Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles
Abstract
1. Introduction
2. Material and Methods
2.1. Ethical Statement
2.2. Capture and Maintenance of Juveniles
2.3. Enzyme Extract and Enzymatic Technique
2.4. Temperature and pH Effect on Enzymatic Activity
2.5. Inhibitor Effect on Enzymatic Activity
2.6. Zymogram Analysis
2.7. Statistical Analysis
3. Results
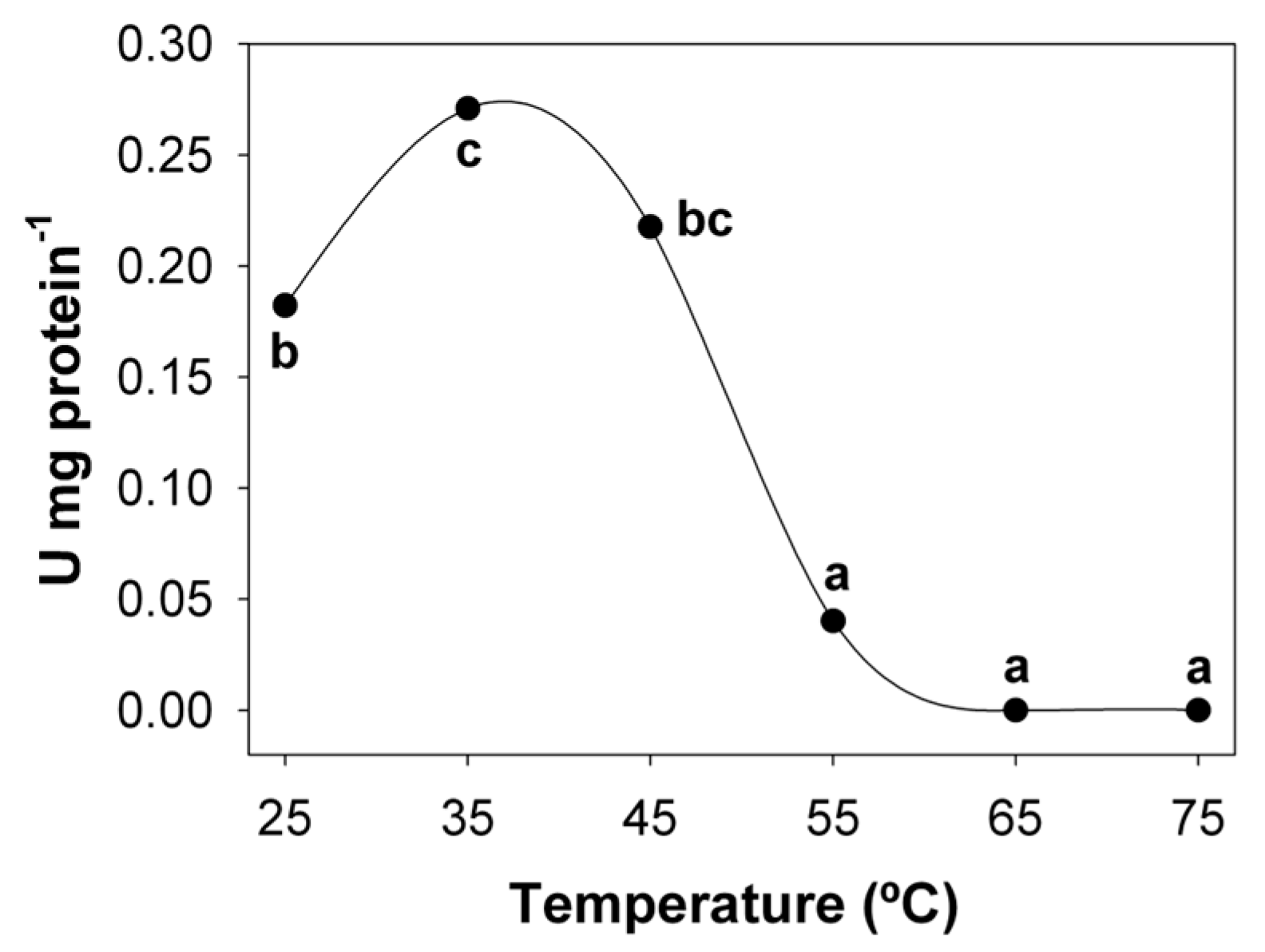
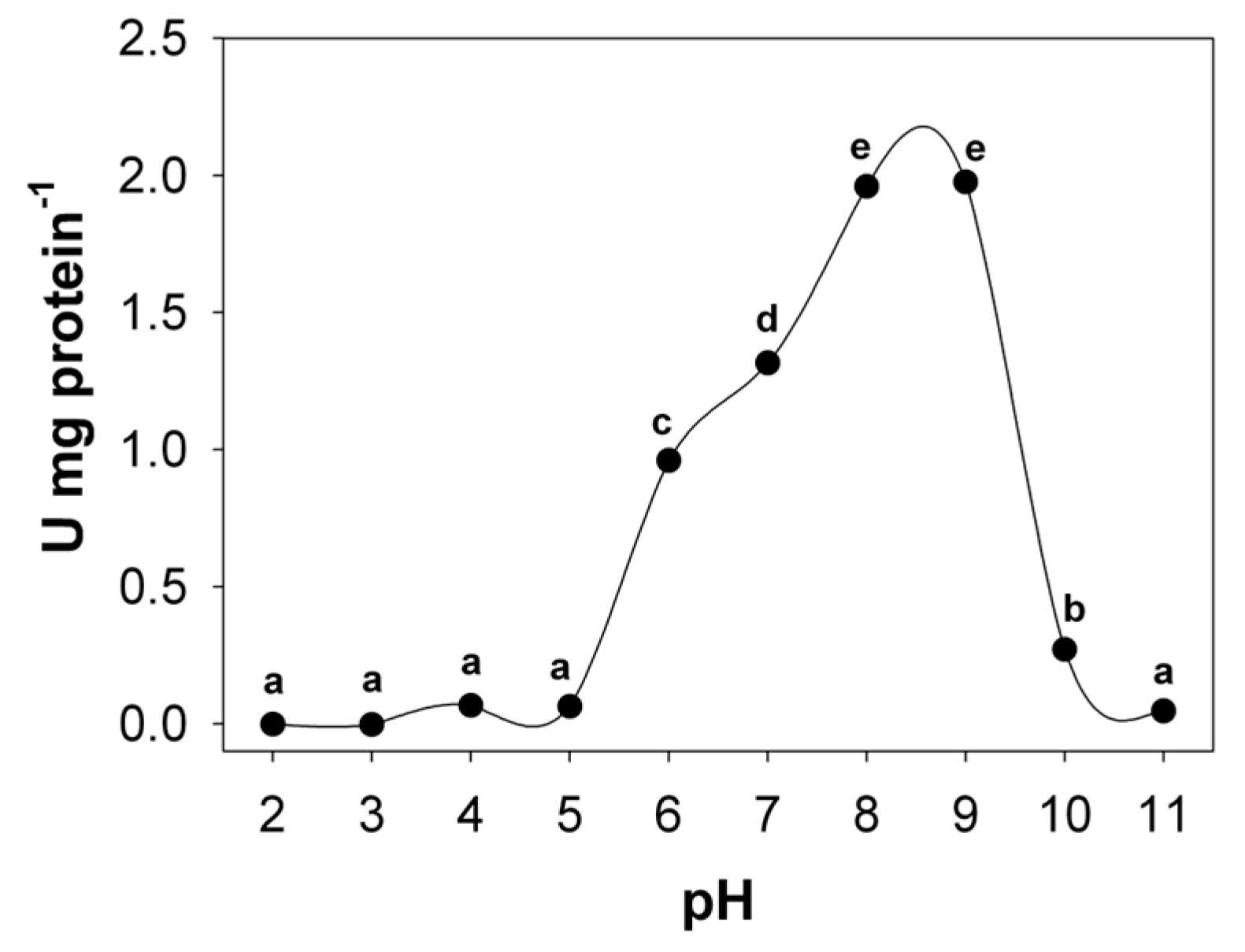
3.1. Temperature and pH Optimum and Stability
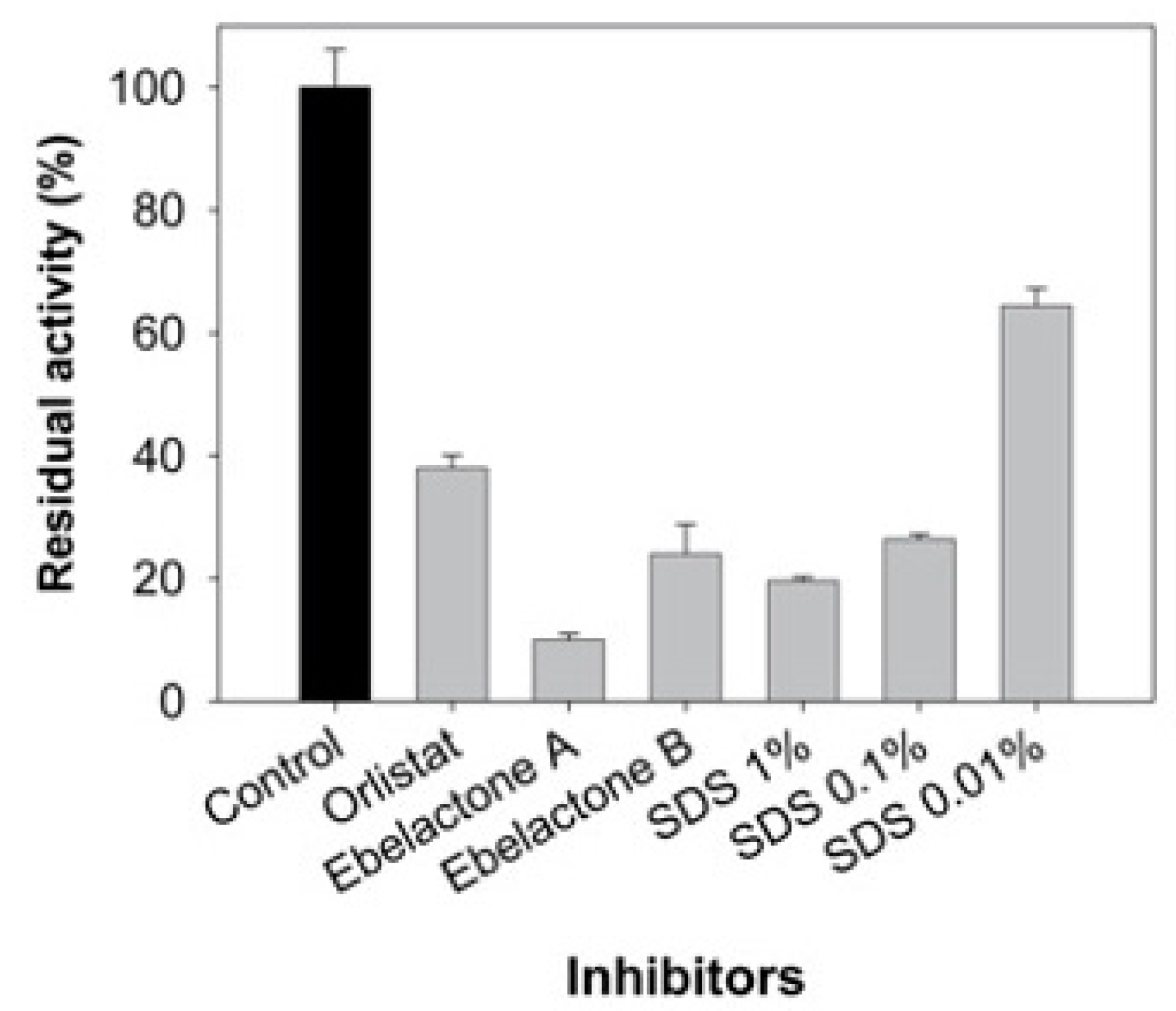
3.2. Inhibitor Effect on Lipase Activity
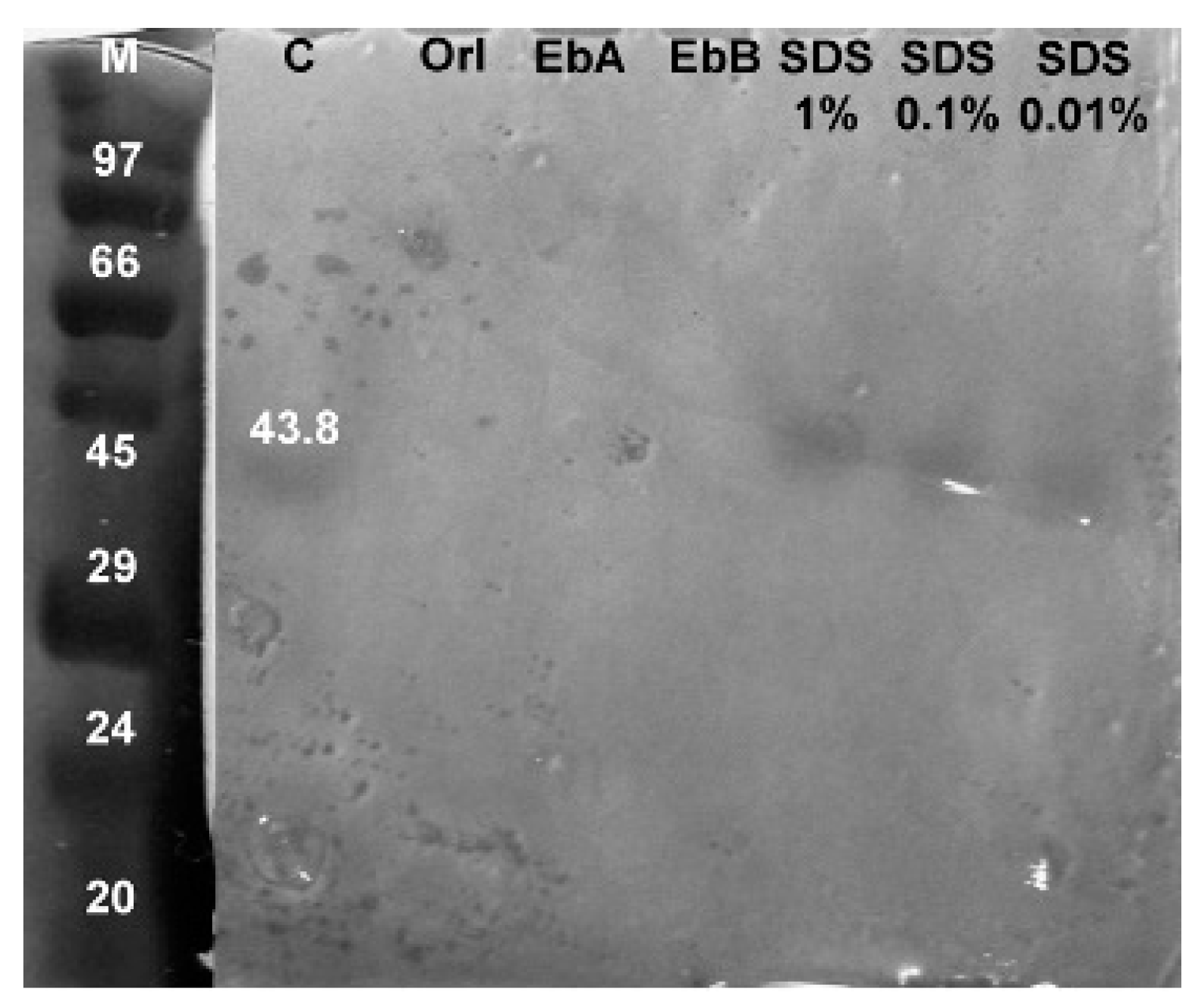
3.3. Zymogram Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alvarez-Lajonchère, L.; Taylor, R.G. Economies of scale for juvenile production of common snook (Centropomus undecimalis Bloch). Aquac. Econ. Manag. 2003, 7, 273–292. [Google Scholar] [CrossRef]
- Tucker, J. Snook culture. World Aquac. Mag. 2003, 34, 42–46. [Google Scholar]
- Ji, H.; Sun, H.T.; Xiong, D.M. Studies on activity, distribution, and zymogram of protease, α-amylase, and lipase in the paddlefish Polyodon spathula. Fish Physiol. Biochem. 2012, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Prasertsan, P.; Prachumratana, T. Comparison and selection of protease and lipase sources from visceral organs of three tuna species. Songklanakarin J. Sci. Technol. 2008, 30, 73–76. [Google Scholar]
- Achionye-Nzeh, C.G.; Obaroh, I.; Adeniyi, V. Lipase activity in the liver and digestive tract of some cichlids (pisces: Cichlidea). Afr. J. Appl. Zool. Environ. Biol. 2005, 7, 136–139. [Google Scholar] [CrossRef]
- Fagbenro, O.; Adedire, O.; Fateru, O.; Oluwabukola, I.; Ogunlana, O.; Akanbi, B.; Fasanmi, T.; Ayo-Amu, P. Digestive enzyme assays in the gut of Oreochromis niloticus Linnaeus 1757, Parachanna (Channa) Obscura Gunther 1861 and Gymnarchus niloticus Cuvier 1829. Anim. Res. Int. 2005, 2, 292–296. [Google Scholar] [CrossRef][Green Version]
- Langeland, T.L.M. Digestive Enzyme Activity in Eurasian Perch (Perca fluviatilis) and Arctic Charr (Salvelinus alpinus). J. Aquac. Res. Dev. 2013, 5, 1–8. [Google Scholar] [CrossRef]
- Sankar, H.S.; Jose, J.; Varadarajan, R.; Bhanu, S.V.; Joy, S.; Philip, B. Functional zonation of different digestive enzymes in Etroplus suratensis and Oreochromis mossambicus. Int. J. Sci. Res. Publ. 2014, 4, 1–10. Available online: http://www.ijsrp.org/research-paper-0514/ijsrp-p29105.pdf?msclkid=b5983ccecf4b11eca17fb6d2c0c1bfef (accessed on 1 May 2022).
- Ran, C.; He, S.; Yang, Y.; Huang, L.; Zhou, Z. A Novel Lipase as Aquafeed Additive for Warm-Water Aquaculture. PLoS ONE 2015, 10, e0132049. [Google Scholar] [CrossRef]
- Murtaza, M.; Abdullah, S.; Hassan, W.; Abbas, K.; Naz, H.; Zia, M.A. Studies on amylase and lipase activity in fishes fed with diet containing different feed ingredients. Punjab Univ. J. Zool. 2016, 31, 165–169. Available online: https://www.semanticscholar.org/paper/Studies-on-amylase-and-lipase-activity-in-fishes-Murtaza-Abdullah/46a199036fa61ad0fc481648c8fddeb5786215a7?msclkid=1aa11afbcf4c11ec9e81520c18d20530 (accessed on 1 May 2022).
- Concha-Frías, B.; Álvarez-González, C.A.; Gaxiola-Cortes, M.G.; Silva-Arancibia, A.E.; Toledo-Agüero, P.H.; Martinez-García, R. Partial characterization of digestive proteases in the Common Snook Centropomus undecimalis. Int. J. Biol. 2016, 8, 1–11. [Google Scholar] [CrossRef][Green Version]
- Jimenez-Martinez, L.D.; Alvarez-González, C.A.; Tovar-Ramírez, D.; Gaxiola, G.; Sanchez-Zamora, A.; Moyano, F.J.; Alarcón, F.J.; Márquez-Couturier, G.; Gisbert, E.; Contreras-Sánchez, W.M.; et al. Digestive enzyme activities during early ontogeny in Common snook (Centropomus undecimalis). Fish Physiol. Biochem. 2011, 38, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Stoytcheva, M.; Montero, G.; Zlatev, R.; Leon, J.A.; Gochev, V. Analytical Methods for Lipases Activity Determination: A Review. Curr. Anal. Chem. 2012, 8, 400–407. [Google Scholar] [CrossRef]
- Thongprajukaew, K.; Kovitvadhi, U.; Engkagul, A.; Rungruangsak-Torrissen, K. Temperature and pH characteristics of amylase and lipase at different developmental stages of Siamese fighting fish (Betta splendens Regan, 1910). Kasetsart J.-Nat. Sci. 2010, 44, 210–219. Available online: https://www.thaiscience.info/Journals/Article/TKJN/10603265.pdf?msclkid=71585ca1cf4c11eca8550b4f3b15ed02 (accessed on 1 May 2022).
- Görgün, S.; Akpınar, M.A. Purification and characterization of lipase from the liver of carp, Cyprinus carpio L. (1758), Living in Lake Tödürge (Sivas, Türkiye). Turk. J. Fish. Aquat. Sci. 2012, 12, 207–215. [Google Scholar] [CrossRef]
- Klahan, R.; Areechon, N.; Yoonpundh, R.; Engkagul, A. Characterization and activity of digestive enzymes in different sizes of Nile tilapia (Oreochromis niloticus L.). Kasetsart J. Nat. Sci. 2009, 43, 143–153. Available online: https://www.thaiscience.info/journals/Article/TKJN/10507053.pdf?msclkid=ae6ce44dcf4c11ecaf75151f970dee88 (accessed on 1 May 2022).
- Gilham, D.; Lehner, R. Techniques to measure lipase and esterase activity in vitro. Methods 2005, 36, 139–147. [Google Scholar] [CrossRef]
- Lewis, D.R.; Liu, D.J. Direct Measurement of Lipase Inhibition by Orlistat Using a Dissolution Linked In Vitro Assay. Clin. Pharmacol. Biopharm. 2012, 1, 1000103. [Google Scholar] [CrossRef]
- Achouri, N.; Tomàs-Gamisans, M.; Triki, S.; Valero, F.; Miled, N.; Fendri, A.; Smichi, N. Dissecting the Interaction Deficiency of a Cartilaginous Fish Digestive Lipase with Pancreatic Colipase: Biochemical and Structural Insights. BioMed Res. Int. 2020, 2020, 3064290. [Google Scholar] [CrossRef]
- Reetz, M.T. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 2002, 6, 145–150. [Google Scholar] [CrossRef]
- Chaurasia, S.P.; Bhandaria, K.; Sharmaa, A.; Dalai, A.K. A review on lipase catalysed synthesis of DHA rich glyceride from fish oils. Int. J. Res. Sci. Innov. 2016, 3, 9–19. [Google Scholar]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Lipases from Mammals and Fishes. Rev. Fish. Sci. 2009, 17, 18–40. [Google Scholar] [CrossRef]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Purification and properties of digestive lipases from Chinook salmon (Oncorhynchus tshawytscha) and New Zealand hoki (Macruronus novaezelandiae). Fish Physiol. Biochem. 2010, 36, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.Y.; Boora, N.; Tyagi, P. Isolation, purification and characterization of secondary structure and kinetic study of lipase from Indian major carp, Catla catla (Catla). Enzym. Eng. 2014, 3, 1–8. [Google Scholar] [CrossRef]
- Rueda-López, S.; Martínez-Montaño, E.; Viana, M.T. Biochemical Characterization and Comparison of Pancreatic Lipases from the Pacific Bluefin Tuna, Thunnus orientalis; Totoaba, Totoaba macdonaldi; and Striped Bass, Morone saxatilis. J. World Aquac. Soc. 2017, 48, 156–165. [Google Scholar] [CrossRef]
- Mardina, V.; Harmawan, T.; Fitriani; Hildayani, G.M.; Yusof, F. Screening of protease and lipase sources from visceral organs of Euthynnusaffinis. IOP Conf. Ser. Mater. Sci. Eng. 2018, 420, 012083. [Google Scholar] [CrossRef]
- Canioni, P.; Julien, R.; Rathelot, J.; Sarda, L. Pancreatic and microbial lipases: A comparison of the interaction of pancreatic colipase with lipases of various origins. Lipids 1977, 12, 393–397. [Google Scholar] [CrossRef]
- Navvabi, A.; Razzaghi, M.; Fernandes, P.; Karami, L.; Homaei, A. Novel lipases discovery specifically from marine organisms for industrial production and practical applications. Process Biochem. 2018, 70, 61–70. [Google Scholar] [CrossRef]
- Villanueva-Gutiérrez, E.; Maldonado-Othón, C.A.; Perez-Velazquez, M.; González-Félix, M.L. Activity and Partial Characterization of Trypsin, Chymotrypsin, and Lipase in the Digestive Tract of Totoaba macdonaldi. J. Aquat. Food Prod. Technol. 2020, 29, 322–334. [Google Scholar] [CrossRef]
- Gjellesvik, D.; Lombardo, D.; Walther, B.T. Pancreatic bile salt dependent lipase from cod (Gadus morhua): Purification and properties. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1992, 1124, 123–134. [Google Scholar] [CrossRef]
- Versaw, W.K.; Cuppett, S.L.; Winters, D.D.; Williams, L.E. An Improved Colorimetric Assay for Bacterial Lipase in Nonfat Dry Milk. J. Food Sci. 1989, 54, 1557–1558. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Stauffer, C.E. Enzyme Assays for Food Scientists; Van Nostand Reinhold: New York, NY, USA, 1989; p. 317. [Google Scholar]
- Islam, M.A.; Parveen, F.; Hossain, K.; Khatun, S.; Karim, M.R.; Kim, G.S.; Absar, N.; Haque, M.S. Purification and biochemical characterization of lipase from the dorsal part of Cirrhinus reba. Thai J. Agric. Sci. 2009, 42, 71–80. Available online: https://www.thaiscience.info/journals/Article/TJAS/10469553.pdf?msclkid=b87f4ffacf4e11ec901022ef25c2cc1e (accessed on 1 May 2022).
- Davis, B.J. Disc electrophoresis. II. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 1964, 121, 404–427. [Google Scholar] [CrossRef] [PubMed]
- Nolasco, H.; Moyano-López, F.; Vega-Villasante, F. Partial characterization of pyloric-duodenal lipase of gilthead seabream (Sparus aurata). Fish Physiol. Biochem. 2011, 37, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, A.; Takano, K.; Kamoi, I. Purification and Properties of Lipase from Tilapia Intestine. Digestive Enzyme of Tilapia VI. Nippon. Suisan Gakk 2001, 67, 78–84. [Google Scholar] [CrossRef]
- González-Félix, M.L.; Santana-Bejarano, E.B.; Perez-Velazquez, M.; Villalba-Villalba, A.G. Partial characterization, quantification and activity of pancreatic lipase in the gastrointestinal tract of Totoaba macdonaldi. Arch. Biol. Sci. 2018, 70, 489–496. [Google Scholar] [CrossRef]
- Page, J.W.; Andrews, J.W.; Murai, T.; Murray, M.W. Hydrogen ion concentration in the gastrointestinal tract of channel catfish. J. Fish Biol. 1976, 8, 225–228. [Google Scholar] [CrossRef]
- Vidaurreta, A.; Cara, B.; Moyano, F.J. Gastrointestinal pH and development of the acid digestion in larvae and early juveniles of Sparus aurata (Pisces: Teleostei). Mar. Biol. 2004, 144, 863–869. [Google Scholar] [CrossRef]
- Horn, M.H.; Gawlicka, A.K.; German, D.P.; Logothetis, E.A.; Cavanagh, J.W.; Boyle, K.S. Structure and function of the stomachless digestive system in three related species of New World silverside fishes (Atherinopsidae) representing herbivory, omnivory, and carnivory. Mar. Biol. 2006, 149, 1237–1245. [Google Scholar] [CrossRef]
- Fard, M.R.S.; Weisheit, C.; Poynton, S. Intestinal pH profile in rainbow trout Oncorhynchus mykiss and microhabitat preference of the flagellate Spironucleus salmonis (Diplomonadida). Dis. Aquat. Org. 2007, 76, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Kuz’Mina, V.V.; Skvortsova, E.G.; Zolotareva, G.V.; Sheptitskiy, V.A. Influence of pH upon the activity of glycosidases and proteinases of intestinal mucosa, chyme and microbiota in fish. Fish Physiol. Biochem. 2011, 37, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, M.M.; Kashinskaya, E.N.; Izvekova, G.I.; Glupov, V.V. pH values and activity of digestive enzymes in the gastrointestinal tract of fish in Lake Chany (West Siberia). J. Ichthyol. 2015, 55, 251–258. [Google Scholar] [CrossRef]
- Areekijseree, M.; Engkagul, A.; Kovitvadhi, U.; Thongpan, A.; Mingmuang, M.; Kovitvadhi, S. Activity profiles at different pH and temperature of cellulases and lipases in freshwater pearl mussel: Hyriopsis (Hyriopsis) bialatus, Simpson 1900. Kasetsart J.-Nat. Sci. 2002, 36, 399–407. Available online: https://www.thaiscience.info/Journals/Article/TKJN/10974126.pdf?msclkid=7464c23acf5111ec9c78738186383a42 (accessed on 1 May 2022).
- Heck, A.M.; Yanovski, J.; Calis, K.A. Orlistat, a New Lipase Inhibitor for the Management of Obesity. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2000, 20, 270–279. [Google Scholar] [CrossRef]
- Bénarouche, A.; Point, V.; Carrière, F.; Cavalier, J.-F. Using the reversible inhibition of gastric lipase by Orlistat for investigating simultaneously lipase adsorption and substrate hydrolysis at the lipid–water interface. Biochimie 2014, 101, 221–231. [Google Scholar] [CrossRef]
- Blackett, P.; Tryggestad, J.; Krishnan, S.; Li, S.; Xu, W.; Alaupovic, P.; Quiroga, C.; Copeland, K. Lipoprotein abnormalities in compound heterozygous lipoprotein lipase deficiency after treatment with a low-fat diet and orlistat. J. Clin. Lipidol. 2013, 7, 132–139. [Google Scholar] [CrossRef]
- Kocełak, P.; Zahorska-Markiewicz, B.; Jonderko, K.; Olszanecka-Glinianowicz, M.; Żak-Gołąb, A.; Holecki, M.; Kamińska, M.; Szymszal, M. Long-term effects of lipase inhibition by orlistat on gastric emptying and orocecal transit time of a solid meal. J. Gastroenterol. 2008, 43, 609–617. [Google Scholar] [CrossRef]
- Olszanecka-Glinianowicz, M.; Dąbrowski, P.; Kocełak, P.; Janowska, J.; Smertka, M.; Jonderko, K.; Chudek, J. Long-term inhibition of intestinal lipase by orlistat improves release of gut hormones increasing satiety in obese women. Pharmacol. Rep. 2013, 65, 666–671. [Google Scholar] [CrossRef]
- Cruz-Hernandez, C.; Oliveira, M.; Pescia, G.; Moulin, J.; Masserey-Elmelegy, I.; Dionisi, F.; Destaillats, F. Lipase inhibitor orlistat decreases incorporation of eicosapentaenoic and docosahexaenoic acids in rat tissues. Nutr. Res. 2010, 30, 134–140. [Google Scholar] [CrossRef]
- Mandal, A.K. Stereocontrolled Total Synthesis of (−)-Ebelactone A. Org. Lett. 2002, 4, 2043–2045. [Google Scholar] [CrossRef] [PubMed]
- Hernández-López, I.A.; Ibarra-Castro, L.; Álvarez-González, C.A.; Martínez-Brown, J.M.; Maytorena-Verdugo, C.I.; Peña-Marín, E.S. Characterization of digestive enzymes during early ontogeny of white Snook (Centropomus viridis). Aquaculture 2021, 535, 736399. [Google Scholar] [CrossRef]
- Heras, J.; Chakraborty, M.; Emerson, J.J.; German, D.P. Genomic and biochemical evidence of dietary adaptation in a marine herbivorous fish. Proc. R. Soc. B Boil. Sci. 2020, 287, 20192327. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, Y.; Ohtaki, H.; Ohtsuka, E.; Kocha, T.; Fukuda, T.; Takeuchi, T.; Aoyagi, T. Effects of Ebelactone B, a Lipase Inhibitor, on Intestinal Fat Absorption in the Rat. J. Enzym. Inhib. 1995, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Smichi, N.; Fendri, A.; Chaâbouni, R.; Ben Rebah, F.; Gargouri, Y.; Miled, N. Purification and Biochemical Characterization of an Acid-Stable Lipase from the Pyloric Caeca of Sardine (Sardinella aurita). Appl. Biochem. Biotechnol. 2010, 162, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, M.K.; Gopakumar, K.; Nair, M.R. Purification of a lipase from the hepatopancreas of oil sardine (Sardinella longiceps linnaeus) and its characteristics and properties. J. Sci. Food Agric. 1985, 36, 191–203. [Google Scholar] [CrossRef]
- Day, R.D.; German, D.P.; Manjakasy, J.M.; Farr, I.; Hansen, M.J.; Tibbetts, I.R. Enzymatic digestion in stomachless fishes: How a simple gut accommodates both herbivory and carnivory. J. Comp. Physiol. B 2011, 181, 603–613. [Google Scholar] [CrossRef]
- German, D.P.; Horn, M.H.; Gawlicka, A. Digestive Enzyme Activities in Herbivorous and Carnivorous Prickleback Fishes (Teleostei: Stichaeidae): Ontogenetic, Dietary, and Phylogenetic Effects. Physiol. Biochem. Zool. 2004, 77, 789–804. [Google Scholar] [CrossRef]
- Almeida, A.P.G.; Zardo, E.L.; Toni, C.; Behr, E.R.; da Silva, L.P.; Vieira, J.P.; Loro, V.L.; Baldisserotto, B. Composition of gastrointestinal content, protease and lipase activities in summer and winter of four freshwater siluriforms (Teleostei: Actinopterygii) with two different feeding habits. Zoologia 2018, 35, 1–8. [Google Scholar] [CrossRef]
- Hariati, A.M.; Yuniarti, A.; Endariani; Kusuma, W.; Wiadnya, D. Albumin and enzyme profiles of dwarf snakehead, Channa gachua caught from River Brantas, East Java. J. Phys. Conf. Ser. 2019, 1146, 012041. [Google Scholar] [CrossRef]
- Ismat, N.; Ashraf, M.; Naeem, M.; Rehman, M. Effect of different feed ingredients on growth and level of intestinal enzyme secretions in juvenile Labeo rohita, Catlacatla, Cirrhinusmrigala and Hypophthalmicthysmolitrix. Int. J. Aquac. 2013, 3, 85–91. [Google Scholar] [CrossRef]
- Diana, A.; Zainal, A.; Hashim, M.; Das, S.K.; Rahim, S.M.; Mazlan, A.G. Enzymatic digestion of stomachless fish Zenarchopterus buffonis. Aquac. Aquar. Conserv. Legis. Bioflux 2016, 9, 695–703. Available online: https://www.cabdirect.org/cabdirect/abstract/20163241214?msclkid=25aab83ccf5811ec9412ab26b3375592 (accessed on 1 May 2022).
- Gabriel, N.N.; Qiang, J.; Ma, X.; Xu, P.; Nakwaya, D. Effects of dietary Aloe vera crude extracts on digestive enzyme activities and muscle proximate composition of GIFT tilapia juveniles. S. Afr. J. Anim. Sci. 2017, 47, 904–913. [Google Scholar] [CrossRef]
| °C | 0 min | 30 min | 60 min | 90 min |
|---|---|---|---|---|
| 25 | 100 ± 0.00 c | 68.16 ± 1.02 d | 154.24 ± 1.71 a | 116.80 ± 1.21 b |
| 35 | 100 ± 0.00 c | 58.24 ± 3.01 d | 116.16 ± 3.69 a | 93.76 ± 2.64 b |
| 45 | 100 ± 0.00 a | 0.00 ± 0.00 c | 6.56 ± 0.09 b | 4.16 ± 0.05 b |
| 55 | 100 ± 0.00 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| pH | 0 min | 30 min | 60 min | 90 min |
|---|---|---|---|---|
| 2 | 100 ± 0.00 a | 0.00 ± 0.04 d | 46.18 ± 3.01 b | 11.34 ± 0.22 c |
| 3 | 100 ± 0.00 a | 12.29 ± 0.82 c | 23.87 ± 1.11 b | 10.74 ± 0.50 c |
| 4 | 100 ± 0.00 a | 80.55 ± 1.04 b | 45.35 ± 1.02 c | 95.59 ± 1.21 a |
| 5 | 100 ± 0.00 c | 122.32 ± 3.04 b | 101.31 ± 1.03 c | 139.98 ± 2.02 a |
| 6 | 100 ± 0.00 d | 139.14 ± 5.01 a | 107.40 ± 4.06 c | 118.74 ± 4.01 b |
| 7 | 100 ± 0.00 c | 144.87 ± 4.02 a | 157.16 ± 12.59 a | 108.95 ± 3.76 b |
| 8 | 100 ± 0.00 d | 137.95 ± 6.01 b | 156.92 ± 8.87 a | 112.05 ± 1.29 c |
| 9 | 100 ± 0.00 a | 70.29 ± 2.01 c | 76.37 ± 1.04 b | 53.34 ± 0.72 d |
| 10 | 100 ± 0.00 a | 101.91 ± 1.02 a | 58.23 ± 1.00 b | 30.31 ± 0.13 c |
| 11 | 100 ± 0.00 a | 36.64 ± 1.03 d | 66.71 ± 1.22 b | 57.04 ± 2.23 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Concha-Frías, B.; Gaxiola-Cortes, M.G.; De la Cruz-Alvarado, F.J.; Jimenez Martinez, L.D.; Peña-Marin, E.S.; Oliva-Arriagada, M.A.; Arias-Moscoso, J.L.; Alvarez-González, C.A. Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles. Fishes 2022, 7, 107. https://doi.org/10.3390/fishes7030107
Concha-Frías B, Gaxiola-Cortes MG, De la Cruz-Alvarado FJ, Jimenez Martinez LD, Peña-Marin ES, Oliva-Arriagada MA, Arias-Moscoso JL, Alvarez-González CA. Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles. Fishes. 2022; 7(3):107. https://doi.org/10.3390/fishes7030107
Chicago/Turabian StyleConcha-Frías, Bartolo, Martha Gabriela Gaxiola-Cortes, Fanny Janet De la Cruz-Alvarado, Luis Daniel Jimenez Martinez, Emyr Saul Peña-Marin, Marcia Angélica Oliva-Arriagada, Joe Luis Arias-Moscoso, and Carlos Alfonso Alvarez-González. 2022. "Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles" Fishes 7, no. 3: 107. https://doi.org/10.3390/fishes7030107
APA StyleConcha-Frías, B., Gaxiola-Cortes, M. G., De la Cruz-Alvarado, F. J., Jimenez Martinez, L. D., Peña-Marin, E. S., Oliva-Arriagada, M. A., Arias-Moscoso, J. L., & Alvarez-González, C. A. (2022). Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles. Fishes, 7(3), 107. https://doi.org/10.3390/fishes7030107








