Larval Retention and Homing Behaviour Shape the Genetic Structure of the Bullet Tuna (Auxis rochei) in the Mediterranean Sea
Abstract
1. Introduction
2. Materials and Methods
3. Results
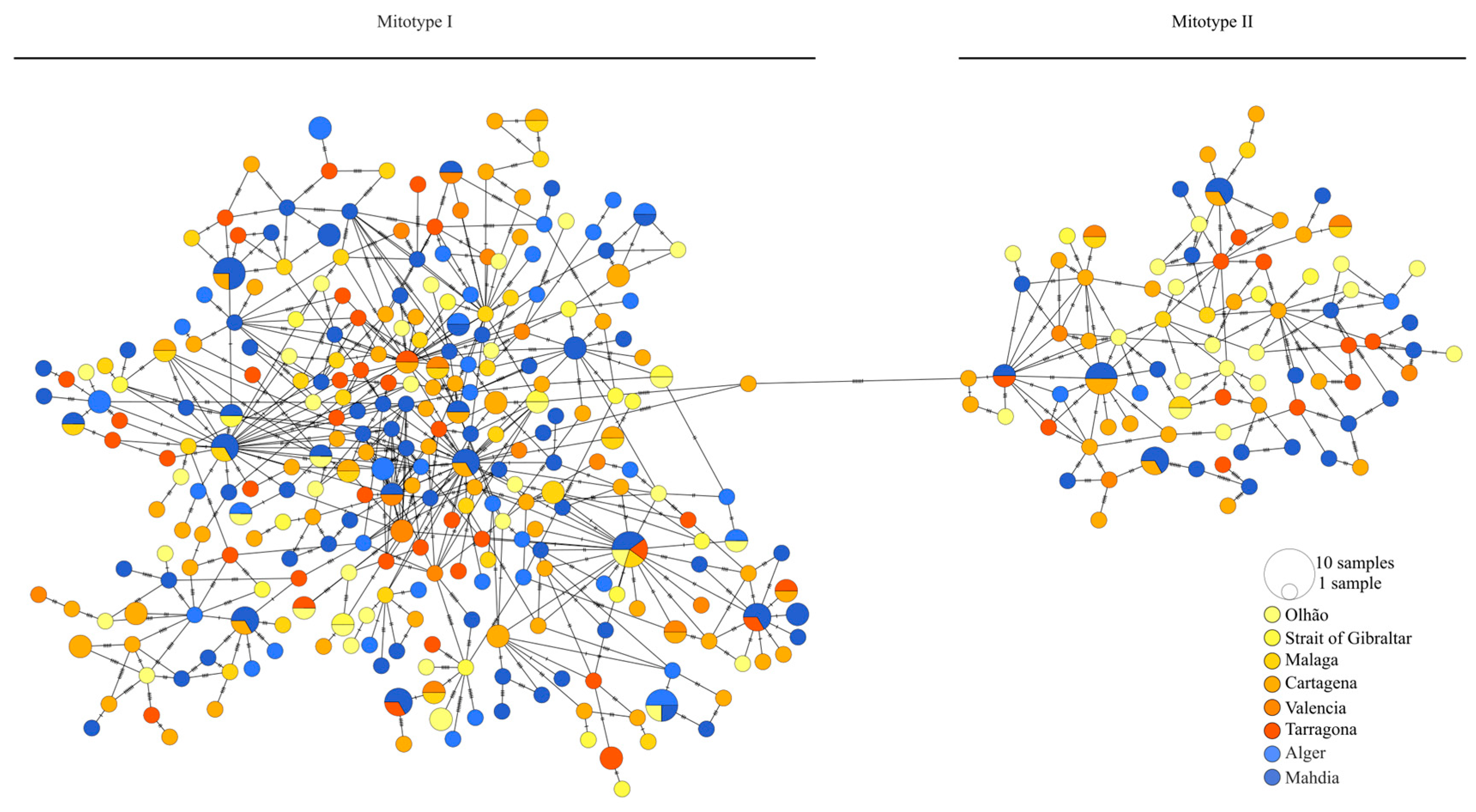
3.1. mtDNA CR Marker
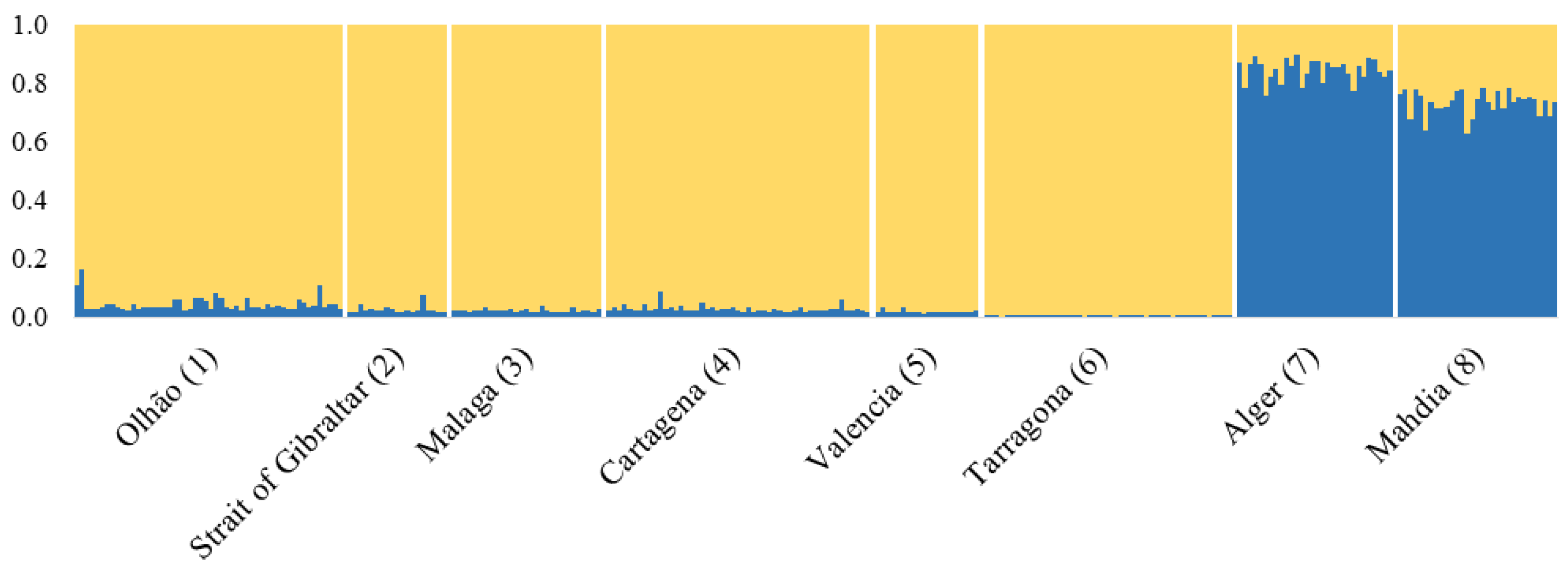
3.2. Microsatellite Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waples, R.S.; Naish, K.A. Genetic and Evolutionary Considerations in Fishery Management: Research Needs for the Future. In The Future of Fisheries Science in North America; Beamish, R., Rothschild, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 427–451. [Google Scholar] [CrossRef]
- Ward, R.D. Population Genetics of Tunas. J. Fish Biol. 1995, 47, 259–280. [Google Scholar] [CrossRef]
- Ward, R.D. Genetics in Fisheries Management. Hydrobiologia 2000, 420, 191–201. [Google Scholar] [CrossRef]
- Palumbi, S.R. Genetic Divergence, Reprodutive Isolation and Marine Speciation. Annu. Rev. Ecol. Syst. 1994, 25, 547–572. [Google Scholar] [CrossRef]
- Pascual, M.; Rives, B.; Schunter, C.; Macpherson, E. Impact of Life History Traits on Gene Flow: A Multispecies Systematic Review across Oceanographic Barriers in the Mediterranean Sea. PLoS ONE 2017, 12, e0176419. [Google Scholar] [CrossRef]
- Patarnello, T.; Volckaert, F.A.M.J.; Castilho, R. Pillars of Hercules: Is the Atlantic-Mediterranean Transition a Phylogeographical Break? Mol. Ecol. 2007, 16, 4426–4444. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, H.; Jorde, P.E.; André, C.; Stenseth, N.C. Fine-Scaled Geographical Population Structuring in a Highly Mobile Marine Species: The Atlantic Cod. Mol. Ecol. 2003, 12, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, K.A.; D’Aloia, C.C.; Crandall, E.D.; Iacchei, M.; Liggins, L.; Puritz, J.B.; Von Der Heyden, S.; Toonen, R.J. A Decade of Seascape Genetics: Contributions to Basic and Applied Marine Connectivity. Mar. Ecol. Prog. Ser. 2016, 554, 1–19. [Google Scholar] [CrossRef]
- Manel, S.; Holderegger, R. Ten Years of Landscape Genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef]
- Rooker, J.R.; Alvarado Bremer, J.R.; Block, B.A.; Dewar, H.; De Metrio, G.; Corriero, A.; Kraus, R.T.; Prince, E.D.; Rodriguez-Marin, E.; Secor, D.H. Life History and Stock Structure of Atlantic Bluefin Tuna (Thunnus thynnus). Rev. Fish. Sci. 2007, 15, 265–310. [Google Scholar] [CrossRef]
- Rooker, J.R.; Secor, D.H.; DeMetrio, G.; Kaufman, A.J.; Ríos, A.B.; Tičina, V. Evidence of Trans-Atlantic Movement and Natal Homing of Bluefin Tuna from Stable Isotopes in Otoliths. Mar. Ecol. Prog. Ser. 2008, 368, 231–239. [Google Scholar] [CrossRef]
- Rooker, J.R.; Arrizabalaga, H.; Fraile, I.; Secor, D.H.; Dettman, D.L.; Abid, N.; Addis, P.; Deguara, S.; Karakulak, F.S.; Kimoto, A.; et al. Crossing the Line: Migratory and Homing Behaviors of Atlantic Bluefin Tuna. Mar. Ecol. Prog. Ser. 2014, 504, 265–276. [Google Scholar] [CrossRef]
- Rodríguez-Ezpeleta, N.; Díaz-Arce, N.; Walter, J.F.; Richardson, D.E.; Rooker, J.R.; Nøttestad, L.; Hanke, A.R.; Franks, J.S.; Deguara, S.; Lauretta, M.V.; et al. Determining Natal Origin for Improved Management of Atlantic Bluefin Tuna. Front. Ecol. Environ. 2019, 17, 439–444. [Google Scholar] [CrossRef]
- Viñas, J.; Alvarado Bremer, J.R.; Pla, C. Phylogeography and Phylogeny of the Epineritic Cosmopolitan Bonitos of the Genus Sarda (Cuvier): Inferred Patterns of Intra- and Inter-Oceanic Connectivity Derived from Nuclear and Mitochondrial DNA Data. J. Biogeogr. 2010, 37, 557–570. [Google Scholar] [CrossRef]
- Viñas, J.; Alvarado Bremer, J.R.; Pla, C. Inter-Oceanic Genetic Differentiation among Albacore (Thunnus alalunga) Populations. Mar. Biol. 2004, 145, 225–232. [Google Scholar] [CrossRef]
- Chow, S.; Takeyama, H. Nuclear and Mitochondrial DNA Analyses Reveal Four Genetically Separated Breeding Units of the Swordfish. J. Fish Biol. 2000, 56, 1087–1098. [Google Scholar] [CrossRef]
- Zardoya, R.; Castilho, R.; Grande, C.; Favre-Krey, L.; Caetano, S.; Marcato, S.; Krey, G.; Patarnello, T. Differential Population Structuring of Two Closely Related Fish Species, the Mackerel (Scomber scombrus) and the Chub Mackerel (Scomber japonicus), in the Mediterranean Sea. Mol. Ecol. 2004, 13, 1785–1798. [Google Scholar] [CrossRef]
- Viñas, J.; Alvarado Bremer, J.R.; Pla, C. Phylogeography of the Atlantic Bonito (Sarda Sarda) in the Northern Mediterranean: The Combined Effects of Historical Vicariance, Population Expansion, Secondary Invasion, and Isolation by Distance. Mol. Phylogenet. Evol. 2004, 33, 32–42. [Google Scholar] [CrossRef]
- Carlsson, J.; McDowell, J.R.; Díaz-Jaimes, P.; Carlsson, J.E.L.; Boles, S.B.; Gold, J.R.; Graves, J.E. Microsatellite and Mitochondrial DNA Analyses of Atlantic Bluefin Tuna (Thunnus thynnus thynnus) Population Structure in the Mediterranean Sea. Mol. Ecol. 2004, 13, 3345–3356. [Google Scholar] [CrossRef]
- Collette, B.B.; Nauen, C.E. FAO Species Catalogue: Vol. 2 Scombrids of the World; Food and Agriculture Organization of the United Nations: Rome, Italy, 1983; Volume 125. [Google Scholar]
- Sabatés, A.; Recasens, L. Seasonal Distribution and Spawning of Small Tunas (Auxis rochei and Sarda sarda) in the Northwestern Mediterranean. Sci. Mar. 2001, 65, 95–100. [Google Scholar] [CrossRef]
- Catanese, G.; Infante, C.; Crespo, A.; Zuasti, E.; Ponce, M.; Funes, V.; Pérez, L.; Manchado, M. Development and Characterization of Eight Microsatellite Markers in Bullet Tuna (Auxis rochei). Mol. Ecol. Notes 2007, 7, 842–844. [Google Scholar] [CrossRef]
- Collette, B.B.; Aadland, C.R. Revision of the Frigate Tunas (Scombridae, Auxis), with Descriptions of Two New Subspecies from the Eastern Pacific. Fish. Res. 1996, 94, 423–441. [Google Scholar]
- ICCAT. Task 1. Nominal Catch Information. Available online: https://www.iccat.int/Data/t1nc_20220131.7z (accessed on 7 February 2022).
- ICCAT. Report of the 2021 ICCAT Small Tuna Species Group Intersessional Meeting. Collect. Vol. Sci. Pap.-ICCAT 2021, 78, 1–58. [Google Scholar]
- Infante, C.; Catanese, G.; Ponce, M.; Manchado, M. Novel Method for the Authentication of Frigate Tunas (Auxis thazard and Auxis rochei) in Commercial Canned Products. J. Agric. Food Chem. 2004, 52, 7435–7443. [Google Scholar] [CrossRef] [PubMed]
- Catanese, G.; Infante, C.; Manchado, M. Complete Mitochondrial DNA Sequences of the Frigate Tuna Auxis thazard and the Bullet Tuna Auxis rochei. DNA Seq.-J. DNA Seq. Mapp. 2008, 19, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Orsi-Relini, L.; Palandri, G.; Garibaldi, F.; Lanteri, L.; Tinti, F. Towards a New Taxonomical Approach to Mediterranean Small Tuna of Genus Auxis. Biol. Mar. Mediter. 2008, 15, 207–210. [Google Scholar]
- Allaya, H.; Ben Faleh, A.; Hattour, A.; Trabelsi, M.; Viñas, J. Disparate Past Demographic Histories of Three Small Scombridae (Actinopterygii) Species in Tunisian Waters. Hydrobiologia 2015, 758, 19–30. [Google Scholar] [CrossRef]
- Ollé, J.; Macías, D.; Saber, S.; Gómez-Vives, M.J.; Pérez-Bielsa, N.; Viñas, J. Genetic Analysis Reveals the Presence of Frigate Tuna (Auxis thazard) in the Bullet Tuna (Auxis rochei) Fishery of the Iberian Peninsula and the Western-Central Mediterranean Sea. Bull. Mar. Sci. 2019, 95, 317–325. [Google Scholar] [CrossRef]
- Alvarado Bremer, J.R.; Mejuto, J.; Greig, T.W.; Ely, B. Global Population Structure of the Swordfish (Xiphias Gladius L.) as Revealed by Analysis of the Mitochondrial DNA Control Region. J. Exp. Mar. Bio. Ecol. 1996, 197, 295–310. [Google Scholar] [CrossRef]
- Ollé, J.; Vilà-Valls, L.; Alvarado-Bremer, J.; Cerdenares, G.; Duong, T.Y.; Hajjej, G.; Lino, P.G.; Muñoz-Lechuga, R.; Sow, F.N.; Diaha, N.C.; et al. Population Genetics Meets Phylogenetics: New Insights into the Relationships among Members of the Genus Euthynnus (Family Scombridae). Hydrobiologia 2021, 849, 47–62. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F. DNA Polymorphism Detectable by Restriction Endonucleases. Genetics 1981, 97, 145–163. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K.; Dudley, J. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets Downloaded From. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Rambaud, A. FigTree; University of Edinburgh: Edinburgh, UK, 2012. [Google Scholar]
- Leigh, J.W.; Bryant, D. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A Complete Re-Implementation of the GENEPOP Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Mostarda, E.; Campo, D.; Castriota, L.; Esposito, V.; Scarabello, M.P.; Andaloro, F. Feeding Habits of the Bullet Tuna Auxis Rochei in the Southern Tyrrhenian Sea. J. Mar. Biol. Assoc. UK 2007, 87, 1007–1012. [Google Scholar] [CrossRef]
- Alvarado Bremer, J.R.; Viñas, J.; Mejuto, J.; Ely, B.; Pla, C. Comparative Phylogeography of Atlantic Bluefin Tuna and Swordfish: The Combined Effects of Vicariance, Secondary Contact, Introgression, and Population Expansion on the Regional Phylogenies of Two Highly Migratory Pelagic Fishes. Mol. Phylogenet. Evol. 2005, 36, 169–187. [Google Scholar] [CrossRef]
- Viñas, J.; Pérez-Serra, A.; Vidal, O.; Alvarado Bremer, J.R.; Pla, C. Genetic Differentiation between Eastern and Western Mediterranean Swordfish Revealed by Phylogeographic Analysis of the Mitochondrial DNA Control Region. ICES J. Mar. Sci. 2010, 67, 1222–1229. [Google Scholar] [CrossRef]
- Nesbø, C.L.; Rueness, E.K.; Iversen, S.A.; Skagen, D.W.; Jakobsen, K.S. Phylogeography and Population History of Atlantic Mackerel (Scomber Scombrus L.): A Genealogical Approach Reveals Genetic Structuring among the Eastern Atlantic Stocks. Proc. R. Soc. B Biol. Sci. 2000, 267, 281–292. [Google Scholar] [CrossRef]
- Alvarado Bremer, J.R.; Stequert, B.; Robertson, N.W.; Ely, B. Genetic Evidence for Inter-Oceanic Subdivision of Bigeye Tuna (Thunnus obesus) Populations. Mar. Biol. 1998, 132, 547–557. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for Ecologists: A Practical Guide to Using and Evaluating Microsatellite Markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Agawa, Y.; Iwaki, M.; Komiya, T.; Honryo, T.; Tamura, K.; Okada, T.; Yagishita, N.; Kobayashi, T.; Sawada, Y. Identification of Male Sex-Linked DNA Sequence of the Cultured Pacific Bluefin Tuna Thunnus Orientalis. Fish. Sci. 2015, 81, 113–121. [Google Scholar] [CrossRef]
- Carlsson, J. Effects of Microsatellite Null Alleles on Assignment Testing. J. Hered. 2008, 99, 616–623. [Google Scholar] [CrossRef]
- Montes, I.; Iriondo, M.; Manzano, C.; Arrizabalaga, H.; Jiménez, E.; Pardo, M.Á.; Goñi, N.; Davies, C.A.; Estonba, A. Worldwide Genetic Structure of Albacore Thunnus alalunga Revealed by Microsatellite DNA Markers. Mar. Ecol. Prog. Ser. 2012, 471, 183–191. [Google Scholar] [CrossRef][Green Version]
- Reglero, P.; Ciannelli, L.; Alvarez-Berastegui, D.; Balbín, R.; López-Jurado, J.L.; Alemany, F. Geographically and Environmentally Driven Spawning Distributions of Tuna Species in the Western Mediterranean Sea. Mar. Ecol. Prog. Ser. 2012, 463, 273–284. [Google Scholar] [CrossRef]
- Russo, S.; Torri, M.; Patti, B.; Reglero, P.; Álvarez-Berastegui, D.; Cuttitta, A.; Sarà, G. Unveiling the Relationship Between Sea Surface Hydrographic Patterns and Tuna Larval Distribution in the Central Mediterranean Sea. Front. Mar. Sci. 2021, 8, 708775. [Google Scholar] [CrossRef]
- Oray, I.; Karakulak, F.S.; Alicli, Z.; Ates, C.; Kahraman, A. First Evidence of Spawning in the Eastern Mediterranean Sea. Preliminary Results of Tuna Larval Survey in 2004. Collect. Vol. Sci. Pap.-ICCAT 2005, 58, 1341–1347. [Google Scholar]
- Macias, D.; Cózar, A.; Garcia-Gorriz, E.; González-Fernández, D.; Stips, A. Surface Water Circulation Develops Seasonally Changing Patterns of Floating Litter Accumulation in the Mediterranean Sea. A Modelling Approach. Mar. Pollut. Bull. 2019, 149, 110619. [Google Scholar] [CrossRef]
- Morote, E.; Olivar, M.P.; Pankhurst, P.M.; Villate, F.; Uriarte, I. Trophic Ecology of Bullet Tuna Auxis Rochei Larvae and Ontogeny of Feeding-Related Organs. Mar. Ecol. Prog. Ser. 2008, 353, 243–254. [Google Scholar] [CrossRef]
- Cuttitta, A.; Torri, M.; Zarrad, R.; Zgozi, S.; Jarboui, O.; Quinci, E.M.; Hamza, M.; Abdulfatah, E.; Haddoud, D.; El Turki, A.; et al. Linking Surface Hydrodynamics to Planktonic Ecosystem: The Case Study of the Ichthyoplanktonic Assemblages in the Central Mediterranean Sea. Hydrobiologia 2018, 821, 191–214. [Google Scholar] [CrossRef]
- Mullins, R.B.; McKeown, N.J.; Sauer, W.H.H.; Shaw, P.W.; Grant, W.S. Genomic Analysis Reveals Multiple Mismatches between Biological and Management Units in Yellowfin Tuna (Thunnus albacares). ICES J. Mar. Sci. 2018, 75, 2145–2152. [Google Scholar] [CrossRef]

| mtDNA CR | Microsatellites | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Area | Location | Study | n | M | h ± SD | π ± SD | n | He | A | Ar | HW |
| Atlantic Iberian Peninsula | |||||||||||
| Olhão | 1 | 52 | 51 | 0.999 ± 0.004 | 0.049 ± 0.024 | 52 | 0.886 ± 0.093 | 22.143 ± 7.058 | 13.338 ± 3.823 | 0.000 *** | |
| Strait of Gibraltar | |||||||||||
| Ceuta † | 1 | 20 | 20 | 1.000 ± 0.016 | 0.041 ± 0.021 | 19 | 0.901 ± 0.056 | 14.290 ± 3.101 | 12.624 ± 2.809 | 0.003 * | |
| East Iberian Peninsula | |||||||||||
| Málaga | 1 | 38 | 37 | 0.999 ± 0.007 | 0.040 ± 0.020 | 30 | 0.905 ± 0.068 | 19.140 ± 5.024 | 13.819 ± 3.352 | 0.000 *** | |
| Cartagena | 1 | 97 | 97 | 1.000 ± 0.002 | 0.046 ± 0.023 | 51 | 0.907 ± 0.057 | 21.429 ± 5.827 | 13.220 ± 3.090 | 0.009 | |
| Valencia | 1 | 21 | 21 | 1.000 ± 0.015 | 0.044 ± 0.023 | 19 | 0.895 ± 0.080 | 15.000 ± 4.163 | 13.125 ± 3.621 | 0.14 | |
| Tarragona | 1 | 49 | 48 | 0.999 ± 0.004 | 0.046 ± 0.023 | 48 | 0.902 ± 0.060 | 21.714 ± 5.823 | 13.593 ± 3.422 | 0.000 *** | |
| North Africa | |||||||||||
| Alger | This study | 45 | 41 | 0.996 ± 0.006 | 0.043 ± 0.021 | 29 | 0.873 ± 0.074 | 15.143 ± 2.545 | 11.779 ± 1.634 | 0.000 *** | |
| Mahdia †† | 2 | 109 | 96 | 0.998 ± 0.002 | 0.041 ± 0.020 | 28 | 0.878 ± 0.050 | 13.429 ± 2.299 | 11.448 ± 1.923 | 0.000 *** | |
| All samples | 431 | 401 | 1.000 ± 0.000 | 0.044 ± 0.022 | 276 | ||||||
| Olhão | Strait of Gibraltar | Málaga | Cartagena | Valencia | Tarragona | Alger | Mahdia | |
|---|---|---|---|---|---|---|---|---|
| Olhão | -- | 0.032 | 0.026 | 0.007 | −0.004 | 0.004 | 0.063 | 0.032 |
| Strait of Gibraltar | 0.003 | -- | 0.002 | 0.010 | 0.010 | 0.006 | 0.043 | 0.054 |
| Málaga | 0.004 | 0.004 | -- | 0.003 | −0.004 | −0.001 | 0.015 | 0.045 |
| Cartagena | 0.004 | 0.002 | 0.001 | -- | −0.010 | −0.002 | 0.038 | 0.033 |
| Valencia | 0.003 | 0.005 | 0.001 | 0.001 | -- | −0.011 | 0.012 | 0.039 |
| Tarragona | 0.007 | 0.002 | 0.005 | 0.003 | 0.007 | -- | 0.035 | 0.030 |
| Alger | 0.02 | 0.022 | 0.019 | 0.019 | 0.021 | 0.019 | -- | 0.098 |
| Mahdia | 0.049 | 0.046 | 0.046 | 0.04 | 0.044 | 0.038 | 0.026 | -- |
| Clusters | mtDNA | Microsatellites | ||
|---|---|---|---|---|
| Fixation Index | p | Fixation Index | p | |
| Iberian Peninsula vs. Alger vs. Mahdia | ɸCT = 0.037 | 0.035 ± 0.005 | FCT = 0.027 | 0.036 ± 0.008 |
| Mediterranean vs. Atlantic | ɸCT = −0.003 | 0.493 ± 0.017 | FCT = −0.004 | 0.403 ± 0.015 |
| Iberian Peninsula vs. North Africa | ɸCT = 0.003 | 0.391 ± 0.015 | FCT = 0.008 | 0.115 ± 0.010 |
| East Iberian Peninsula vs. Olhão vs. Mahdia vs. Alger | ɸCT = 0.036 | 0.020 ± 0.005 | FCT = 0.017 | 0.097 ± 0.009 |
| Global | ɸST = 0.028 | 0.000 ± 0.000 | FST = 0.015 | 0.000 ± 0.000 |
| Aro 3-37 | Aro 2-40 | Aro 1-59 | Aro 2-15 | Aro 4-13 | Aro 1-3 | Aro 1-10 | Total per Location | |
|---|---|---|---|---|---|---|---|---|
| Olhão | 0.578 | 0.397 | 0.416 | 0.045 | 0.020 | 0.000 | 0.000 | 0.000 |
| Strait of Gibraltar | 0.791 | 0.916 | 0.539 | 0.296 | 0.017 | 0.000 | 0.125 | 0.003 |
| Málaga | 0.235 | 0.091 | 0.456 | 0.978 | 0.000 | 0.471 | 0.012 | 0.000 |
| Cartagena | 0.702 | 0.515 | 0.196 | 0.149 | 0.006 | 0.631 | 0.009 | 0.009 |
| Valencia | 0.795 | 0.871 | 0.580 | 0.043 | 0.069 | 0.777 | 0.057 | 0.140 |
| Tarragona | 0.780 | 0.130 | 0.021 | 0.614 | 0.000 | 0.028 | 0.000 | 0.000 |
| Alger | 0.132 | 0.921 | 0.565 | 0.000 | 0.000 | 0.023 | 0.006 | 0.000 |
| Mahdia | 0.075 | 0.846 | 0.137 | 0.485 | 0.000 | 0.360 | 0.024 | 0.000 |
| Total per locus | 0.499 | 0.673 | 0.151 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ollé-Vilanova, J.; Pérez-Bielsa, N.; Araguas, R.M.; Sanz, N.; Saber, S.; Macías, D.; Viñas, J. Larval Retention and Homing Behaviour Shape the Genetic Structure of the Bullet Tuna (Auxis rochei) in the Mediterranean Sea. Fishes 2022, 7, 300. https://doi.org/10.3390/fishes7050300
Ollé-Vilanova J, Pérez-Bielsa N, Araguas RM, Sanz N, Saber S, Macías D, Viñas J. Larval Retention and Homing Behaviour Shape the Genetic Structure of the Bullet Tuna (Auxis rochei) in the Mediterranean Sea. Fishes. 2022; 7(5):300. https://doi.org/10.3390/fishes7050300
Chicago/Turabian StyleOllé-Vilanova, Judith, Núria Pérez-Bielsa, Rosa Maria Araguas, Núria Sanz, Sámar Saber, David Macías, and Jordi Viñas. 2022. "Larval Retention and Homing Behaviour Shape the Genetic Structure of the Bullet Tuna (Auxis rochei) in the Mediterranean Sea" Fishes 7, no. 5: 300. https://doi.org/10.3390/fishes7050300
APA StyleOllé-Vilanova, J., Pérez-Bielsa, N., Araguas, R. M., Sanz, N., Saber, S., Macías, D., & Viñas, J. (2022). Larval Retention and Homing Behaviour Shape the Genetic Structure of the Bullet Tuna (Auxis rochei) in the Mediterranean Sea. Fishes, 7(5), 300. https://doi.org/10.3390/fishes7050300