Abstract
Nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) family members are innate immune sensors involved in the recognition of highly conserved pathogen-associated molecular patterns (PAMPs). Apoptosis-associated speck-like protein (ASC) is a critical adaptor molecule in multiple inflammasome protein complexes, mediating inflammation and host defense. Caspase1, an inflammatory caspase, has been documented to play important roles in the innate immune system. In this study, we identified and characterized NLRC3-like, ASC, and Caspase1 (referred to as LmNLRC3L, LmASC, and LmCaspase1) from the spotted sea bass (Lateolabrax maculatus). A sequence analysis revealed that LmNLRC3L, LmASC, and LmCaspase1 shared similar features with their fish counterparts. LmNLRC3L contained a FISNA domain, a NACHT domain, and four LRR motifs, followed by a C-terminal fish-specific B30.2 domain. LmASC possessed a PYRIN domain for interacting with inflammasome sensor proteins, as well as a CARD domain. LmCaspase1 had a CARD domain at its N-terminus and a CASC domain at its C-terminus. These three genes were ubiquitously distributed in the liver, spleen, head kidney, gill, intestine, skin, muscle, and brain. They share similar expression patterns, and all demonstrate the highest level of expression in the gill. We analyzed the expression changes in genes in the spleen, gill, and head kidney after stimulation experiments in vivo. After lipopolysaccharide (LPS) stimulation, the expression levels of these three genes were significantly upregulated in the short term, followed by significant downregulation at 48 and 72 h in some examined tissues. Following Edwardsiella tarda infection, these three genes were upregulated in various tissues. However, the expressions of these three genes were not affected by polyinosinic-polycytidylic acid (poly (I:C)) stimulation. Overall, our results indicate that these three genes are involved in the immune response against bacterial infection in the spotted sea bass, providing the foothold for understanding the immune function and mechanism of the fish inflammasome.
Key Contribution:
NLRC3-like, ASC, and Caspase1 could be regulated by PAMPs and bacteria, indicating that they may be involved in the immune response of the spotted sea bass.
1. Introduction
Innate immunity serves as the first line of defense against pathogen invasion, triggered by the recognition of pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs), initiating rapid defense mechanisms [1]. Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are cytosolic PRRs that mediate the immune response against a wide range of pathogens [2]. Based on their N-terminal structural domains, mammalian NLRs can be categorized into five subfamilies: NLRA, NLRB, NLRC, NLRP, and NLRX [3]. NLRC3 is a member of the NLRC subfamily [4] and can interact with apoptosis-associated speck-like protein (ASC) and pro-caspase1 to modulate the maturation and release of Caspase1 [5,6]. ASC, a crucial adaptor molecule in multiple inflammasome protein complexes, contains a pyrin domain (PYD) and a caspase recruitment domain (CARD). Caspase1 is an inflammatory caspase [7] that cleaves the pro-interleukins (IL)-1β and pro-IL-18 to produce mature IL-1β (mIL-1β) and IL-18 (mIL-18) [8,9]. It has been established that NLRC3, ASC and Caspase1 play significant roles in the innate immune responses of the host.
It is now understood that teleost fish possess the NLRC3 or NLRC3-like gene [10]. Teleost fish NLRC3 shares similar motifs with mammalian NLRC3, although it lacks the CARD domain but contains additional domain(s) such as FISNA, PRY/SPRY (B30.2), and SPRY_PRY_SNTX [11]. Through genomic or transcriptomic sequencing, NLRC3 or NLRC3-like (NLRC3L) have been identified in several teleost fish species, including channel catfish (Ictalurus punctatus) [12], Japanese flounder (Paralichthys olivaceus) [13], zebrafish (Danio rerio) [14], miiuy croaker (Miichthys miiuy) [15], Asian seabass (Lates calcarifer) [16], blunt snout bream (Megalobrama amblycephala) [17], rainbow trout (Oncorhynchus mykiss) [18], turbot (Scophthalmus maximus L.) [19], goldfish (Carassius auratus L.) [20], and Nile Tilapia (Oreochromis niloticus) [21]. ASC has also been characterized in some teleost fish species, including zebrafish [22], mandarin fish (Siniperca chuatsi) [23], Japanese flounder [24], goldfish [25], orange-spotted grouper (Epinephelus coioides) [26], turbot [27], and Japanese medaka (Oryzias latipes) [28]. Moreover, Caspase1 has been characterized in gilthead seabream (Sparus aurata) [29], sea bass (Dicentrarchus labrax) [7], orange-spotted grouper [26], Japanese flounder [30], murrel (Channa striatus) [31], and miiuy croaker [32]. In particular, more than one ASC gene has been found in some fish, for example, three ASC genes were found in Japanese medaka [28]. It has been found that goldfish NLRC3L could interact with ASC to regulate the inflammasome pathway [20], indicating that fish NLRC3 or NLRC3L, ASC, and Caspase1 may have similar functions to their mammalian counterparts. However, the immune roles of these genes vary in different teleost fish species. For instance, zebrafish NLRC3L has been reported to suppress inflammation [33], while flounder NLRC3 or NLRC3L positively regulate the expression of proinflammatory cytokines [11]. Nonetheless, further information from diverse teleost fish species is required to elucidate the functions of these genes.
In this study, NLRC3-like, ASC, and Caspase1 were identified in the spotted sea bass (Lateolabrax maculatus), and their expressions in normal tissues, as well as in tissues following Edwardsiella tarda infection and PAMPs stimulation, were investigated. Our findings provide a basis for understanding the immune roles of these three genes in the spotted sea bass.
2. Materials and Methods
2.1. Fish
Healthy spotted sea bass (Lateolabrax maculatus) (100 ± 10 g) were purchased from a fish farm near Hangzhou city (Zhejiang Province, China), maintained under laboratory conditions at 26 ± 2 °C in freshwater, and fed with commercial feed for sea bass once a day for at least 2 weeks prior to the experimental procedures. Healthy fish without any pathological signs were then selected for the experiments, and the fish were anesthetized with 0.05% MS-222 before the experiments and dissection.
2.2. RNA Extraction and cDNA Synthesis
The total RNA of each tissue was extracted with a Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The concentration and quality of the total RNA were measured using a NanoDrop 2000c. The first-strand cDNA was reverse-synthesized from 5 μg of total RNA using the primer Oligo (dT)18 with the Hifair® II 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Yeasen, Shanghai, China), according to our previous methods [34].
2.3. Gene Cloning
Partial sequences of the NLRC3L, ASC, and Caspase1 of spotted sea bass were obtained by searching our previous study on the transcriptome data of spotted sea bass (unpublished), using homologous genes from other teleost fish via local BLAST. The local blast was performed using UltraEdit and BioEdit software. Then, the GSPs (gene specific primers) were designed using Primer 5.0, and a PCR was performed to clone the sequences. After that, a RACE-PCR was constructed using GSP, APG/AP (Anchoring Primer), UPM (Universal Primer Mix), and NUP (Nested Universal Primer) to obtain the full cDNA sequences of the genes with LATaq Mix DNA Polymerase (Takara, Shiga, Japan). The PCR products were separated on an agarose gel via electrophoresis, purified, and ligated into the pMD19-T vector (Takara, Shiga, Japan), and the pMD19-T were transformed into E. coli DH5α cells (SangonBiotech, Shanghai, China). Finally, positive clones were screened and sequenced on an automatic DNA sequencer (SangonBiotech, Shanghai, China). All primers used for gene cloning are listed in Table 1. The primers were synthesized by GENEWIZ (Suzou, China) and the annealing/melting temperatures and product sizes were confirmed according to the primer information sheet provided by the company.

Table 1.
Primers used in this study.
2.4. LPS and Poly (I:C) Stimulation
A total of 90 fish (100 ± 10 g) were randomly divided into three groups: the lipopolysaccharide (LPS)-stimulated group, in which the fish were intraperitoneally (i.p.) injected with 500 μL LPS (5 mg/kg body weight), the polyinosinic-polycytidylic acid (poly (I:C))-stimulated group, in which the fish were i.p. injected with 500 μL Poly (I:C) (5 mg/kg body weight), and the control group, in which the fish were injected with the same amount of PBS. At 6, 12, 24, 48, and 72 h post injection (hpi), tissues of three fish from each group were collected, including the spleen, head kidney, and gill. Each tissue was homogenized in 1 mL of Trizol reagent, using the QIAGEN® Tissue Lyser II system, and reverse-transcribed into cDNA using Hifair® II 1st Strand cDNA Synthesis SuperMix (gDNA digester plus) (YEASEN), according to the method described previously. The synthesized cDNA samples were stored at −20 °C until use.
2.5. E. tarda Culture and Infection
The E. tarda used for the infection experiment were cultured in tryptic soy agar (TSA) medium at 28 °C overnight and prepared according to protocols previously described in the literature [35,36,37]. A single colony was transferred to a tryptic soy broth (TSB) medium and shaken at 170 rpm at 28 °C overnight. The bacterial culture was diluted 1:10 (v/v) with fresh medium and cultured for 2 h to reach the logarithmic phase. The bacteria were counted via spectrophotometry and plating dilutions [36].
For E. tarda infection, 60 fish were randomly divided into two groups: the bacterial infection group and the control group, in which fish were i.p. injected with 500 μL of the E. tarda solution (1 × 105 CFU/mL) and 500 μL PBS, respectively. At 6 h, 12 h, 24 h, and 48 h post injection, fish tissues, including the liver, spleen, head kidney, gill, intestine, skin, muscle, and brain, were collected from 5 fish in each group. Each tissue was homogenized in 1 mL of Trizol reagent, using the QIAGEN® Tissue Lyser II system, and reverse-transcribed into cDNA using Hifair® II 1st Strand cDNA Synthesis SuperMix (gDNA digester plus) (Yeasen, Shanghai, China), according to the method described previously.
2.6. qPCR for Expression Analysis
The primers for qPCR analysis are provided in Table 1. The primers were tested using RT-PCR, and the resultant PCR amplicons were ligated into the pMD-19T vector (Takara, Shiga, Japan). The ligation reaction was transformed into E. coli DH5α cells, and positive clones were screened for plasmid preparation using the plasmid Mini Kit I (OMEGA, Omega, GA, USA). Three clones were sequenced to confirm primer specificity. After sequence verification, the plasmid DNA was diluted in a 10-fold series. Gene copy numbers were calculated and used for establishing the standard curve for the qPCR quantitation of gene expression. The qPCR reaction was set up as follows: 5 μL of SYBR® Green PreMix Ex Taq™ II (Yeasen, Shanghai, China), 1 μL of cDNA template (20-fold dilution), 0.2 μL of forward primer (10 μM), 0.2 μL of reverse primer (10 μM), and 3.6 μL of H2O. The qPCR reactions were performed using the Roche Light Cycler® 96 system (Roche), using the following conditions: 1 cycle of 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 62 °C for 30 s, 72 °C for 10 s, followed by 1 cycle of 95 °C for 10 s, 65 °C for 60 s, and 97 °C for 1 s. Elongation factor 1-alpha (EF-1α) was used as an internal reference gene, and the fold change in gene expression was calculated by comparing the expression level of the experimental group with that of the respective control group (defined as 1).
2.7. Bioinformatics Analysis
The cDNA sequences of the NLRC3L, ASC, and Caspase1 of spotted sea bass were confirmed using BLAST in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 16 July 2023). The theoretical molecular weights and pI values were predicted using the Peptide Mass program on the ExPASy website (https://www.expasy.org/, accessed on 16 July 2023). Multiple sequence alignment was performed with the ClustalW program and shaded using GeneDoc 2.7. The protein domains were predicted using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl.de/smart/set_mode.cgi?NORMAL=1, accessed on 16 July 2023). A gene synteny analysis was performed using the genomic data deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/43909?genome_assembly_id=437727, accessed on 16 July 2023) for spotted sea bass and the Ensembl database for other species (https://www.ensembl.org/index.html, accessed on 16 July 2023). A neighbor-joining tree was constructed from pairwise Poisson correction distances with 1000 bootstrap replications in MEGA5.0 software.
2.8. Statistical Analysis of Data
The SPSS 17.0 software package was used for statistical analysis of data generated via quantitative real-time PCR. For the challenge experiments, the expression data were analyzedvia Student’s t-test, with p-values < 0.05 and p < 0.01, indicating statistically significant differences between the experimental and control groups.
3. Results
3.1. Sequence Features of NLRC3L, ASC, and Caspase1 of Spotted Sea Bass
The full cDNA sequences of NLRC3L, ASC, and Caspase1 from the spotted sea bass were obtained via RT-PCR and RACE-PCR methods and submitted to the GenBank database with accession numbers OQ470527 (LmNLRC3L), OQ470528 (LmASC) and OQ470529 (LmCaspase1).
The LmNLRC3L cDNA was 3755 bp long, including a 5′-untranslated region (UTR) of 108 bp, an open reading frame (ORF) of 3327 bp, and a 3′-UTR of 320 bp containing a poly (A) tail (Figure S1). The ORF of LmNLRC3L was predicted to encode a protein of 1108 amino acids, with a molecular weight of approximately 124.5 kDa and a theoretical pI of 5.71. The LmNLRC3L protein possessed multiple conserved domains: a fish-specific NACHT-associated domain (FISNA, residues site 199–271), a NACHT domain (residues site 281–449) and four LRR motifs (residues site 788–896), followed by a C-terminal fish-specific B30.2 domain (PRY/SPRY domain, residues site 920–1103) (Figure 1A). A multiple sequence alignment revealed that the most non-conserved regions among the fish and mammalian NLRC3 proteins were primarily located in the N-terminus. However, the functional domains, such as the FISNA domain, B30.2 domain, and NACHT domain, were relatively conserved among fish and mammalian NLRC3 (Figure 1A). Phylogenetic analysis revealed the formation of the two main clades, NLRC3 and NLRC3L. Fish NLRC3 clustered with mammalian NLRC3, while LmNLRC3L was clustered with NLRC3L from Asian seabass (Lates calcarifer), supported by a bootstrap value of 100% (Figure 1B).

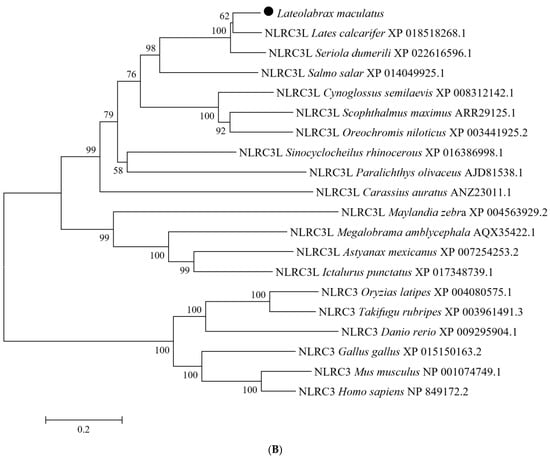
Figure 1.
Multiple sequence alignment (A) and phylogenetic tree analysis (B) of LmNLRC3L. (A) the predicted structural domains of LmNLRC3L are marked above the sequence. (B) the phylogenetic tree was constructed using the NJ method and run for 10,000 replications; The percentages of bootstrap values for branches are indicated and the symbols (●) indicate LmNLRC3L.
The cDNA sequence of LmASC was 825 bp in length and consisted of a 5′-UTR of 59 bp, an ORF of 618 bp, and a 3′-UTR of 150 bp containing a poly(A) tail (Figure S2). The ORF of LmASC was predicted to encode a protein of 205 amino acids, with a molecular weight of approximately 22 kDa and a theoretical pI of 5.86. Through a multiple sequence alignment, it was found that LmASC possessed a conserved N-terminal pyrin domain (residues site 6–86) and a C-terminal CARD domain (residues site 116–204) (Figure 2A). A synteny analysis revealed that the FLI1B, ETV2, ASC, GRAMD1A, SCN1BA, and MAG genes of the spotted sea bass were arranged in a tandem manner (Figure 2B). These linked genes were found to be conserved in turbot, goldfish, and Asian sea bass. A phylogenetic tree analysis demonstrated that LmASC clustered with fish ASC (Figure 2C).
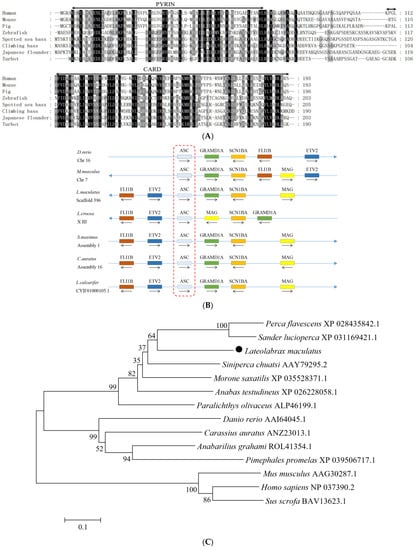
Figure 2.
Multiple sequence alignment (A), gene synteny (B), and phylogenetic tree analysis (C) of LmASC. (A) The predicted structural domains of LmASC are marked above the sequence. (B) The genome sequence data of seven species were obtained and retrieved from the NCBI and Ensembl databases; arrows indicate transcription orientations. (C) the phylogenetic tree was constructed using the NJ method and run for 10,000 replications; the percentages of the bootstrap values for branches are indicated, and the symbols (●) indicate LmASC.
The cDNA sequence of LmCaspase1 was 2228 bp in length, including a 5′-UTR of 324 bp, an open reading frame (ORF) of 1185 bp, and a 3′-UTR of 719 bp containing a poly (A) tail (Figure S3). The ORF of LmCaspase1 was predicted to encode a protein of 394 amino acids, with a molecular weight of approximately 44 kDa and a theoretical pI of 5.87. Similar to other inflammatory caspases, the LmCaspase1 protein possessed a typical caspase recruitment domain (CARD, residues site 1–89) at the N-terminus, a CASC domain (residues site 128–392) at the C-terminus, and the conserved pentapeptide active-site motif (QACRG) (Figure 3A). A synteny analysis revealed that BUD23, STX1A, CASPASE1, OTOL1B, SPTSSB, and NMD3 were arranged in tandem in the spotted sea bass, European seabass, gilthead seabream, and turbot (Figure 3B). A phylogenetic tree analysis demonstrated the clustering of LmCaspase1 with fish Caspase1 (Figure 3C).
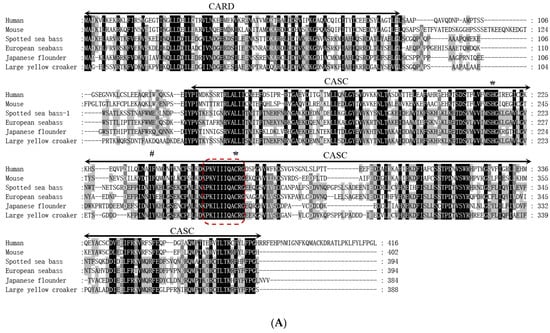
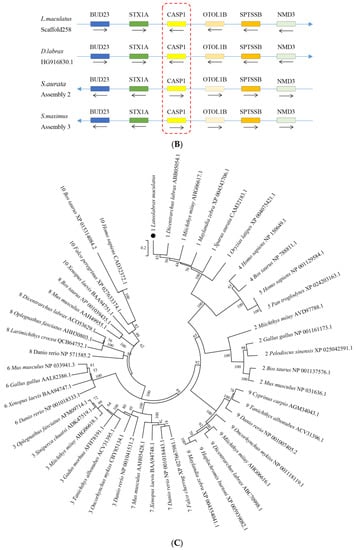
Figure 3.
Multiple sequence alignment (A), gene synteny (B), and phylogenetic tree analysis (C) of LmCaspase1. (A) The predicted structural domains of LmCaspase1 are marked above the sequence and the conserved pentapeptide active-site motif (QACRG) is boxed in red. (B) The genome sequence data were obtained and retrieved from the NCBI and Ensembl databases; arrows indicate transcription orientations. (C) The phylogenetic tree was constructed using the NJ method and run for 10,000 replications; The percentages of the bootstrap values for branches are indicated, and the symbols (●) indicate LmCaspase1. The numbers in front of the branches’ names represent caspase types.
3.2. Tissue Distributions of NLRC3L, ASC, and Caspase1 of Spotted Sea Bass
The mRNA expression of LmNLRC3L, LmASC, and LmCaspase1 was detected in various tissues through qPCR analysis. As depicted in Figure 4, these three genes exhibited constitutive expression in all examined tissues and displayed similar expression patterns. Among the tissues, the gill (mucosal immune tissue) showed the highest expression levels of all three genes, followed by the spleen and head kidney (systemic immune tissues), while the expression levels in the liver, brain, intestine, and skin were relatively lower.
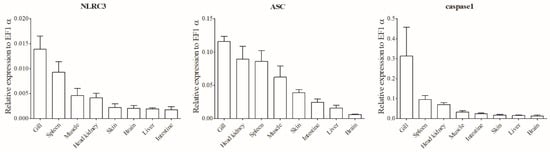
Figure 4.
Expression analysis of LmNLRC3L, LmASC and LmCaspase1 in tissues. The mRNA levels were determined via qPCR. The relative expression levels of the three genes were expressed as arbitrary units normalized against the expression levels of EF-1α. The results are expressed as mean + SEMs (N = 3).
3.3. Expressions of NLRC3L, ASC, and Caspase1 of Spotted Sea Bass Following LPS Stimulation
The mRNA expressions of the three genes in the HK, spleen, and gill of spotted sea bass were determined using a qPCR following stimulation with LPS, the main component of the outer membrane of Gram-negative bacteria [36]. As shown in Figure 5, the expressions of LmNLRC3L, LmASC, and LmCaspase1 were upregulated in the spleen at 6 h after LPS stimulation. In addition, the expression levels of LmNLRC3L and LmASC were upregulated in HK at 6 h after LPS stimulation. After further stimulation, some genes were downregulated in the selected tissues. For example, LmNLRC3L was downregulated in HK at 24 h post LPS stimulation, and LmCaspase1 was downregulated in the gill at 48 h.
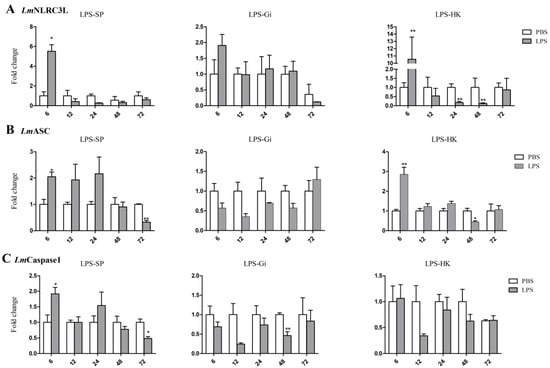
Figure 5.
Expression analysis of LmNLRC3L (A), LmASC (B), and LmCaspase1 (C) after LPS stimulation in the spleen, gill, and head kidney. The relative expression level of the target gene was normalized to that of EF1α. The data are shown as means + SEMs (N = 3). * p < 0.05 and ** p < 0.01 indicate significant differences.
3.4. Expressions of NLRC3L, ASC, and Caspase1 of Spotted Sea Bass Following Poly (I:C) Stimulation
Poly (I:C) is a synthetic double-stranded RNA analog of a viral nucleotide and can activate an antiviral response in fish [36]. We assessed its effects on the NLRC3L, ASC, and Caspase1 expression of spotted sea bass in the HK, spleen, and gill via qPCR (Figure 6). At 6 h post poly(I:C) stimulation, only LmNLRC3L was up regulated in HK. At 48 h post poly(I:C) stimulation, the expression of LmASC was downregulated in the HK and gill. Downregulations of LmASC and LmCaspase1 were also observed in the spleen at 72 h after poly(I:C) stimulation.
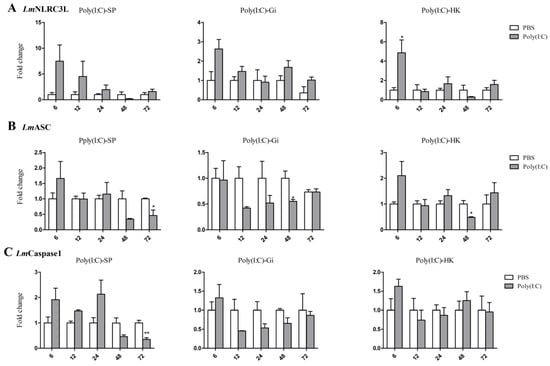
Figure 6.
Expression analysis of LmNLRC3L (A), LmASC (B), and LmCaspase1 (C) after Poly (I:C) stimulation in the spleen, gill, and head kidney. The relative expression level of the target gene was normalized to that of EF1α. The data are shown as means + SEMs (N = 3). * p < 0.05 and ** p < 0.01 indicate significant differences.
3.5. Expressions of NLRC3L, ASC, and Caspase1 of Spotted Sea Bass Following E. tarda Infection
E. tarda is an intracellular bacterial pathogen that infects a wide range of fish species [36]. LmNLRC3L, LmASC, and LmCaspase1 expression levels in the HK, gill, and spleen after E. tarda infection were analyzed via qPCR. The results showed that these three genes had similar expressional changes in all selected tissues (Figure 7). LmNLRC3L, LmASC, and LmCaspase1 were significantly upregulated in the HK at 6 h and 24 h post-E. tarda infection. LmNLRC3L was also upregulated in the HK at 12 h post infection. LmNLRC3L was upregulated in the gill and spleen at 6 h and in the gill at 48 h after bacterial infection. LmCaspase1 was significantly upregulated in the HK at 48 h after E. tarda infection.
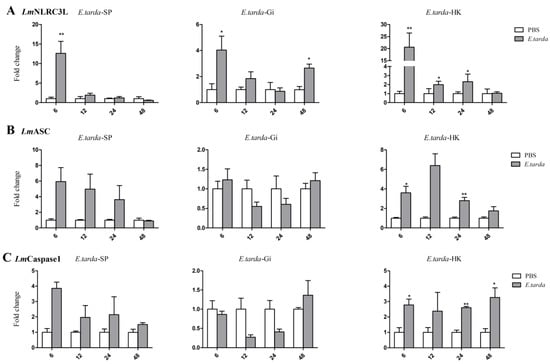
Figure 7.
Expression analysis of LmNLRC3L (A), LmASC (B), and LmCaspase1 (C) after Edwardsiella tarda challenge in the spleen, gill, and head kidney. The relative expression level of the target gene was normalized to that of EF1α. The data are shown as means + SEMs (N = 3). * p < 0.05 and ** p < 0.01 indicate significant differences.
4. Discussion
It has been stablished that mammalian NLRC3 can interact with ASC and Caspase1 to provide essential functions in inflammation and innate immune responses [5]. Also, NLRC3 or NLRC3L, ASC, and Caspase1 have been demonstrated to be involved in the immune responses of several fish species. In this study, NLRC3L, ASC, and Caspase1 were characterized in the spotted sea bass, representing the first investigation of these genes in this species. These three genes shared similar features to their fish counterparts (Figure 1, Figure 2 and Figure 3) [10,28,30]. Notably, LmNLRC3L contained a conserved fish-specific B30.2 domain, which is important in anti-viral activities and for directing the interaction with Caspase1 to modulate IL-1β production [11]. Both LmASC and LmCaspase1 possessed the CARD domain, which is required for their binding [6]. These results confirm that these three genes are indeed homologs of NLRC3L, ASC, and Caspase1. Conserved motifs in these genes suggest they can interact, similar to their mammalian counterparts. It is noteworthy that different isoforms of these three genes have been found in some fish, so whether they have different isoforms in spotted sea bass requires further study.
Over the years, the tissue expression profiles of NLRC3 or NLRC3L, ASC, and Caspase1 have been examined in various fish species. These three genes consistently exhibited similar expression patterns across different fish species, including constitutive distribution in all examined tissues and high levels of expression in immune tissues. For instance, NLRC3/NLRC3L showed dominant expression in the gill of channel catfish [38], blunt snout bream [17], and Asian seabass [16]. ASC and Caspase1 were highly expressed in the gill of Japanese flounder [24,30] and highly expressed in the gill and HK of orange-spotted grouper [26] and zebrafish [39]. ASC and NLRC3L were also highly expressed in the spleen, intestine, and gill of goldfish [20,25]. Consistent with the literature, our results found that LmNLRC3L, LmASC, and LmCaspase1 were highly expressed in the gill, HK, and spleen. The gill is a crucial mucosal immune tissue in fish that comes into contact with waterborne pathogens. High levels of expressions of LmNLRC3L, LmASC, and LmCaspase1 in the gill suggest a potential cooperative relationship in combating waterborne pathogens. High levels of expression of these three genes in other immune tissues indicate their important roles in the immune response of spotted sea bass.
The expression of NLRC3 or NLRC3L, ASC, and Caspase1 can be regulated by pathogen-associated molecular patterns (PAMPs). Rainbow trout NLRC3 was found to be upregulated after LPS stimulation [18], while Asian seabass NLRC3 showed upregulation in all tested tissues following poly(I:C) infection [16]. Our findings are consistent with the above studies. We also observed that some genes were significantly regulated by LPS but remained unresponsive to poly(I:C) stimulation, suggesting that these three genes might be more sensitive to LPS than poly(I:C). In addition, these genes were significantly down-regulated in the late stages of LPS and poly(I:C) stimulation. This could be due to the body upregulating negative feedback regulation to avoid excessive inflammation.
Furthermore, the expression levels of NLRC3 or NLRC3L, ASC, and Caspase1 can be regulated by bacterial infection. Japanese flounder NLRC3 was upregulated at 6 h post-E. tarda stimulation [16]. In channel catfish, NLRC3 exhibited a 72-fold increase in the liver at 12 h after E. tarda infection, followed by a return to normal levels [38]. Consistent with these findings, our results showed that the NLRC3L, ASC, and Caspase1 of spotted sea bass were upregulated following E. tarda infection, aligning with previous studies. However, it is worth nothing that Japanese flounder ASC was downregulated in the gill at 8 and 12 h post-E. tarda infection [24], and turbot ASC expression was decreased in the gill after E. tarda infection [27]. These findings differ from ours, as LmASC was significantly upregulated after E. tarda stimulation in the spotted sea bass. These divergent results suggest that the three genes may exhibit varied expression patterns after bacterial infection. The upregulation of these three genes following E. tarda stimulation indicates their immune roles in the spotted sea bass.
Interestingly, NLRC3L has been demonstrated to play completely opposite effects in different teleost fish species. For example, NLRC3L in zebrafish has been reported to inhibit inflammation [33], while flounder NLRC3 or NLRC3L positively regulated the expression of proinflammatory cytokines [11]. Therefore, the immunological roles and mechanisms of action of these genes in spotted sea bass must be elucidated in future studies.
5. Conclusions
Overall, the present study documented NLRC3L, ASC, and Caspase1 identification in spotted sea bass. The sequence features of these genes were analyzed, and their expression patterns in normal tissues and tissues following PAMP stimulation and bacterial infection were investigated. Our results demonstrate that PAMPs and bacterial stimuli can regulate these three genes, indicating their important roles in the immune response of spotted sea bass. Collectively, these genes may participate in the immune response of spotted sea bass through their interactions. However, further studies are necessary to elucidate the relationships among these genes and their potential biological functions following pathogen invasion.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8070378/s1, Figures S1–S3: cDNA and deduced amino acid sequence of LmNLRC3L (S1), LmASC (S2), and LmCaspase1 (S3). The ORF is shown in upper case and the 5′-UTR and 3′-UTR sequences are in lower case. The translation initiation codon, stop codon, and polyadenylation signal are shown in bold.
Author Contributions
Z.S. and S.Y.: investigation, methodology, data curation, and writing-original draft preparation. Q.G.: conceptualization, funding acquisition, project administration, supervision, writing—review and editing. Z.Q.: supervision, writing—review and editing. Z.L.: writing—review and editing. S.Z. and D.L.: investigation and methodology. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (grant number: 2018YFD0900605) and the Major Projects of Natural Science Research for University and Colleges in Jiangsu Province (grant no. 21KJA240001).
Institutional Review Board Statement
All fish experiments were conducted under the national regulations on laboratory animals of China and were reviewed and approved by the ethics committee of laboratory animals of Shanghai Ocean University (SHOU-DW-2019-012).
Data Availability Statement
All the data generated or used during the study appear in the submitted article.
Acknowledgments
We thank Haixia Xie, Institute of Hydrobiology, Chinese Academy of Sciences, for providing Edwardsiella tarda.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of the work described in this manuscript.
References
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.L.; Hao, G.X.; Geng, X.Y.; Zhan, W.B.; Sun, J.S. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immunol. 2016, 50, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Pontillo, A.; Crovella, S. NOD-Like Receptors: A Tail from Plants to Mammals Through Invertebrates. Curr. Protein Pept. Sc. 2017, 18, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Motyan, J.A.; Bagossi, P.; Benko, S.; Tozser, J. A molecular model of the full-length human NOD-like receptor family CARD domain containing 5 (NLRC5) protein. BMC Bioinform. 2013, 14, 275. [Google Scholar] [CrossRef] [PubMed]
- Gultekin, Y.; Eren, E.; Ozoren, N. Overexpressed NLRC3 Acts as an Anti-Inflammatory Cytosolic Protein. J. Innate Immun. 2015, 7, 25–36. [Google Scholar] [CrossRef]
- Eren, E.; Berber, M.; Ozoren, N. NLRC3 protein inhibits inflammation by disrupting NALP3 inflammasome assembly via competition with the adaptor protein ASC for pro-caspase-1 binding. J. Biol. Chem. 2017, 292, 12691–12701. [Google Scholar] [CrossRef]
- Reis, M.I.R.; do Vale, A.; Pereira, P.J.B.; Azevedo, J.E.; dos Santos, N.M.S. Caspase-1 and IL-1β Processing in a Teleost Fish. PLoS ONE 2012, 7, e50450. [Google Scholar] [CrossRef]
- Ghayur, T.; Banerjee, S.; Hugunin, M.; Butler, D.; Herzog, L.; Carter, A.; Quintal, L.; Sekut, L.; Talanian, R.; Paskind, M.; et al. Caspase-1 processes IFN-γ-inducing factor and regulates LPS-induced IFN-γ production. Nature 1997, 386, 619–623. [Google Scholar] [CrossRef]
- Vanaja, S.K.; Rathinam, V.A.K.; Fitzgerald, K.A. Mechanisms of inflammasome activation: Recent advances and novel insights. Trends Cell Biol. 2015, 25, 308–315. [Google Scholar] [CrossRef]
- Chuphal, B.; Rai, U.; Roy, B. Teleost NOD-like receptors and their downstream signaling pathways: A brief review. Fish Shellfish Immunol. Rep. 2022, 3, 100056. [Google Scholar] [CrossRef]
- Chang, M.X.; Xiong, F.; Wu, X.M.; Hu, Y.W. The expanding and function of NLRC3 or NLRC3-like in teleost fish: Recent advances and novel insights. Dev. Comp. Immunol. 2021, 114, 103859. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.X.; Abernathy, J.W.; Wang, S.L.; Li, P.; Kucuktas, H.; Liu, H.; Peatman, E.; Liu, Z.J. NOD-like subfamily of the nucleotide-binding domain and leucine-rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunol. 2009, 33, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Unajak, S.; Santos, M.D.; Hikima, J.; Jung, T.S.; Kondo, H.; Hirono, I.; Aoki, T. Molecular characterization, expression and functional analysis of a nuclear oligomerization domain proteins subfamily C (NLRC) in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 31, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Shiau, C.E.; Monk, K.R.; Joo, W.; Talbot, W.S. An Anti-inflammatory NOD-like Receptor Is Required for Microglia Development. Cell Rep. 2013, 5, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Li, J.R.; Kong, L.C.; Gao, Y.H.; Wu, C.Q.; Xu, T.J. Characterization of NLR-A subfamily members in miiuy croaker and comparative genomics revealed NLRX1 underwent duplication and lose in actinopterygii. Fish Shellfish Immunol. 2015, 47, 397–406. [Google Scholar] [CrossRef]
- Paria, A.; Deepika, A.; Sreedharan, K.; Makesh, M.; Chaudhari, A.; Purushothaman, C.S.; Thirunavukkarasu, A.R.; Rajendran, K.V. Identification of Nod like receptor C3 (NLRC3) in Asian seabass, Lates calcarifer: Characterisation, ontogeny and expression analysis after experimental infection and ligand stimulation. Fish Shellfish Immunol. 2016, 55, 602–612. [Google Scholar] [CrossRef]
- Zhou, F.J.; Zhan, Q.F.; Ding, Z.J.; Su, L.N.; Fan, J.; Cui, L.; Chen, N.; Wang, W.M.; Liu, H. A NLRC3-like gene from blunt snout bream (Megalobrama amblycephala): Molecular characterization, expression and association with resistance to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2017, 63, 213–219. [Google Scholar] [CrossRef]
- Alvarez, C.A.; Ramirez-Cepeda, F.; Santana, P.; Torres, E.; Cortes, J.; Guzman, F.; Schmitt, P.; Mercado, L. Insights into the diversity of NOD-like receptors: Identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol. Immunol. 2017, 87, 102–113. [Google Scholar] [CrossRef]
- Hou, Z.M.; Ye, Z.; Zhang, D.D.; Gao, C.B.; Su, B.F.; Song, L.; Tan, F.H.; Song, H.H.; Wang, Y.; Li, C. Characterization and expression profiling of NOD-like receptor C3 (NLRC3) in mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 2017, 66, 231–239. [Google Scholar] [CrossRef]
- Xie, J.S.; Belosevic, M. Characterization and functional assessment of the NLRC3-like molecule of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2018, 79, 1–10. [Google Scholar] [CrossRef]
- Gao, F.Y.; Pang, J.C.; Lu, M.X.; Yang, X.L.; Zhu, H.P.; Ke, X.L.; Liu, Z.G.; Cao, J.M.; Wang, M. Molecular characterization, expression and functional analysis of NOD1, NOD2 and NLRC3 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2018, 73, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Huang, Y.; Cao, X.C.; Yin, X.Y.; Jin, X.Y.; Liu, S.; Jiang, J.S.; Jiang, W.; Xiao, T.S.; Zhou, R.B.; et al. Functional and structural characterization of zebrafish ASC. FEBS J. 2018, 285, 2691–2707. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.N.; Wang, J.; La, H.H.; Yin, Z.X.; He, W.; Weng, S.P.; Yu, X.Q.; Chan, S.M.; He, J.G. Molecular cloning and expression analysis of the ASC gene from mandarin fish and its regulation of NF-κB activation. Dev. Comp. Immunol. 2008, 32, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.L.; Peng, W.J.; Hao, G.X.; Geng, X.Y.; Zhan, W.B.; Sun, J.S. Cloning and characterization of apoptosis-associated speck-like protein containing a CARD domain (ASC) gene from Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2016, 54, 294–301. [Google Scholar] [CrossRef]
- Xie, J.S.; Belosevic, M. Functional characterization of apoptosis-associated speck-like protein (ASC) of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2016, 65, 201–210. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.T.; Li, C.; Zhang, Y.; Wan, L.Q.; Wei, J.G.; Qin, Q.W. Characterization of orange-spotted grouper (Epinephelus coioides) ASC and caspase-1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2020, 97, 58–71. [Google Scholar] [CrossRef]
- Wang, W.H.; Tan, J.C.; Wang, Z.; Zhang, Y.X.; Liu, Q.; Yang, D.H. Characterization of the inflammasome component SmASC in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2020, 100, 324–333. [Google Scholar] [CrossRef]
- Morimoto, N.; Okamura, Y.; Kono, T.; Sakai, M.; Hikima, J. Characterization and expression analysis of tandemly-replicated asc genes in the Japanese medaka, Oryzias latipes. Dev. Comp. Immunol. 2021, 115, 103894. [Google Scholar] [CrossRef]
- Lopez-Castejon, G.; Sepulcre, M.P.; Mulero, I.; Pelegrin, P.; Meseguer, J.; Mulero, V. Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: The first identification of an inflammatory caspase in fish. Mol. Immunol. 2008, 45, 49–57. [Google Scholar] [CrossRef]
- Li, S.; Peng, W.J.; Li, J.F.; Hao, G.X.; Geng, X.Y.; Sun, J.S. Characterization of Japanese flounder (Paralichthys olivaceus) Caspase1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2017, 67, 536–545. [Google Scholar] [CrossRef]
- Kumaresan, V.; Ravichandran, G.; Nizam, F.; Dhayanithi, N.B.; Arasu, M.V.; Al-Dhabi, N.A.; Harikrishnan, R.; Arockiaraj, J. Multifunctional murrel caspase 1, 2, 3, 8 and 9: Conservation, uniqueness and their pathogen-induced expression pattern. Fish Shellfish Immunol. 2016, 49, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.P.; Wang, R.X.; Xu, T.J. Three representative subtypes of caspase in miiuy croaker: Genomic organization, evolution and immune responses to bacterial challenge. Fish Shellfish Immunol. 2014, 40, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.F.; Luo, G.Y.; Liang, R.; Qiu, C.L.; Yang, J.W.; Xie, L.L.; Zhang, K.L.; Tian, Y.; Wang, D.C.; Song, S.; et al. Negative Regulator Nlrc3-like Maintain the Balanced Innate Immune Response During Mycobacterial Infection in Zebrafish. Front. Immunol. 2022, 13, 893611. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.S.; Qin, Y.T.; Liu, D.J.; Wang, B.J.; Jia, Z.; Wang, J.Y.; Gao, Q.; Zou, J.; Pang, Y. The evolution and functional characterization of CXC chemokines and receptors in lamprey. Dev. Comp. Immunol. 2021, 116, 103905. [Google Scholar] [CrossRef]
- Yi, L.; Nie, P.; Yu, H.B.; Xie, H.X. Regulation of Type III Secretion of Translocon and Effector Proteins by the EsaB/EsaL/EsaM Complex in Edwardsiella tarda. Infect. Immun. 2017, 85, e00322-1. [Google Scholar] [CrossRef]
- Li, X.; Yuan, S.Y.; Sun, Z.S.; Lei, L.N.; Wan, S.; Wang, J.Y.; Zou, J.; Gao, Q. Gene identification and functional analysis of peptidoglycan recognition protein from the spotted sea bass (Lateolabrax maculatus). Fish Shellfish Immunol. 2020, 106, 1014–1024. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Q.; Wang, J.Y.; Tian, J.Y.; Song, Y.J.; Xie, H.X.; Chang, M.X.; Nie, P.; Gao, Q.; Zou, J. Transcriptomic responses of S100 family to bacterial and viral infection in zebrafish. Fish Shellfish Immunol. 2019, 94, 685–696. [Google Scholar] [CrossRef]
- Li, M.; Wang, Q.L.; Lu, Y.; Chen, S.L.; Li, Q.; Sha, Z.X. Expression profiles of NODs in channel catfish (Ictalurus punctatus) after infection with Edwardsiella tarda, Aeromonas hydrophila, Streptococcus iniae and channel catfish hemorrhage reovirus. Fish Shellfish Immunol. 2012, 33, 1033–1041. [Google Scholar] [CrossRef]
- Li, J.Y.; Gao, K.; Shao, T.; Fan, D.D.; Hu, C.B.; Sun, C.C.; Dong, W.R.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. Characterization of an NLRP1 Inflammasome from Zebrafish Reveals a Unique Sequential Activation Mechanism Underlying Inflammatory Caspases in Ancient Vertebrates. J. Immunol. 2018, 201, 1946–1966. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).