Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequences for Microsatellite Identification
2.2. Characterization of Microsatellites in Siamese Fighting Fish Genome
2.3. Microsatellite Marker Development
2.4. In Silico Polymorphism Screening of Siamese Fighting Fish Species
2.5. In Silico Cross-Species Transferability
2.6. Specimen Collection and DNA Extraction
2.7. Microsatellite Genotyping of Siamese Fighting Fish
2.8. Microsatellite Marker Polymorphism Testing
2.9. Cross-Species Amplification Test of Microsatellite Markers
3. Results
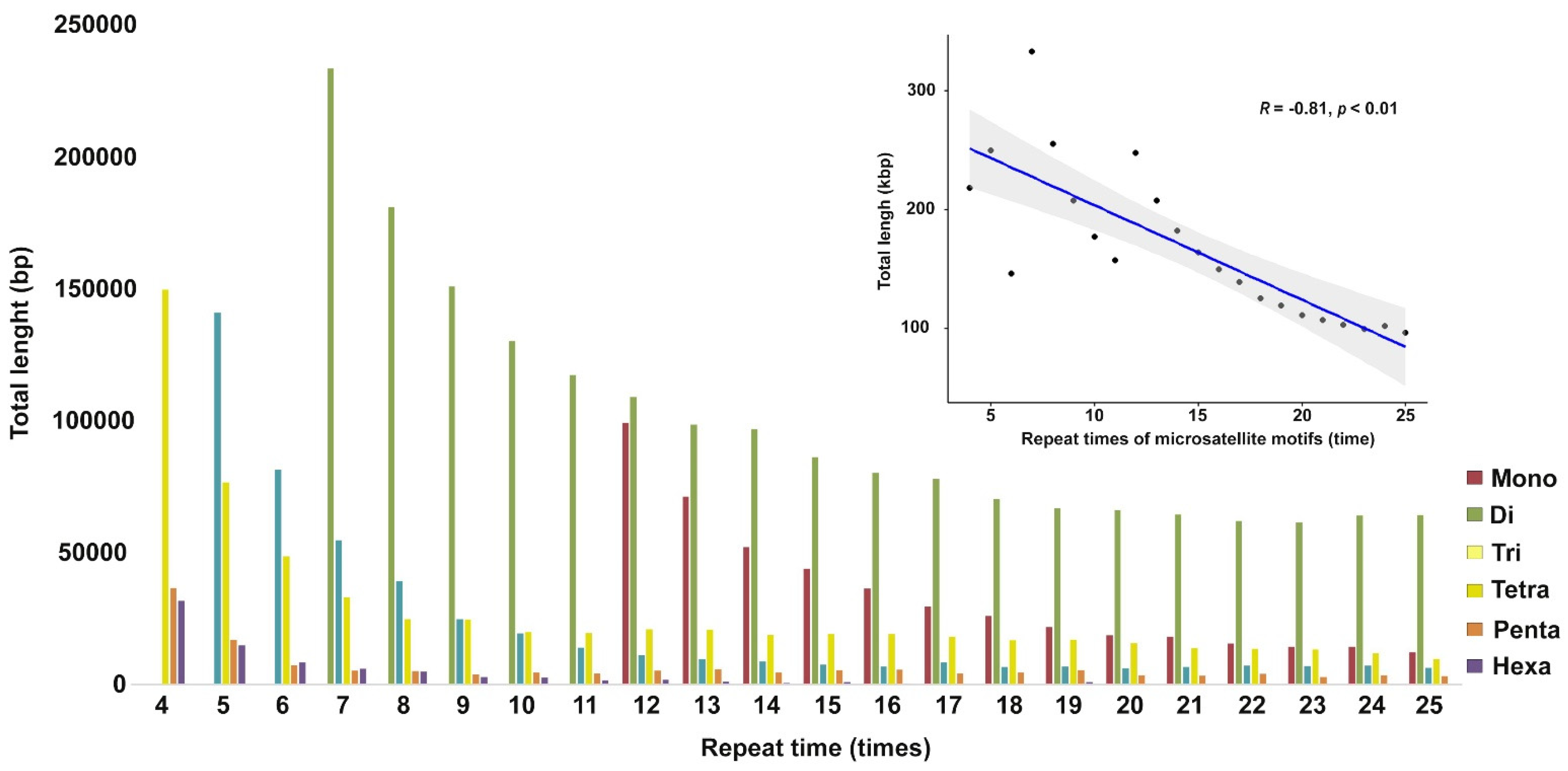
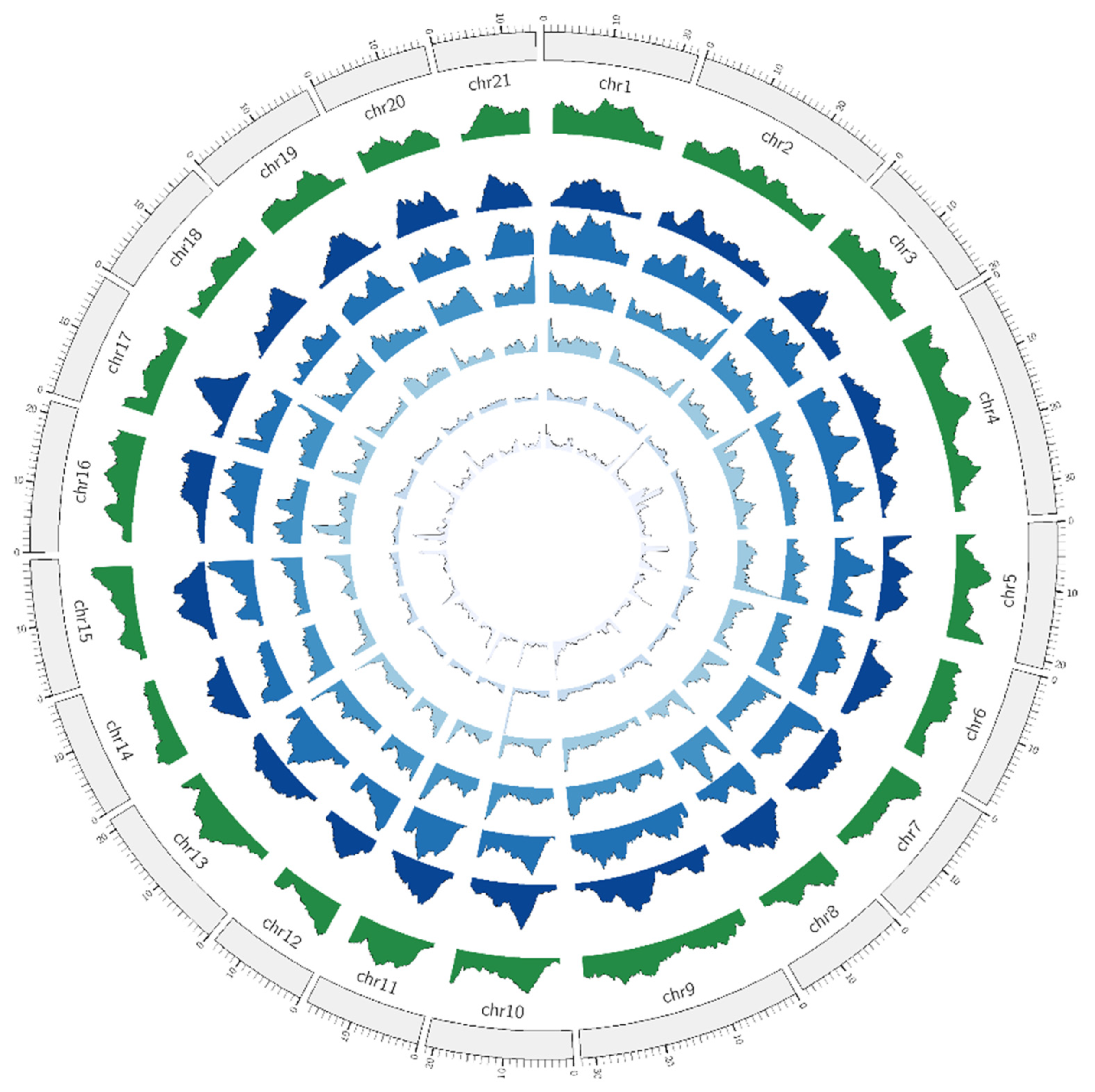
3.1. Microsatellite Distribution in Siamese Fighting Fish Genome
3.2. Polymorphic Microsatellite Identification through In Silico Genome Sequence Comparison and a PCR-Based Assay
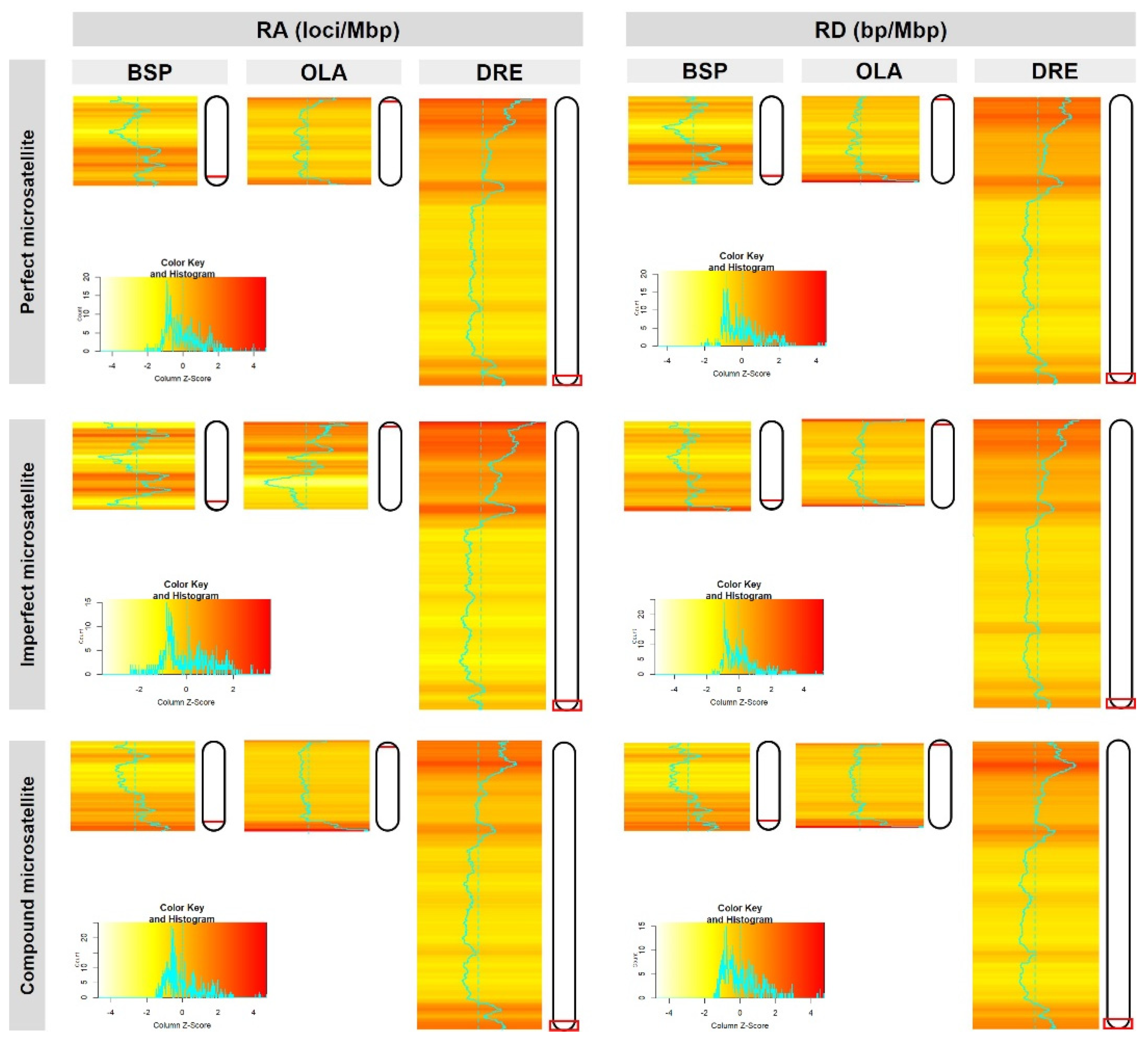
3.3. In Silico Cross-Species Transferability and Cross-Species Amplification
4. Discussion
4.1. Coincidence of Microsatellite Density and in Sex-Determining Regions of Siamese Fighting Fish
4.2. Microsatellite Marker Development for Genetic Diversity Studies, Conservation, and Future Breeding Programs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srikulnath, K.; Singchat, W.; Laopichienpong, N.; Ahmad, S.F.; Jehangir, M.; Subpayakom, N.; Suntronpong, A.; Jangtarwan, K.; Pongsanarm, T.; Panthum, T.; et al. Overview of the betta fish genome regarding species radiation, parental care, behavioral aggression, and pigmentation model relevant to humans. Genes Genom. 2021, 43, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Regan, C.T. The Asiatic fishes of the family Anabantidae. Proc. Zool. Soc. Lond. 1909, 79, 767–787. [Google Scholar]
- U.S. Fish and Wildlife Service. Siamese Fighting Fish (Betta splendens) Ecological Risk Screening Summary; U.S. Fish and Wildlife Service: Washington, DC, USA, 2019. [Google Scholar]
- Monvises, A.; Nuangsaeng, B.; Sriwattanarothai, N.; Panijpan, B. The Siamese fighting fish: Well-known generally but little-known scientifically. Sci. Asia 2009, 35, 8–16. [Google Scholar] [CrossRef]
- Department of Fisheries. Fisheries Single Window Data System; Fish Quarantine and Inspection Division, Department of Fisheries: Bangkok, Thailand, 2019. [Google Scholar]
- Verbeek, P.; Iwamoto, T.; Murakami, N. Variable stress-responsiveness in wild type and domesticated fighting fish. Physiol. Behav. 2008, 93, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.M.; Vranken, N.; Hoge, C.; Lichak, M.R.; Norovich, A.L.; Francis, K.X.; Camacho-Garcia, J.; Bista, I.; Wood, J.; McCarthy, S.; et al. Genomic consequences of domestication of the Siamese fighting fish. Sci. Adv. 2022, 8, eabm4950. [Google Scholar] [CrossRef] [PubMed]
- Panthum, T.; Jaisamut, K.; Singchat, W.; Ahmad, S.F.; Kongkaew, L.; Wongloet, W.; Dokkaew, S.; Kraichak, E.; Muangmai, N.; Duengkae, P.; et al. Something fishy about Siamese fighting fish (Betta splendens) sex: Polygenic sex determination or a newly emerged sex-determining region? Cells 2022, 11, 1764. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, F.; Lee, M.; Yue, G.H. Whole-genome resequencing infers genomic basis of giant phenotype in Siamese fighting fish (Betta splendens). Zool. Res. 2022, 43, 78–80. [Google Scholar] [CrossRef]
- Saint-Pé, K.; Blanchet, S.; Tissot, L.; Poulet, N.; Plasseraud, O.; Loot, G.; Veyssière, C.; Prunier, J.G. Genetic admixture between captive-bred and wild individuals affects patterns of dispersal in a brown trout (Salmo trutta) population. Conserv. Genet. 2018, 19, 1269–1279. [Google Scholar] [CrossRef]
- Beer, S.D.; Cornett, S.; Austerman, P.; Trometer, B.; Hoffman, T.; Bartron, M.L. Genetic diversity, admixture, and hatchery influence in Brook Trout (Salvelinus fontinalis) throughout western New York State. Ecol. Evol. 2019, 9, 7455–7479. [Google Scholar] [CrossRef]
- Rüber, L.; Britz, R.; Kullander, S.O.; Zardoya, R. Evolutionary and biogeographic patterns of the Badidae (Teleostei: Perciformes) inferred from mitochondrial and nuclear DNA sequence data. Mol. Phylogenet. Evol. 2004, 32, 1010–1022. [Google Scholar] [CrossRef]
- Tanpitayacoop, C.; Na-Nakorn, U. Genetic variation of Betta spp. in Thailand by random amplified polymorphic DNA (RAPD) method. In Proceedings of the 43rd Kasetsart University Annual Conference, Bangkok, Thailand, 1–4 February 2005; Subject: Fisheries. 2005; pp. 185–192. [Google Scholar]
- Chailertrit, V.; Swatdipong, A.; Peyachoknagul, S.; Salaenoi, J.; Srikulnath, K. Isolation and characterization of novel microsatellite markers from Siamese fighting fish (Betta splendens, Osphronemidae, Anabantoidei) and their transferability to related species, B. smaragdina and B. imbellis. Genet. Mol. Res. 2014, 13, 7157–7162. [Google Scholar] [CrossRef] [PubMed]
- Kowasupat, C.; Panijpan, B.; Laosinchai, P.; Ruenwongsa, P.; Phongdara, A.; Wanna, W.; Senapin, S.; Phiwsaiya, K. Biodiversity of the Betta smaragdina (Teleostei: Perciformes) in the northeast region of Thailand as determined by mitochondrial COI and nuclear ITS1 gene sequences. Meta Gene 2014, 2, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Panijpan, B.; Kowasupat, C.; Laosinchai, P.; Ruenwongsa, P.; Phongdara, A.; Senapin, S.; Wanna, W.; Phiwsaiya, K.; Kuhne, J.; Fasquel, F. Southeast Asian mouth-brooding Betta fighting fish (Teleostei: Perciformes) species and their phylogenetic relationships based on mitochondrial COI and nuclear ITS1 DNA sequences and analyses. Meta Gene 2014, 2, 862–879. [Google Scholar] [CrossRef] [PubMed]
- Prakhongcheep, O.; Muangmai, N.; Peyachoknagul, S.; Srikulnath, K. Complete mitochondrial genome of mouthbrooding fighting fish (Betta pi) compared with bubble nesting fighting fish (B. splendens). Mitochondrial DNA B Resour. 2017, 3, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Ponjarat, J.; Areesirisuk, P.; Prakhongcheep, O.; Dokkaew, S.; Sillapaprayoon, S.; Muangmai, N.; Peyachoknagul, S.; Srikulnath, K. Complete mitochondrial genome of two mouthbrooding fighting fishes, Betta apollon and B. simplex (Teleostei: Osphronemidae). Mitochondrial DNA B Resour. 2019, 4, 672–674. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Laopichienpong, N.; Singchat, W.; Suntronpong, A.; Pongsanarm, T.; Panthum, T.; Ariyaraphong, N.; Bulan, J.; Pansrikaew, T.; Jangtarwan, K.; et al. Next-generation sequencing yields complete mitochondrial genome assembly of peaceful betta fish, Betta imbellis (Teleostei: Osphronemidae). Mitochondrial DNA B Resour. 2020, 5, 3856–3858. [Google Scholar] [CrossRef]
- Singchat, W.; Ahmad, S.F.; Laopichienpong, N.; Suntronpong, A.; Pongsanarm, T.; Panthum, T.; Ariyaraphong, N.; Subpayakom, N.; Dokkaew, S.; Muangmai, N.; et al. Complete mitochondrial genome of Mahachai betta, Betta mahachaiensis (Teleostei: Osphronemidae). Mitochondrial DNA B Resour. 2020, 5, 3059–3061. [Google Scholar] [CrossRef]
- Fahmi, M.R.; Kusrini, E.; Hayuningtiyas, E.P.; Sinansari, S.; Gustiano, R. DNA barcoding using COI gene sequences of wild betta fighting fish from Indonesia: Phylogeny, status and diversity. Indones. Fish. Res. J. 2020, 26, 97–105. [Google Scholar] [CrossRef]
- Laopichienpong, N.; Ahmad, S.F.; Singchat, W.; Suntronpong, A.; Pongsanarm, T.; Jangtarwan, K.; Bulan, J.; Pansrikaew, T.; Panthum, T.; Ariyaraphong, N.; et al. Complete mitochondrial genome of Mekong fighting fish, Betta smaragdina (Teleostei: Osphronemidae). Mitochondrial DNA B Resour. 2021, 6, 776–778. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, D.; Yu, S.; Li, Z.; Cao, Y.; Miao, Z.; Qian, H.; Tang, K. Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics 2004, 20, 1081–1086. [Google Scholar] [CrossRef]
- Goldstein, D.B.; Ruiz Linares, A.; Cavalli-Sforza, L.L.; Feldman, M.W. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. USA 1995, 92, 6723–6727. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Briñez, R.B.; Caraballo, O.X.; Salazar, V.M. Genetic diversity of six populations of red hybrid tilapia, using microsatellites genetic markers. Rev. MVZ Cordoba 2011, 16, 2491–2498. [Google Scholar]
- Danish, M.; Chauhan, R.; Kanyal, P.; Khati, A.; Chauhan, S. A review on molecular markers and their application in fisheries and aquaculture. Natl. J. Life Sci. 2015, 12, 47–55. [Google Scholar]
- Chen, H.; Li, X.; Wang, Y.; Zhu, C.; Huang, H.; Yang, W.; Li, G. De novo transcriptomic characterization enables novel microsatellite identification and marker development in Betta splendens. Life 2021, 11, 803. [Google Scholar] [CrossRef]
- Bleidorn, C. Third generation sequencing: Technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers. 2016, 14, 1–8. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef]
- Kim, C.; Guo, H.; Kong, W.; Chandnani, R.; Shuang, L.S.; Paterson, A.H. Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci. 2016, 242, 14–22. [Google Scholar] [CrossRef]
- Hunter, M.E.; Hoban, S.M.; Bruford, M.W.; Segelbacher, G.; Bernatchez, L. Next-generation conservation genetics and biodiversity monitoring. Evol. Appl. 2018, 11, 1029–1034. [Google Scholar] [CrossRef]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Koomgun, T.; Prasongmaneerut, T.; Thongchum, R.; Singchat, W.; Tawichasri, P.; Fukayama, T.; Sillapaprayoon, S.; Kraichak, E.; Muangmai, N.; et al. Take one step backward to move forward: Assessment of genetic diversity and population structure of captive Asian woolly-necked storks (Ciconia episcopus). PLoS ONE 2019, 14, e0223726. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Kamsongkram, P.; Subpayakom, N.; Sillapaprayoon, S.; Muangmai, N.; Kongphoemph, A.; Wongsodchuen, A.; Intapan, S.; Chamchumroon, W.; Safoowong, M.; et al. Predictive genetic plan for a captive population of the Chinese goral (Naemorhedus griseus) and prescriptive action for ex situ and in situ conservation management in Thailand. PLoS ONE 2020, 15, e0234064. [Google Scholar] [CrossRef]
- Hata, A.; Nunome, M.; Suwanasopee, T.; Duengkae, P.; Chaiwatana, S.; Chamchumroon, W.; Suzuki, T.; Koonawootrittriron, S.; Matsuda, Y.; Srikulnath, K. Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens. Sci. Rep. 2021, 11, 2035. [Google Scholar] [CrossRef]
- Vasemägi, A.; Nilsson, J.; Primmer, C.R. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Mol. Biol. Evol. 2005, 22, 1067–1076. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Li, Z.; Zhang, G.; Ma, Z. Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet. Mol. Res. 2011, 10, 1462–1470. [Google Scholar] [CrossRef]
- Hung, C.M.; Yu, A.Y.; Lai, Y.T.; Shaner, P.J. Developing informative microsatellite markers for non-model species using reference mapping against a model species’ genome. Sci. Rep. 2016, 6, 23087. [Google Scholar] [CrossRef]
- Ariede, R.B.; Freitas, M.V.; Hata, M.E.; Mastrochirico-Filho, V.A.; Pilarski, F.; Batlouni, S.R.; Porto-Foresti, F.; Hashimoto, D.T. Microsatellites associated with growth performance and analysis of resistance to Aeromonas hydrophila in Tambaqui Colossoma macropomum. Front. Genet. 2018, 9, 3. [Google Scholar] [CrossRef]
- Olschewsky, A.; Hinrichs, D. An overview of the use of genotyping techniques for assessing genetic diversity in local farm animal breeds. Animals 2021, 11, 2016. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B.; Hancock, J. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Martins, W.S.; Lucas, D.C.; Neves, K.F.; Bertioli, D.J. WebSat--a web software for microsatellite marker development. Bioinformation 2009, 3, 282–283. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, Z.; Wang, C.; Zhang, X.; Li, J.; Yue, B. Characterization of perfect microsatellite based on genome-wide and chromosome level in Rhesus monkey (Macaca mulatta). Gene 2016, 592, 269–275. [Google Scholar] [CrossRef]
- Song, X.; Yang, T.; Yan, X.; Zheng, F.; Xu, X.; Zhou, C. Comparison of microsatellite distribution patterns in twenty-nine beetle genomes. Gene 2020, 757, 144919. [Google Scholar] [CrossRef]
- Singh, P.; Nath, R.; Venkatesh, V. Comparative genome-wide characterization of microsatellites in Candida albicans and Candida dubliniensis leading to the development of species-specific marker. Public Health Genom. 2021, 24, 1–13. [Google Scholar] [CrossRef]
- Zhou, C.; Li, F.; Wen, Q.; Price, M.; Yang, N.; Yue, B. Characterization of microsatellites in the endangered snow leopard based on the chromosome-level genome. Mamm. Res. 2021, 66, 385–398. [Google Scholar] [CrossRef]
- Fan, G.; Chan, J.; Ma, K.; Yang, B.; Zhang, H.; Yang, X.; Shi, C.; Chun-Hin Law, H.; Ren, Z.; Xu, Q.; et al. Chromosome-level reference genome of the Siamese fighting fish Betta splendens, a model species for the study of aggression. Gigascience 2018, 7, giy087. [Google Scholar] [CrossRef]
- Rhie, A.; McCarthy, S.A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J.; et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 2021, 592, 737–746. [Google Scholar] [CrossRef]
- Kassambara, A.; Kassambara, M.A. R Package ‘ggpubr’. 2020. Available online: https://cran.Rproject.org/web/packages/ggpubr/ggpubr.pdf (accessed on 5 May 2022).
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Hamilton, F. An Account of the Fishes Found in the River Ganges and Its Branches; Archibald Constable and Co.: Edinburgh, Scotland, 1822. [Google Scholar]
- Temminck, C.J.; Schlegel, H. Pisces. Fauna Japonica, Sive Descriptio Animalium, quae in Itinere per Japoniam … Suscepto annis 1823–1830 Collegit, Notis, Observationibus et Adumbrationibus Illustravit Ph. Fr. de Siebold. Lugduni Batavorum [Leiden] (A. Arnz et soc.); London, England, 1846; pp. 173–269. Available online: https://www.biodiversitylibrary.org/bibliography/124951(1846) (accessed on 16 August 2022).
- Winchell, A. Description of a gar-pike, supposed to be new--Lepidosteus (Cylindrosteus) oculatus. Proc. Acad. Nat. Sci. USA 1864, 16, 183–185. [Google Scholar]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Meglécz, E.; Pech, N.; Gilles, A.; Dubut, V.; Hingamp, P.; Trilles, A.; Grenier, R.; Martin, J.F. QDD version 3.1: A user-friendly computer program for microsatellite selection and primer design revisited: Experimental validation of variables determining genotyping success rate. Mol. Ecol. Resour. 2014, 14, 1302–1313. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef]
- Merritt, B.J.; Culley, T.M.; Avanesyan, A.; Stokes, R.; Brzyski, J. An empirical review: Characteristics of plant microsatellite markers that confer higher levels of genetic variation. Appl. Plant Sci. 2015, 3, 1500025. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.; Weng, Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef]
- Cai, G.; Leadbetter, C.W.; Muehlbauer, M.F.; Molnar, T.J.; Hillman, B.I. Genome-wide microsatellite identification in the fungus Anisogramma anomala using Illumina sequencing and genome assembly. PLoS ONE 2013, 8, e82408. [Google Scholar] [CrossRef]
- Castoe, T.A.; Poole, A.W.; Gu, W.; Jason de Koning, A.P.; Daza, J.M.; Smith, E.N.; Pollock, D.D. Rapid identification of thousands of copperhead snake (Agkistrodon contortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol. Ecol. Resour. 2010, 10, 341–347. [Google Scholar] [CrossRef]
- Meglécz, E.; Costedoat, C.; Dubut, V.; Gilles, A.; Malausa, T.; Pech, N.; Martin, J.F. QDD: A user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 2010, 26, 403–404. [Google Scholar] [CrossRef]
- Yu, J.N.; Won, C.; Jun, J.; Lim, Y.; Kwak, M. Fast and cost-effective mining of microsatellite markers using NGS technology: An example of a Korean water deer Hydropotes inermis argyropus. PLoS ONE 2011, 6, e26933. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 May 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Willems, T.; Zielinski, D.; Yuan, J.; Gordon, A.; Gymrek, M.; Erlich, Y. Genome-wide profiling of heritable and de novo STR variations. Nat. Methods 2017, 14, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Kowasupat, C.; Panijpan, B.; Ruenwongsa, P.; Sriwattanarothai, N. Betta mahachaiensis, a new species of bubble-nesting fighting fish (Teleostei: Osphronemidae) from Samut Sakhon Province, Thailand. Zootaxa 2012, 3522, 49–60. [Google Scholar] [CrossRef]
- Ladiges, W. Betta smaragdina nov. spec. Die Aquar. und Terr. Z. 1972, 25, 190–191. [Google Scholar]
- Ladiges, W. Betta imbellis nov. spec., der Friedliche Kampffisch. Die Aquar. und Terr. Z. 1975, 28, 262–264. [Google Scholar]
- Perugia, A. Di alcuni pesci raccolti in Sumatra dal Dott. Elio Modigliani. In Annali del Museo Civico di Storia Naturale Giacoma Doria, Genova; The Museum: Genoa, Italy, 1893; Volume 13, pp. 241–247. [Google Scholar]
- Witte, K.; Schmidt, J. Betta brownorum, a new species of anabantoids (Teleostei: Belontiidae) from northwestern Borneo, with a key to the genus. Ichthyol. Explor. Freshw. 1992, 2, 305–330. [Google Scholar]
- Schaller, D. Betta tussyae spec. nov., ein neuer Kampffisch aus Malaysia (vorläufige Mitteilung). Die Aquar. und Terr. Z. 1985, 38, 348–350. [Google Scholar]
- Tan, H.; Tan, S. Redescription of the Malayan fighting fish Betta pugnax (Teleostei: Belontiidae), and description of Betta pulchra, new species from Peninsular Malaysia. Raffles Bull. Zool. 1996, 44, 419–434. [Google Scholar]
- Ng, P.K.; Kottelat, M. Betta livida, a new fighting fish (Teleostei: Belontiidae) from blackwater swamps in Peninsular Malaysia. Ichthyol. Explor. Freshw. 1992, 3, 177–182. [Google Scholar]
- Kottelat, M.; Ng, P.K. Diagnose of five new species of fighting fishes from Banka and Borneo (Teleostei: Belontiidae). Ichthyol. Explor. Freshw. 1994, 5, 65–78. [Google Scholar]
- Sauvage, H. Note sur une collection de poissons recueillie à Pérak, presqu’île de Malacca. Bull. Soc. Zool. Fr. 1884, 9, 216–220. [Google Scholar]
- Hui, T.H.; Ng, P.K. Six new species of fighting fish (Teleostei: Osphronemidae: Betta) from Borneo. Ichthyol. Explor. Freshw. 2006, 17, 97. [Google Scholar]
- Ng, P.K.; Kottelat, M. Revision of the Betta-waseri species group (Teleostei, Belontiidae). Raffles Bull. Zool. 1994, 42, 593–611. [Google Scholar]
- Danecek, P.; McCarthy, S.A. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics 2017, 33, 2037–2039. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015, 14, 13981–13997. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph. D. Thesis, University of Dublin, Dublin, Ireland, 2001. [Google Scholar]
- Kowasupat, C.; Panijpan, B.; Ruenwongsa, P.; Jeenthong, T. Betta siamorientalis, a new species of bubble-nest building fighting fish (Teleostei: Osphronemidae) from eastern Thailand. Vertebr. Zool. 2012, 62, 387–397. [Google Scholar]
- Tan, H. Description of two new species of the Betta waseri group (Teleostei: Osphronemidae). Ichthyol. Explor. Freshw. 1998, 8, 281–287. [Google Scholar]
- Schindler, I.; Schmidt, J. Betta pallida spec. nov., a new fighting fish from southern Thailand (Teleostei: Be lontiidae). Z. für Fischkunde 2004, 7, 1–4. [Google Scholar]
- Schindler, I.; Schmidt, J. Review of the mouthbrooding Betta (Teleostei, Osphronemidae) from Thailand, with descriptions of two new species. Z. für Fischkunde 2006, 8, 47–69. [Google Scholar]
- Cantor, T.E. Catalogue of Malayan fishes. J. Asiat. Soc. Bengal 1849, 18, i-xii+983–i-xii+1443. [Google Scholar]
- Avvaru, A.K.; Sharma, D.; Verma, A.; Mishra, R.K.; Sowpati, D.T. MSDB: A comprehensive, annotated database of microsatellites. Nucleic Acids Res. 2020, 48, D155–D159. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genom. 2021, 22, 421. [Google Scholar] [CrossRef]
- Brandström, M.; Ellegren, H. Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res. 2008, 18, 881–887. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Lemos, F.; Monteiro, G.A.; Prazeres, D.M. Recombination frequency in plasmid DNA containing direct repeats--predictive correlation with repeat and intervening sequence length. Plasmid 2008, 60, 159–165. [Google Scholar] [CrossRef]
- Linnaeus, C. Systema Naturae, per regna tria naturae secundum classes, ordines, genera, species cum characteribus, differentiis, synonymis, locis. Editio Decima, Reformata; Laurentius Salvius: Stockholm, Sweden, 1758; Volume 1, pp. 1–824. [Google Scholar]
- Temminck, C.J.; Schlegel, H. Pisces. Fauna Japonica, Sive Descriptio Animalium, Quae in Itinere per Japoniam … Suscepto annis 1823–1830 Collegit, Notis, Observationibus et Adumbrationibus Illustravit Ph. Fr. de Siebold. Lugduni Batavorum [Leiden] (A. Arnz et soc.); London, England, 1850; pp. 270–324. Available online: https://www.biodiversitylibrary.org/bibliography/124951(1850) (accessed on 16 August 2022).
- de Filippi, F. Nouvelles espèces de poissons. Rev. Mag. Zool. 1853, 5, 164–171. [Google Scholar]
- Walbaum, J.J. Petri Artedi sueci genera Piscium inquibus systema totum ichthyologiae proponitur cum classibus, ordinibus, generum characteribus, specierum diffentiis, observationibus plumiris. Ichthyologiae Pars III; Grypeswaldiae, Impensis Ant, Ferdin, Röse, 1792. Available online: https://www.abebooks.com/first-edition/Petri-Artedi-sueci-Genera-piscium-quibus/15466141115/bd (accessed on 16 August 2022).
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef]
- Merkel, A.; Gemmell, N.J. Detecting microsatellites in genome data: Variance in definitions and bioinformatic approaches cause systematic bias. Evol. Bioinform. Online 2008, 4, EBO-S420. [Google Scholar] [CrossRef]
- Leclercq, S.; Rivals, E.; Jarne, P. Detecting microsatellites within genomes: Significant variation among algorithms. BMC Bioinformatics 2007, 8, 125. [Google Scholar] [CrossRef]
- Cuvier, G.; Valenciennes, A. Histoire naturelle des poissons. Tome second. Livre Troisième. Des poissons de la famille des perches, ou des percoïdes. Hist. Nat. Poisson. 1828, 2, 1–490. [Google Scholar]
- Przewalski, N.M. From Zaisanthrough Khami to the Tibet and Upper Reaches of the Yellow River: Third Journey in the Central Asia (1879–1880); Imperial Russian Geographic Society: St. Petersburg, Russia, 1883. [Google Scholar]
- Abel, C. On the supposed unicorn of the Himalayas. Philos. Mag. J. 1826, 68, 232–234. [Google Scholar]
- Qi, W.H.; Jiang, X.M.; Du, L.M.; Xiao, G.S.; Hu, T.Z.; Yue, B.S.; Quan, Q.M. Genome-wide survey and analysis of microsatellite sequences in bovid species. PLoS ONE 2015, 10, e0133667. [Google Scholar] [CrossRef]
- Gomes, N.M.; Shay, J.W.; Wright, W.E. Telomere biology in Metazoa. FEBS Lett. 2010, 584, 3741–3751. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, chromatin and evolution of satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Bellott, D.W.; Skaletsky, H.; Pyntikova, T.; Mardis, E.R.; Graves, T.; Kremitzki, C.; Brown, L.G.; Rozen, S.; Warren, W.C.; Wilson, R.K.; et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 2010, 466, 612–616. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Ichikawa, K.; Tomioka, S.; Suzuki, Y.; Nakamura, R.; Doi, K.; Yoshimura, J.; Kumagai, M.; Inoue, Y.; Uchida, Y.; Irie, N.; et al. Centromere evolution and CpG methylation during vertebrate speciation. Nat. Commun. 2017, 8, 1833. [Google Scholar] [CrossRef]
- The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [Google Scholar] [CrossRef]
- Herpin, A.; Schartl, M. Plasticity of gene-regulatory networks controlling sex determination: Of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 2015, 16, 1260–1274. [Google Scholar] [CrossRef]
- Brunner, B.; Hornung, U.; Shan, Z.; Nanda, I.; Kondo, M.; Zend-Ajusch, E.; Haaf, T.; Ropers, H.H.; Shima, A.; Schmid, M.; et al. Genomic organization and expression of the doublesex-related gene cluster in vertebrates and detection of putative regulatory regions for DMRT1. Genomics 2001, 77, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, G.; Han, H.; Li, Y.; Li, J.; Wang, J.; Cao, G.; Li, X. Genome collinearity analysis illuminates the evolution of donkey chromosome 1 and horse chromosome 5 in perissodactyls: A comparative study. BMC Genom. 2021, 22, 665. [Google Scholar] [CrossRef] [PubMed]
- Nagabhushana, A.; Mishra, R.K. Finding clues to the riddle of sex determination in zebrafish. J. Biosci. 2016, 41, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Andrews, C. The ornamental fish trade and fish conservation. J. Fish. Biol. 1990, 37, 53–59. [Google Scholar] [CrossRef]
- Griffin, M.; Coetzee, C.G. Annotated Checklist and Provisional National Conservation Status of Namibian Mammals; Directorate of Scientific Services, Ministry of Environment and Tourism: Windhoek, Namibia, 2005. [Google Scholar]
- Lim, K.G.; Kwoh, C.K.; Hsu, L.Y.; Wirawan, A. Review of tandem repeat search tools: A systematic approach to evaluating algorithmic performance. Brief Bioinform. 2013, 14, 67–81. [Google Scholar] [CrossRef]
- Domanic, N.O.; Preparata, F.P. A novel approach to the detection of genomic approximate tandem repeats in the Levenshtein metric. J. Comput. Biol. 2007, 14, 873–891. [Google Scholar] [CrossRef]
- Vukosavljev, M.; Esselink, G.D.; van ’t Westende, W.P.; Cox, P.; Visser, R.G.; Arens, P.; Smulders, M.J. Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol. Ecol. Resour. 2015, 15, 17–27. [Google Scholar] [CrossRef]
- Lopez, L.; Barreiro, R.; Fischer, M.; Koch, M.A. Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants. BMC Genom. 2015, 16, 781. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, W.; Yuan, S.; Zheng, Y.; Zeng, X. Microsatellite development from genome skimming and transcriptome sequencing: Comparison of strategies and lessons from frog species. BMC Genom. 2018, 19, 886. [Google Scholar] [CrossRef]
- Zhou, Y.; Tong, J.; Wang, J.; Yu, X. Development of microsatellite markers and genetic diversity in wild and cultured populations of black carp (Mylopharyngodon piceus) along the Yangtze River. Aquacult. Int. 2020, 28, 1867–1882. [Google Scholar] [CrossRef]
- Fazzi-Gomes, P.F.; Aguiar, J.D.P.; Marques, D.; Fonseca Cabral, G.; Moreira, F.C.; Rodrigues, M.D.N.; Silva, C.S.; Hamoy, I.; Santos, S. Novel microsatellite markers used for determining genetic diversity and tracing of wild and farmed populations of the amazonian giant fish Arapaima gigas. Genes 2021, 12, 1324. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, V.C.F.; Yamachita, A.L.; de Souza, F.P.; de Godoy, S.M.; de Lima, E.C.S.; Feliciano, D.C.; de Pádua Pereira, U.; Povh, J.A.; Ayres, D.R.; Bignardi, A.B. Development of microsatellite markers and evaluation of genetic diversity of the Amazonian ornamental fish Pterophyllum scalare. Aquacult. Int. 2021, 29, 2435–2449. [Google Scholar] [CrossRef]
- Wang, X.; Weng, Z.; Yang, Y.; Hua, S.; Zhang, H.; Meng, Z. Genetic evaluation of black sea bream (Acanthopagrus schlegelii) stock enhancement in the South China sea based on microsatellite DNA markers. Fishes 2021, 6, 47. [Google Scholar] [CrossRef]
- Hedrick, P.W. A standardized genetic differentiation measure. Evolution 2005, 59, 1633–1638. [Google Scholar] [CrossRef]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central–marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef]
- González-Castellano, I.; Perina, A.; González-Tizón, A.M.; Torrecilla, Z.; Martínez-Lage, A. Isolation and characterization of 21 polymorphic microsatellite loci for the rockpool shrimp Palaemon elegans using Illumina MiSeq sequencing. Sci. Rep. 2018, 8, 17197. [Google Scholar] [CrossRef]
- Barbará, T.; Palma-Silva, C.; Paggi, G.M.; Bered, F.; Fay, M.F.; Lexer, C. Cross-species transfer of nuclear microsatellite markers: Potential and limitations. Mol. Ecol. 2007, 16, 3759–3767. [Google Scholar] [CrossRef]
- Yue, G.H.; Kovacs, B.; Orban, L. A new problem with cross-species amplification of microsatellites: Generation of non-homologous products. Dongwuxue Yanjiu 2010, 31, 131–140. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Patterson, L.B.; Parichy, D.M. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet. 2013, 9, e1003561. [Google Scholar] [CrossRef]
- Kolvenbach, C.M.; Dworschak, G.C.; Frese, S.; Japp, A.S.; Schuster, P.; Wenzlitschke, N.; Yilmaz, Ö.; Lopes, F.M.; Pryalukhin, A.; Schierbaum, L.; et al. Rare variants in BNC2 are implicated in autosomal-dominant congenital lower urinary-tract obstruction. Am. J. Hum. Genet. 2019, 104, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Rual, J.F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005, 437, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Tijssen, M.R.; Cvejic, A.; Joshi, A.; Hannah, R.L.; Ferreira, R.; Forrai, A.; Bellissimo, D.C.; Oram, S.H.; Smethurst, P.A.; Wilson, N.K.; et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 2011, 20, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Melvin, V.S.; Feng, W.; Hernandez-Lagunas, L.; Artinger, K.B.; Williams, T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Dev. Dyn. 2013, 242, 817–831. [Google Scholar] [CrossRef]
- Mei, X.; Westfall, T.A.; Zhang, Q.; Sheffield, V.C.; Bassuk, A.G.; Slusarski, D.C. Functional characterization of Prickle2 and BBS7 identify overlapping phenotypes yet distinct mechanisms. Dev. Biol. 2014, 392, 245–255. [Google Scholar] [CrossRef]
- Lee, S.; Page-McCaw, P.; Gamse, J.T. Kctd12 and Ulk2 partner to regulate dendritogenesis and behavior in the habenular nuclei. PLoS ONE 2014, 9, e110280. [Google Scholar] [CrossRef]
- Taylor, R.W.; Qi, J.Y.; Talaga, A.K.; Ma, T.P.; Pan, L.; Bartholomew, C.R.; Klionsky, D.J.; Moens, C.B.; Gamse, J.T. Asymmetric inhibition of Ulk2 causes left-right differences in habenular neuropil formation. J. Neurosci. 2011, 31, 9869–9878. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.; Palma, K.; Armijo, L.; Mione, M.; Signore, I.A.; Morales, C.; Guerrero, N.; Meynard, M.M.; Pérez, R.; Suazo, J.; et al. Daam1a mediates asymmetric habenular morphogenesis by regulating dendritic and axonal outgrowth. Development 2013, 140, 3997–4007. [Google Scholar] [CrossRef]
- Cayuso, J.; Dzementsei, A.; Fischer, J.C.; Karemore, G.; Caviglia, S.; Bartholdson, J.; Wright, G.J.; Ober, E.A. EphrinB1/EphB3b coordinate bidirectional epithelial-mesenchymal interactions controlling liver morphogenesis and laterality. Dev. Cell. 2016, 39, 316–328. [Google Scholar] [CrossRef]
- Cavodeassi, F.; Ivanovitch, K.; Wilson, S.W. Eph/Ephrin signalling maintains eye field segregation from adjacent neural plate territories during forebrain morphogenesis. Dev. 2013, 140, 4193–4202. [Google Scholar] [CrossRef]
| Species | Common Name | Breeding Type | Location | Latitude | Specimens |
|---|---|---|---|---|---|
| Betta splendens | Siamese fighting fish | Bubble nesting | Bangkok | 13.7563° N, 100.5018° E | 10 |
| Chumphon | 10.4930° N, 99.1800° E | 5 | |||
| Surat Thani | 9.1342° N, 99.3334° E | 6 | |||
| Betta mahachaiensis | Mahachai betta | Bubble nesting | Samut Sakhon | 13.5498° N, 100.2741° E | 2 |
| Betta siamorientalis | Eastern wild betta | Bubble nesting | Chachoengsao | 13.6904° N, 101.0780° E | 2 |
| Betta imbellis | Peaceful betta | Bubble nesting | Krabi | 8.0863° N, 98.9063° E | 2 |
| Betta smaragdina | Mekong fighting fish | Bubble nesting | Mukdahan | 16.5436° N, 104.7024° E | 2 |
| Betta prima | Three-lined mouth-brooder | Mouth-brooders | Chanthaburi | 12.6112° N, 102.1038° E | 2 |
| Betta simplex | Simple mouth-brooder | Mouth-brooders | Unknown | - | 2 |
| Betta pi | Pi betta | Mouth-brooders | Unknown | - | 2 |
| Betta pallida | Pallida betta | Mouth-brooders | Unknown | - | 1 |
| Betta apollon | Apollon betta | Mouth-brooders | Unknown | - | 2 |
| Betta ferox | Ferox betta | Mouth-brooders | Unknown | - | 2 |
| Betta pugnax | Penang betta | Mouth-brooders | Phatthalung | 7.6167° N, 100.0740° E | 2 |
| Locus | Primer Sequence (5’ to 3’) | Repeat | Ta (°C) | Size Range (bp) | Chromosome | Gene | Genomic Region | Source |
|---|---|---|---|---|---|---|---|---|
| BettaMS4 | F: GTTTCATCAGGAGCAGCAGCATAA R: CTGTTTGATGGCCGACTTTT | ) | 59 | 259–315 | 12 | Noncoding gene | - | [13] |
| BettaMS5 | F: GTTTCGTCACCTTCTGAGCAAACA R: AAATGCGCTGGGTAGACTTG | ) | 59 | 198–218 | 3 | ush2a | Between intron and exon | [13] |
| BettaMS8 | F: CGTGAGCTGCAAAGAAAACA R: GCTGTTGCACATGAATCCAG | ) | 57 | 223 | 14 | tcf7 | Intron | [13] |
| BettaMS15 | F: ACTGTAACCGGGCTGTTCTG R: AACGCACCCAGAAACAAATC | ) | 57 | 216–225 | 22 | dlgap2a | Intron | [13] |
| BettaMS17 | F: AAGCAGGTCTTTCACCTCCA R: TCACCCTGCGTCTAAGTCAA | ) | 61 | 194–221 | 16 | Noncoding gene | - | [13] |
| BettaMS23 | F: GTTTGAGAGAAATGGGTTCTTCG R: TCACTACGCTGCCAAATCAG | ) | 55 | 277–296 | 4 | LOC114853122+ | Intron | [13] |
| BettaMS25 | F: GTTTGGGTAAAACCCAACTCTGG R: AACGTCACGTGGAACAGATG | ) | 55 | 194–224 | 15 | ctu2 | Between intron and exon | [13] |
| BettaMS40 | F: CAGTACATTTGACTGATCGCAGA R: CAGGATGCTTCCTTGGGTAA | ) | 57 | 136–165 | 12 | Noncoding gene | - | [13] |
| BettaMS10.1 | F: TCTGAGGAAGGAGGCGATTA R: GCGTGCACTGAAGCATAAAG | ) | 55 | 280–313 | 9 | slc20a2 | Between intron and exon | This study |
| BettaMS14.1 | F: GGGCTGCACCTTAAACTCAT R: GTCCACTGGGCTGATGTTCT | 55 | 324–396 | 2 | LOC114850832 + | Between intron and exon | This study | |
| BettaMS2.2 | F: ATTCCTTTCTGCCGCTAA R: AAAGAGGGCACTAAGCCA | 50 | 165–199 | 22 | meis2a | Intron | This study | |
| BettaMS14.2 | F: CCCGGTTTCTTGTCATTC R: CGCTGATGGAAATTGAGT | 50 | 228 | 21 | Noncoding gene | - | This study |
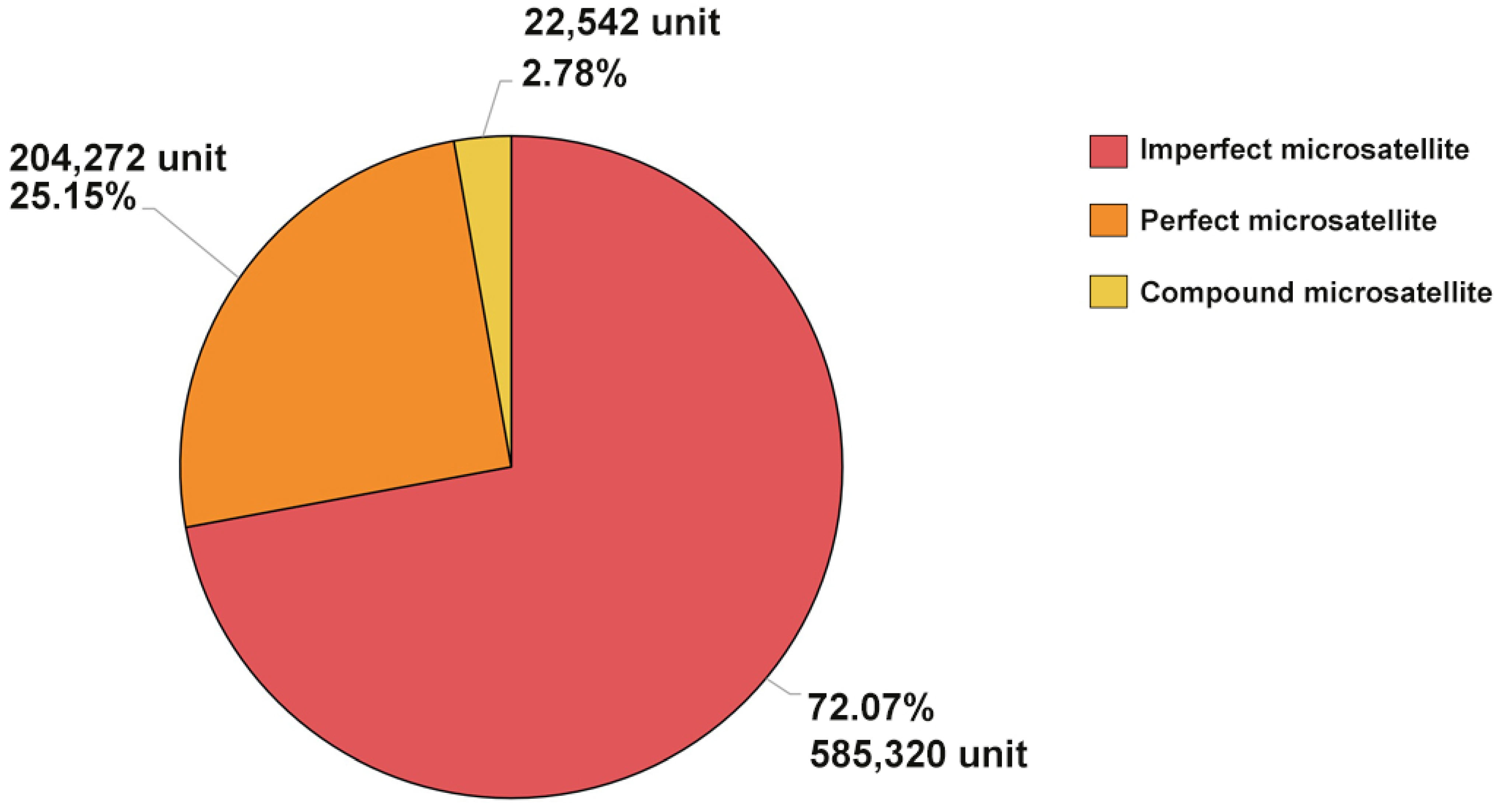
| Description | Perfect Microsatellite | Imperfect Microsatellite | Compound Microsatellite | |
|---|---|---|---|---|
| Total number of microsatellites | Unit | 204,272 | 585,320 | 22,542 |
| Total length of microsatellites | bp | 7,010,790 | 21,999,074 | 1,692,553 |
| Average of microsatellites | Total microsatellite length/total microsatellite count (bp) | 34.33 | 37.58 | 75.08 |
| Microsatellites per sequence | Total microsatellite counts/sequence counts | 2918 | 8,362 | 322 |
| Percentage of sequence covered by microsatellites | Total microsatellite length/total sequence length (%) | 1.59 | 4.98 | 0.004 |
| Relative abundance | Total microsatellites/total valid length (loci/Mb) | 462.82 | 1326.18 | 51.07 |
| Relative density | Total microsatellite length/total valid length (bp/Mb) | 15,884.54 | 49,843.92 | 3,834.86 |
| Experiment | Na | Ho | He | PIC | |
|---|---|---|---|---|---|
| This study | Mean | 11.500 | 0.417 | 0.667 | 0.659 |
| S.E. | 4.330 | 0.203 | 0.223 | 0.221 | |
| Chailertrit et al., 2014 | Mean | 11.125 | 0.570 | 0.779 | 0.762 |
| S.E. | 1.529 | 0.093 | 0.068 | 0.070 | |
| Both | Mean | 11.250 | 0.519 | 0.742 | 0.728 |
| S.E. | 1.643 | 0.089 | 0.082 | 0.082 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wattanadilokchatkun, P.; Panthum, T.; Jaisamut, K.; Ahmad, S.F.; Dokkaew, S.; Muangmai, N.; Duengkae, P.; Singchat, W.; Srikulnath, K. Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity. Fishes 2022, 7, 251. https://doi.org/10.3390/fishes7050251
Wattanadilokchatkun P, Panthum T, Jaisamut K, Ahmad SF, Dokkaew S, Muangmai N, Duengkae P, Singchat W, Srikulnath K. Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity. Fishes. 2022; 7(5):251. https://doi.org/10.3390/fishes7050251
Chicago/Turabian StyleWattanadilokchatkun, Pish, Thitipong Panthum, Kitipong Jaisamut, Syed Farhan Ahmad, Sahabhop Dokkaew, Narongrit Muangmai, Prateep Duengkae, Worapong Singchat, and Kornsorn Srikulnath. 2022. "Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity" Fishes 7, no. 5: 251. https://doi.org/10.3390/fishes7050251
APA StyleWattanadilokchatkun, P., Panthum, T., Jaisamut, K., Ahmad, S. F., Dokkaew, S., Muangmai, N., Duengkae, P., Singchat, W., & Srikulnath, K. (2022). Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity. Fishes, 7(5), 251. https://doi.org/10.3390/fishes7050251









