNA was removed using the Riboclear plus kit (GeneAll Biotechnology, Seoul, Republic of Korea), and the purity and concentration of isolated RNA were evaluated using a NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using the PrimeScript 1st strand cDNA Synthesis Kit (Takara, Japan). RT-qPCR was performed using TB Green Premix Ex Taq (Takara, Kusatsu, Japan) on a CFX96 system (Bio-Rad, Hercules, CA, USA) at the Core-Facility Center, Dong-Eui University (Busan, Republic of Korea). Relative quantification was calculated using the 2−ΔΔCT method and normalized to the β-actin gene. Primers used for gene expression analysis are shown in Table 2.

Table 2.
Gene-specific primers used to quantify relative gene expression.
2.6. Statistical Analysis
All data were analyzed by one-way analysis of variance (ANOVA) using IBM’s Statistical Package for the Social Sciences software (SPSS Inc., version 17.0, Chicago, IL, USA) followed by Duncan’s multiple range test. Statistical significance was accepted at a p-value < 0.05 unless otherwise noted.
3. Results and Discussion
3.1. Bacterial Isolation and Identification
Three species of Bacillus (B. velezensis, AJBV; B. subtilis, AJBS; B. licheniformis, AJBL) were identified through 16S rRNA sequence analysis. The three selected Bacillus species shared high similarities with B. velezensis CR-502T, B. subtilis NCIB 3610T, and B. licheniformis ATCC 14580T, respectively (Figure 1a). Hemolytic activity was not observed in any of the isolated Bacillus strains (Figure 1b).

Figure 1.
Phylogenetic tree and hemolytic activity analysis of the isolated strains. (a) The phylogenetic tree was constructed by the neighbor-joining method using Molecular Evolutionary Genetics Analysis version 7 (MEGA7) with 1000 bootstrap repetitions, after which the 16S rRNA sequences were aligned by the Clustal W program. NCBI accession numbers are presented in parentheses. (b) The hemolytic activity was measured using a blood agar plate, and Pseudomonas aeruginosa had α-hemolytic activity, Staphylococcus aureus had β-hemolytic activity, and Lactiplantibacillus plantarum having γ (non)-hemolytic activity were used as controls. AJBV, Bacillus velezensis; AJBS, B. subtilis; AJBL, B. licheniformis.
Previous studies have reported that the use of probiotics in aquaculture can provide beneficial effects on the growth and immunity of aquacultured animals [10]. Among many probiotic candidates, Bacillus can form spores that can survive in harsh environments such as low pH and high temperature, are non-pathogenic and non-toxic to the host, and produce various antibacterial substances [15]. As such, they have been evaluated as preferred candidates over other genera of probiotics [15]. The reason for selecting the Bacillus species as probiotic candidates for eel larvae in this study is because of their ability to improve feed utilization, growth, and disease resistance. A variety of enzymes produced by Bacillus make them efficient symbionts to aid the metabolism of a wide range of lipids, proteins, and carbohydrates. Probiotics, therefore, may increase digestive enzyme activity that can contribute to fish growth [16,17]. In addition, the antibacterial substances they produce inhibit the growth of pathogenic microorganisms, thereby excluding pathogens through competition for nutrients and space, enhancing resistance to pathogens by upregulating the host’s immunity, and subsequently increasing the survival rates of fish [18,19]. Therefore, in this study, three species of host-associated Bacillus were selected.
3.2. Growth and Survival Rate
The growth and survival rates of eel larvae supplemented with the artificial diet for 30 days are shown in Figure 2. The average total length (TL) of eel larvae was 10.44 ± 0.87 mm with no significant differences (p > 0.05) among the groups. Similarly, there were no significant differences in mean body depth (BD, average 1.15 ± 0.09 mm) and TL/BD values (average 11.11 ± 1.23%). Regarding survival rates, the AJBV group (24.00 ± 12.16%) increased compared to controls (14.90 ± 1.27%), but no significant differences were observed. Survival rates of the AJBS and AJBL groups were 17.80 ± 3.11% and 14.10 ± 1.56%, respectively, and there was no significant difference compared to the controls (Figure 2e).
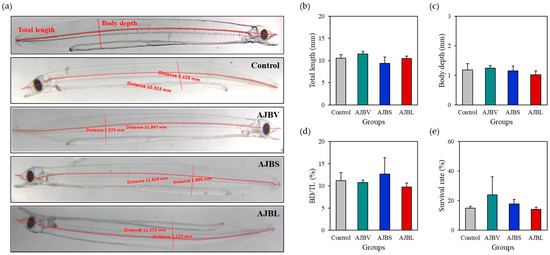
Figure 2.
Growth and survival rate analysis of eel larvae fed a diet containing host-associated Bacillus species (AJBV, Bacillus velezensis; AJBS, B. subtilis; AJBL, B. licheniformis). (a) Total length and body depth were measured using the ZEN pro-2012 (blue edition) program on a stereo microscope. (b) Total length (TL), (c) body depth (BD), (d) BD/TL value, and (e) survival rate of eel larvae at 30 days after hatching are shown as group averages. The data are represented as the means ± standard deviation of three replicates (30 fish/replicate).
In a previous study using Bacillus as a probiotic in the aquaculture of olive flounder (Paralichthys olivaceus) [10,20,21], rockfish (Sebastes schlegelii) [13], red seabream (Pagrus major) [22,23], Nile tilapia (Oreochromis niloticus) [24], and whiteleg shrimp (Litopenaeus vannamei) [25], it was reported that fish growth and immunity increased. However, in this study, the three species of host-associated Bacillus did not affect the growth and survival rates of eel larvae. Although they did not provide beneficial effects on growth and survival rates, they also had no harmful effects. Perhaps under adverse conditions, Bacillus can interfere with the survival of pathogenic strains present in the body and breeding water and can prevent mass mortality due to pathogen infection. In this regard, further studies related to the antibacterial activity of the isolated strains, an extension of the feeding trial period, and an analysis of the effect of using multiple strains need to be conducted to uncover the long-term benefits of probiotics and better understand how the microbiome is important.
3.3. Gene Expression Analysis
The gene expression profile of eel larvae fed a diet containing host-associated Bacillus at 30 days after hatching is shown in Figure 3. Compared to the controls, no significant differences were found in the relative expression levels of growth (growth hormone, growth hormone receptor 1, and insulin-like growth factor II-2) and digestive enzyme (amylase, trypsin, and lipase).
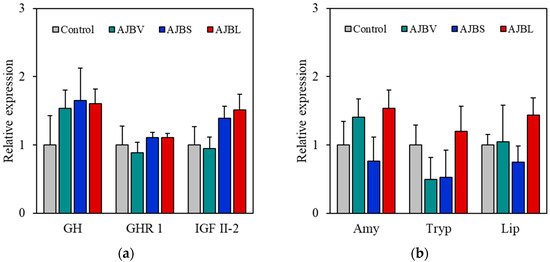
Figure 3.
Gene expression profiles of eel larvae fed a diet containing host-associated Bacillus species (AJBV, Bacillus velezensis; AJBS, B. subtilis; AJBL, B. licheniformis). Comparison of gene expression 30 days after hatching was determined by RT-qPCR analysis. Levels of (a) growth (growth hormone, GH; growth hormone receptor 1, GHR 1; insulin-like growth factor II-2, IGF II-2) and (b) digestive enzyme (amylase, Amy; trypsin, Tryp; lipase, Lip) relative gene expression were quantified relative to β-actin transcription. The data are represented as the means ± standard deviation of three replicates (5 fish/replicate).
Many previous studies have reported that supplementation of probiotics in feed contributes to increased expression of growth-related hormones and digestive enzyme genes in farmed fish [26,27]. However, in this study, supplementation with the three strains of host-associated Bacillus did not significantly affect eel larval growth and digestive enzyme gene expression. The development of the digestive system of eel larvae is very delayed compared to other fish larvae [28]. Their stomach develops during metamorphosis (269–311 days after hatching), meaning that food ingested during the early larval stage enters the intestine directly [29,30]. Ingested food does not remain in the intestine long enough for digestion and absorption of nutrients and is excreted immediately. In addition, the supplemented host-associated Bacillus cannot colonize the host’s intestines because they are excreted together. For this reason, short-term host-associated Bacillus supplementation may not be effective for eel larvae. However, supplying fermented feed using host-associated Bacillus to replace the digestive process in the stomach or supplementing the diet with host-associated Bacillus long after the stomach has developed (after 311 days) may have a positive effect on eel growth and survival rates.
4. Conclusions
In conclusion, feed supplementation with host-associated Bacillus did not significantly affect the growth and survival rates of eel larvae reared on an artificial diet in the early stages of life. Further research will evaluate the growth and survival rate of eel larvae by supplying fermented feed using the three species of host-associated Bacillus isolated in this study.
Author Contributions
Conceptualization, W.J.J., S.-K.K. and J.M.L.; methodology, W.J.J., S.-K.K., S.Y.P., D.P.K., Y.-J.H., H.K., S.-J.L. and M.G.S.; validation, W.J.J., S.-K.K., S.Y.P., D.P.K., Y.-J.H., H.K. and S.-J.L.; investigation, S.Y.P., D.P.K., Y.-J.H., H.K. and S.-J.L.; writing—original draft preparation, W.J.J. and J.M.L.; writing—review and editing, W.J.J. and J.M.L.; supervision, E.-W.L., S.L. and J.M.L.; project administration, E.-W.L., S.L. and J.M.L.; funding acquisition, E.-W.L., S.L. and J.M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the National Institute of Fisheries Science, Republic of Korea (R2023026).
Institutional Review Board Statement
All experiments were performed following the guidelines of the Korean Association for Laboratory Animals (approval no. 2022-NIFS-IACUC-8).
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kagawa, H.; Tanaka, H. The first success of glass eel production in the world: Basic biology on fish reproduction advances new applied technology in aquaculture. Fish Physiol. Biochem. 2005, 31, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Kurokawa, T. Influence of salinity on morphological deformities in cultured larvae of Japanese eel, Anguilla japonica, at completion of yolk resorption. Aquaculture 2009, 293, 113–118. [Google Scholar] [CrossRef]
- Jang, W.J.; Kim, S.K. Effect of Bacillus sp. supplementation diet on survival rate and microbiota composition in artificially produced eel larvae (Anguilla japonica). Front. Microbiol. 2022, 13, 891070. [Google Scholar] [CrossRef]
- Kurokawa, T.; Okamoto, T. Influence of water temperature on morphological deformities in cultured larvae of Japanese eel, Anguilla japonica, at completion of yolk resorption. J. World Aquac. Soc. 2008, 39, 726–735. [Google Scholar] [CrossRef]
- Harrison, A.J.; Walker, A.M. A review of glass eel migratory behaviour, sampling techniques and abundance estimates in estuaries: Implications for assessing recruitment, local production and exploitation. Rev. Fish Biol. Fish. 2014, 24, 967–983. [Google Scholar] [CrossRef]
- Hibiya, T. Success in collecting fully matured eel eggs. Aquaculture 1970, 3, 12–15. [Google Scholar]
- Yamamoto, K.; Yamauchi, K. Sexual maturation of Japanese eel and production of eel larvae in the aquarium. Nature 1974, 251, 220–222. [Google Scholar] [CrossRef]
- Shin, M.G.; Kim, S.K. Histological development of the digestive system in artificially produced Anguilla japonica larvae. Korean J. Fish. Aquat. Sci. 2021, 54, 298–310. [Google Scholar] [CrossRef]
- Tsukamoto, K. Aquaculture production of glass eels as a possible conservation measure for freshwater eels. In Proceedings of the 144th Annual Meeting of the American Fisheries Society 2014, Quebec City, QC, Canada, 17–21 August 2014. [Google Scholar]
- Hasan, M.T.; Jang, W.J. Effects of immunostimulants, prebiotics, probiotics, synbiotics, and potentially immunoreactive feed additives on olive flounder (Paralichthys olivaceus) aquaculture: A review. Rev. Fish. Sci. Aquac. 2019, 27, 417–437. [Google Scholar] [CrossRef]
- Mladineo, I.; Bušelić, I. Autochthonous bacterial isolates successfully stimulate in vitro peripheral blood leukocytes of the European sea bass (Dicentrarchus labrax). Front. Microbiol. 2016, 7, 1244. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Sun, Y.Z. Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 2018, 9, 2429. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.J.; Lee, S.J. Characterization of a Bacillus sp. KRF-7 isolated from the intestine of rockfish and effects of dietary supplementation with mannan oligosaccharide in rockfish aquaculture. Fish Shellfish Immunol. 2021, 119, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Van Doan, H.; Hoseinifar, S.H. Host-associated probiotics: A key factor in sustainable aquaculture. Rev. Fish. Sci. Aquac. 2020, 28, 16–42. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.; Abarike, E.D. A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.J.; Lee, J.M. Effects of probiotic supplementation of a plant-based protein diet on intestinal microbial diversity, digestive enzyme activity, intestinal structure, and immunity in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2019, 92, 719–727. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S. Dietary administration of Bacillus subtilis HAINUP40 enhances growth, digestive enzyme activities, innate immune responses and disease resistance of tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2017, 60, 326–333. [Google Scholar] [CrossRef]
- Cha, J.H.; Rahimnejad, S. Evaluations of Bacillus spp. as dietary additives on growth performance, innate immunity and disease resistance of olive flounder (Paralichthys olivaceus) against Streptococcus iniae and as water additives. Aquaculture 2013, 402, 50–57. [Google Scholar] [CrossRef]
- Cerezuela, R.; Guardiola, F.A. Increases in immune parameters by inulin and Bacillus subtilis dietary administration to gilthead seabream (Sparus aurata L.) did not correlate with disease resistance to Photobacterium damselae. Fish Shellfish Immunol. 2012, 32, 1032–1040. [Google Scholar] [CrossRef]
- Hasan, M.T.; Jang, W.J. Synergistic effects of dietary Bacillus sp. SJ-10 plus β-glucooligosaccharides as a synbiotic on growth performance, innate immunity and streptococcosis resistance in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2018, 82, 544–553. [Google Scholar] [CrossRef]
- Jang, W.J.; Hasan, M.T. Comparison of spore or vegetative Bacillus sp. supplementation on physiological changes and gut microbiota of the olive flounder (Paralichthys olivaceus). Aquaculture 2021, 535, 736355. [Google Scholar] [CrossRef]
- Jang, W.J.; Lee, K.B. Characteristics and biological control functions of Bacillus sp. PM8313 as a host-associated probiotic in red sea bream (Pagrus major) aquaculture. Anim. Nutr. 2023, 12, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.J.; Jeon, M.H. Dietary supplementation of Bacillus sp. PM8313 with β-glucan modulates the intestinal microbiota of red sea bream (Pagrus major) to increase growth, immunity, and disease resistance. Front. Immunol. 2022, 13, 960554. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Hamidoghli, A. Effects of Bacillus subtilis WB60 and Lactococcus lactis on growth, immune responses, histology and gene expression in Nile tilapia, Oreochromis niloticus. Microorganisms 2020, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Hamidoghli, A. Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 2020, 8, 281. [Google Scholar] [CrossRef]
- Olmos, J.; López, L.M. Bacillus subtilis effects on growth performance and health status of Totoaba macdonaldi fed with high levels of soy protein concentrate. Animals 2022, 12, 3422. [Google Scholar] [CrossRef]
- Ringø, E.; Harikrishnan, R.; Soltani, M.; Ghosh, K. The effect of gut microbiota and probiotics on metabolism in fish and shrimp. Animals 2022, 12, 3016. [Google Scholar] [CrossRef]
- Kim, S.K.; Lee, B.I. Development of the slurry type diet for the growing leptocephalus, eel larvae (Anguilla japonica). J. Fish. Mar. Sci. Edu. 2014, 26, 1209–1216. [Google Scholar]
- Kurokawa, T.; Koshio, M. Distribution of pepsinogen- and ghrelin-producing cells in the digestive tract of Japanese eel (Anguilla japonica) during metamorphosis and the adult stage. Gen. Comp. Endocrinol. 2011, 173, 475–482. [Google Scholar] [CrossRef]
- Murashita, K.; Furuita, H. Partial characterization and ontogenetic development of pancreatic digestive enzymes in Japanese eel Anguilla japonica larvae. Fish Physiol. Biochem. 2013, 39, 895–905. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).