Identification and Characterization of Immunoglobulin T Heavy Chain in Large Yellow Croaker (Larimichthys crocea)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Infection and Tissue Preparation
2.2. Complete cDNA Cloning and Sequence Analysis of LcIgT
2.3. Tissue Distribution of LcIgT by RT-PCR
2.4. LcIgT Expression Analysis after Infection with C. irritans
2.5. Western Blotting Analysis of LcIgT Expression
2.6. Statistical Analysis
3. Results
3.1. Identification and Characterization of LcIgT cDNA
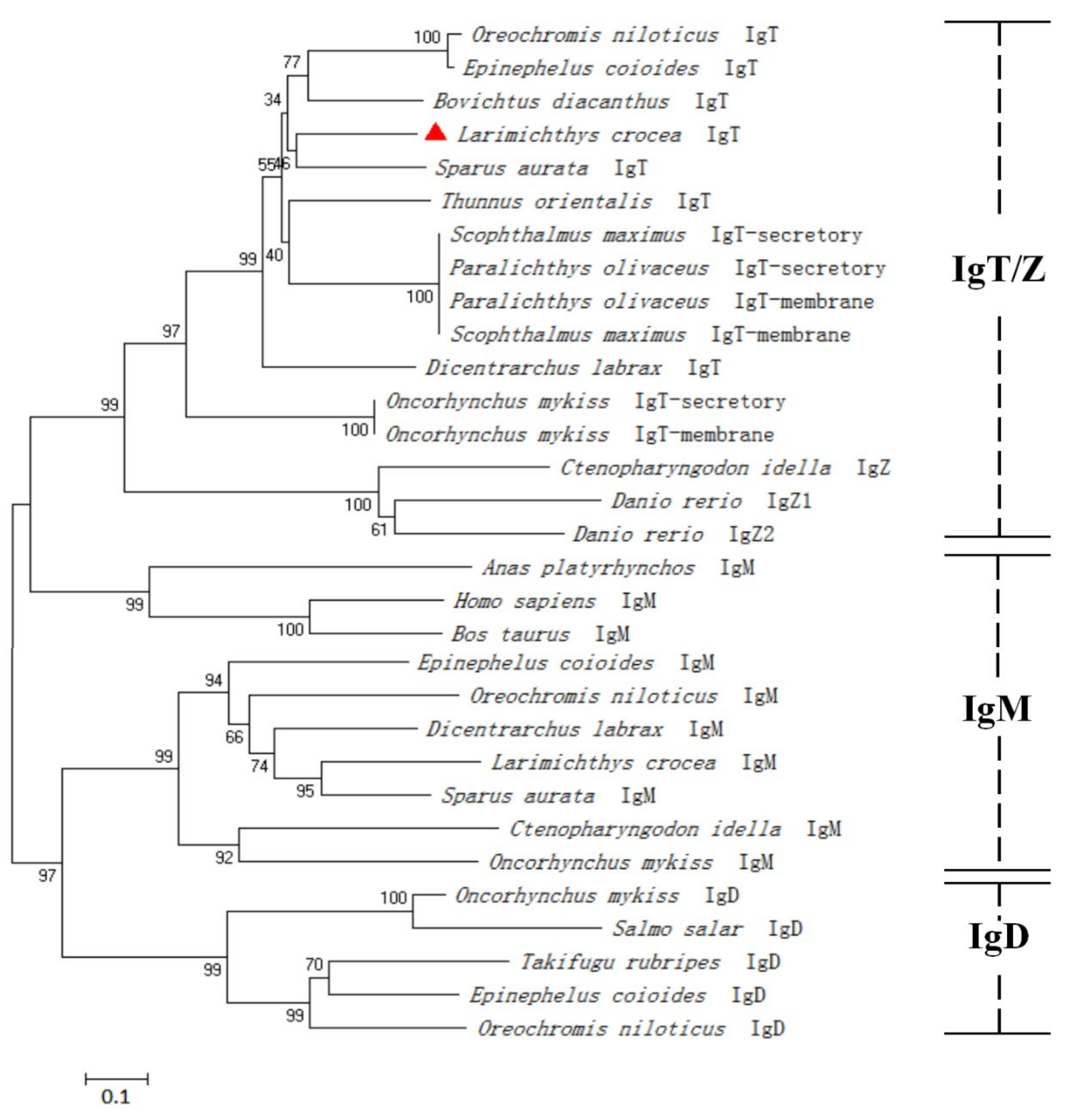
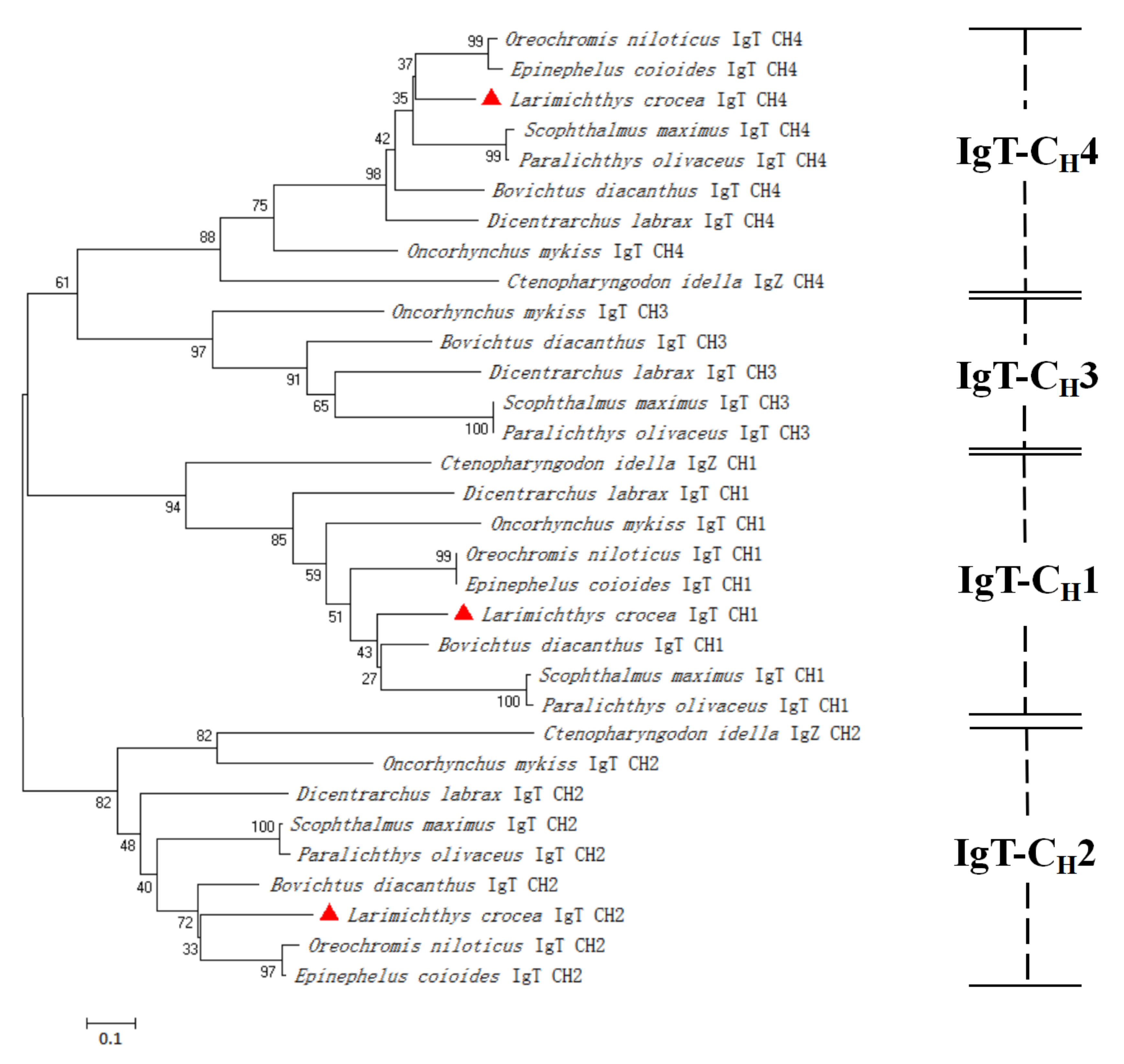
3.2. Multiple Sequence Alignment and Phylogenetic Analysis of LcIgT
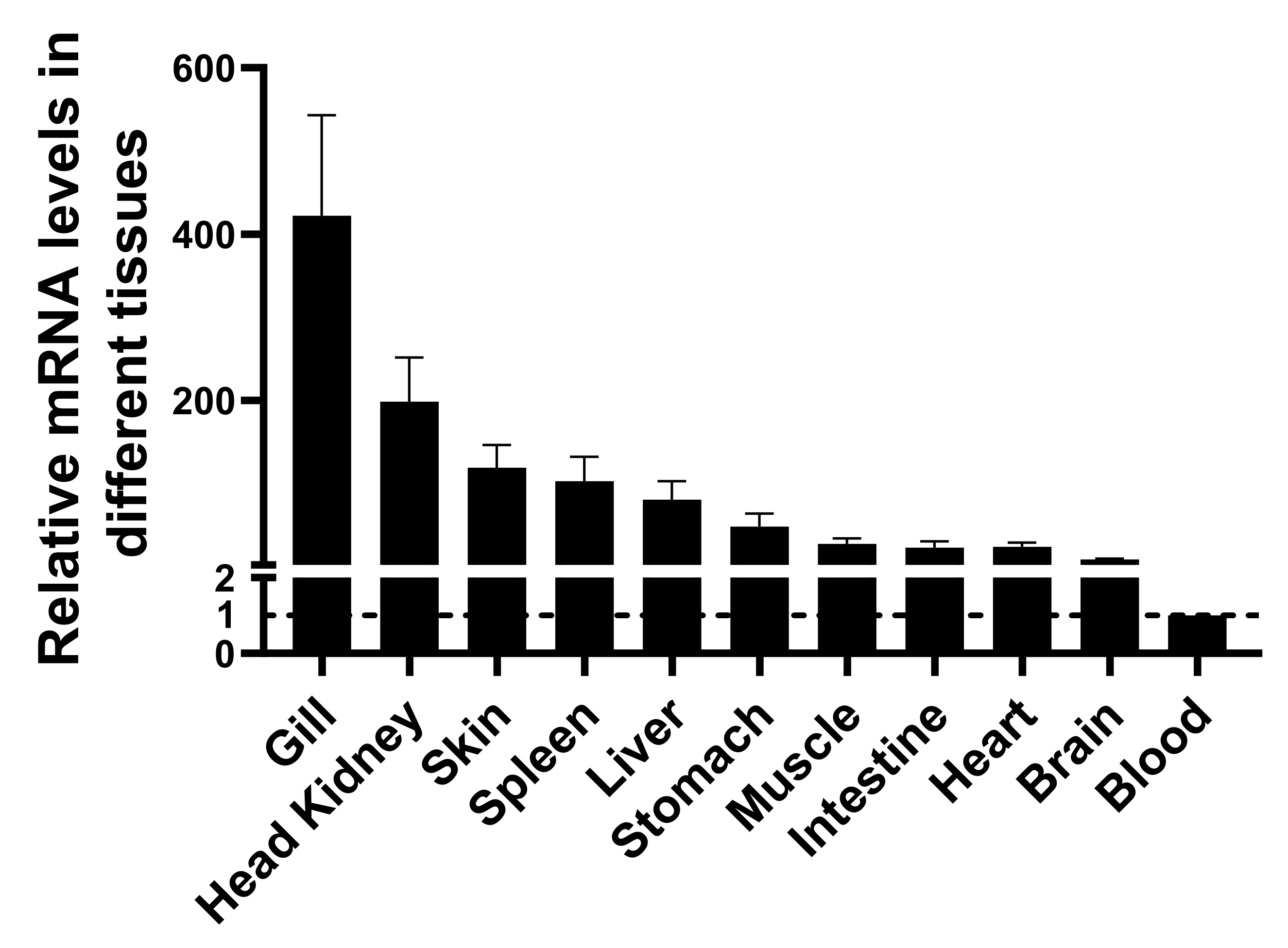
3.3. Tissue Distribution of LcIgT by RT-PCR
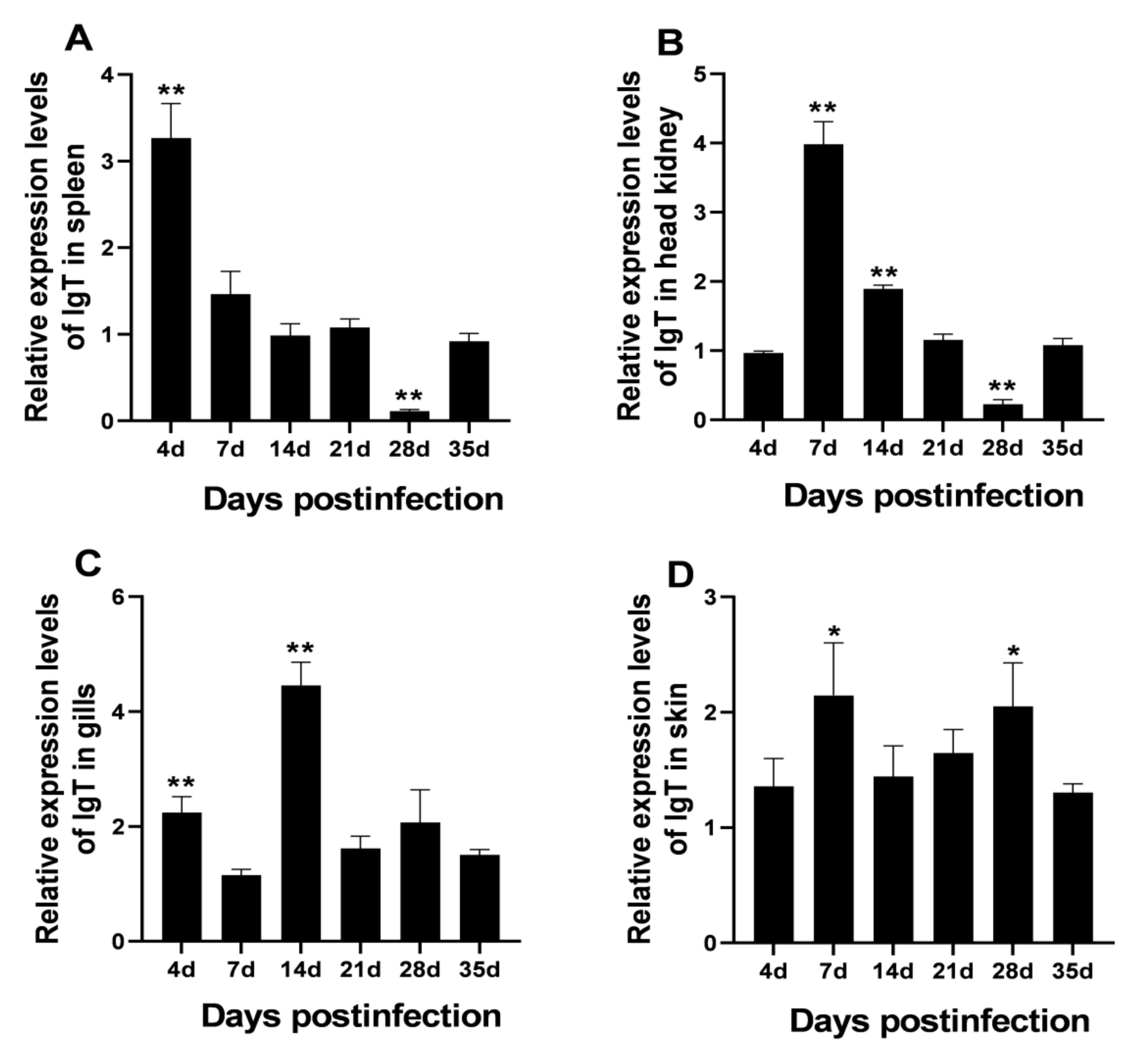
3.4. Modulation of LcIgT Gene Expression after C. irritans Infection
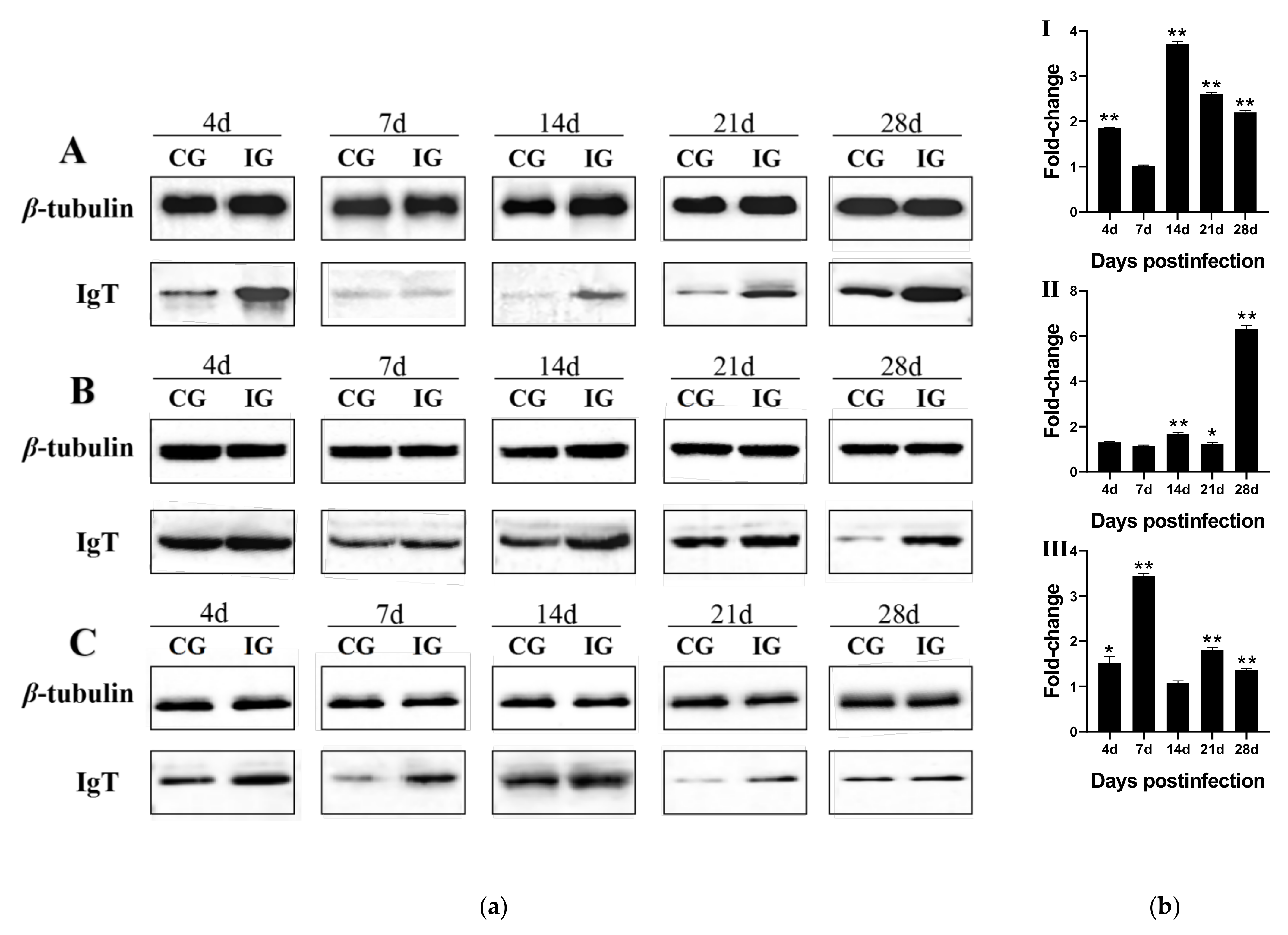
3.5. Expression Change at Protein Levels of LcIgT after C. irritans Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Schroeder, H.J.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125 (Suppl. 2), S41–S52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunyer, J.O. Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 2013, 14, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, C.M.; Richard, A.S.; Sigel, M.M. IgM antibodies in fish mucus. Proc. Soc. Exp. Biol. Med. 1971, 136, 1122–1124. [Google Scholar] [CrossRef] [PubMed]
- Sunyer, J.O. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets 2012, 12, 200–212. [Google Scholar] [CrossRef]
- Edholm, E.S.; Bengten, E.; Wilson, M. Insights into the function of IgD. Dev. Comp. Immunol. 2011, 35, 1309–1316. [Google Scholar] [CrossRef]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.L.; Xiang, L.X.; Shao, J.Z. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: Implications for a distinct B cell receptor in lower vertebrates. Mol. Immunol. 2010, 47, 738–746. [Google Scholar] [CrossRef]
- Fillatreau, S.; Six, A.; Magadan, S.; Castro, R.; Sunyer, J.O.; Boudinot, P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Edholm, E.S.; Bengten, E.; Stafford, J.L.; Sahoo, M.; Taylor, E.B.; Miller, N.W.; Wilson, M. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J. Immunol. 2010, 185, 4082–4094. [Google Scholar] [CrossRef] [Green Version]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar] [CrossRef]
- Savan, R.; Aman, A.; Sato, K.; Yamaguchi, R.; Sakai, M. Discovery of a new class of immunoglobulin heavy chain from fugu. Eur. J. Immunol. 2005, 35, 3320–3331. [Google Scholar] [CrossRef] [PubMed]
- Ryo, S.; Wijdeven, R.H.; Tyagi, A.; Hermsen, T.; Kono, T.; Karunasagar, I.; Rombout, J.H.; Sakai, M.; Verburg-van Kemenade, B.M.; Savan, R. Common carp have two subclasses of bonyfish specific antibody IgZ showing differential expression in response to infection. Dev. Comp. Immunol. 2010, 34, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Tang, X.; Zhan, W.; Xing, J.; Sheng, X. Immunoglobulin Tau Heavy Chain (IgT) in Flounder, Paralichthys olivaceus: Molecular Cloning, Characterization, and Expression Analyses. Int. J. Mol. Sci. 2016, 17, 1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Du, Y.; Sheng, X.; Xing, J.; Zhan, W. Molecular cloning and expression analyses of immunoglobulin tau heavy chain (IgT) in turbot, Scophthalmus maximus. Vet. Immunol. Immunopathol. 2018, 203, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, F.; Stocchi, V.; Nunez-Ortiz, N.; Randelli, E.; Gerdol, M.; Pallavicini, A.; Facchiano, A.; Bernini, C.; Guerra, L.; Scapigliati, G.; et al. Immunoglobulin T from sea bass (Dicentrarchus labrax L.): Molecular characterization, tissue localization and expression after nodavirus infection. BMC Mol. Biol. 2017, 18, 8. [Google Scholar] [CrossRef] [Green Version]
- Mashoof, S.; Pohlenz, C.; Chen, P.L.; Gatlin, D., 3rd; Buentello, A.; Criscitiello, M.F. Expressed IgH mu and tau transcripts share diversity segment in ranched Thunnus orientalis. Dev. Comp. Immunol. 2014, 43, 76–86. [Google Scholar] [CrossRef]
- Tadiso, T.M.; Lie, K.K.; Hordvik, I. Molecular cloning of IgT from Atlantic salmon, and analysis of the relative expression of tau, mu, and delta in different tissues. Vet. Immunol. Immunopathol. 2011, 139, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Zhang, X.J.; Chen, D.D.; Sunyer, J.O.; Zhang, Y.A. Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2017, 70, 94–105. [Google Scholar] [CrossRef] [Green Version]
- Bengten, E.; Clem, L.W.; Miller, N.W.; Warr, G.W.; Wilson, M. Channel catfish immunoglobulins: Repertoire and expression. Dev. Comp. Immunol. 2006, 30, 77–92. [Google Scholar] [CrossRef]
- Magadan-Mompo, S.; Sanchez-Espinel, C.; Gambon-Deza, F. Immunoglobulin heavy chains in medaka (Oryzias latipes). BMC Evol. Biol. 2011, 11, 165. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Wang, T.; Guo, Y.; Zhao, Z.; Li, N.; Zhao, Y. The immunoglobulin gene loci in the teleost Gasterosteus aculeatus. Fish Shellfish Immunol. 2010, 28, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, S.; Buonocore, F.; Albanese, F.; Scapigliati, G.; Gerdol, M.; Oreste, U.; Coscia, M.R. New insights into evolution of IgT genes coming from Antarctic teleosts. Mar. Genom. 2015, 24 Pt 1, 55–68. [Google Scholar] [CrossRef]
- Schatz, D.G.; Swanson, P.C. V(D)J recombination: Mechanisms of initiation. Annu. Rev. Genet. 2011, 45, 167–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambon-Deza, F.; Sanchez-Espinel, C.; Magadan-Mompo, S. Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev. Comp. Immunol. 2010, 34, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Yasuike, M.; de Boer, J.; von Schalburg, K.R.; Cooper, G.A.; McKinnel, L.; Messmer, A.; So, S.; Davidson, W.S.; Koop, B.F. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genom. 2010, 11, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savan, R.; Aman, A.; Nakao, M.; Watanuki, H.; Sakai, M. Discovery of a novel immunoglobulin heavy chain gene chimera from common carp (Cyprinus carpio L.). Immunogenetics 2005, 57, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef]
- Xu, Z.; Parra, D.; Gomez, D.; Salinas, I.; Zhang, Y.A.; von Gersdorff Jørgensen, L.; Heinecke, R.D.; Buchmann, K.; LaPatra, S.; Sunyer, J.O. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Takizawa, F.; Parra, D.; Gómez, D.; von Gersdorff Jørgensen, L.; LaPatra, S.E.; Sunyer, J.O. Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat. Commun. 2016, 7, 10728. [Google Scholar] [CrossRef] [Green Version]
- Dong, F.; Yin, G.M.; Meng, K.F.; Xu, H.Y.; Liu, X.; Wang, Q.C.; Xu, Z. IgT Plays a Predominant Role in the Antibacterial Immunity of Rainbow Trout Olfactory Organs. Front. Immunol. 2020, 11, 583740. [Google Scholar] [CrossRef]
- Mu, Y.; Li, M.; Ding, F.; Ding, Y.; Ao, J.; Hu, S.; Chen, X. De novo characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PLoS ONE 2014, 9, e97471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.H.; Lin, K.B.; Wang, X.W. Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J. Fish Dis. 2003, 26, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Yujia, S.; Lingmin, Z.; Xu, X.; Huang, L.; Qin, Y.; Su, Y.; Yi, G.; Yan, Q. Mechanistic insight into the roles of Pseudomonas plecoglossicida clpV gene in host-pathogen interactions with Larimichthys crocea by dual RNA-seq. Fish Shellfish Immunol. 2019, 93, 344–353. [Google Scholar]
- Li, C.; Wang, S.; Ren, Q.; He, T.; Chen, X. An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12 degrees C in cultured large yellow croaker (Larimichthys crocea) in China. J. Fish Dis. 2020, 43, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Ke, Q.; Chen, L.; Zhou, Z.; Pu, F.; Zhao, J.; Bai, H.; Peng, W.; Xu, P. Constructing a High-Density Genetic Linkage Map for Large Yellow Croaker (Larimichthys crocea) and Mapping Resistance Trait Against Ciliate Parasite Cryptocaryon irritans. Mar. Biotechnol. 2019, 21, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Ding, Y.; Ao, J.; Chen, X. Three isotypes of immunoglobulin light chains in large yellow croaker, Pseudosciaena crocea: Molecular cloning, characterization, and expression analysis. Fish Shellfish Immunol. 2011, 30, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Chen, X.; Ao, J. The up-regulation of large yellow croaker secretory IgM heavy chain at early phase of immune response. Fish Physiol. Biochem. 2010, 36, 483–490. [Google Scholar] [CrossRef]
- Ao, J.; Mu, Y.; Xiang, L.X.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Mu, P.; Li, X.; Ren, Q.; Zhang, X.Y.; Mu, Y.; Chen, X. Functions of TNF-alpha1 and TNF-alpha2 in large yellow croaker (Larimichthys crocea) in monocyte/macrophage activation. Dev. Comp. Immunol. 2020, 105, 103576. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fu, Q.; Wei, Z.; Chen, Y.; Xie, J.; Zhang, X.; He, T.; Chen, X. Development of monoclonal antibody against IgT of a perciform fish, large yellow croaker (Larimichthys crocea) and characterization of IgT(+) B cells. Dev. Comp. Immunol. 2021, 119, 104027. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Liu, W.; Wu, K.; Wang, W.; Zhang, X. sIgZ exhibited maternal transmission in embryonic development and played a prominent role in mucosal immune response of Megalabrama amblycephala. Fish Shellfish Immunol. 2016, 54, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Coscia, M.R.; Simoniello, P.; Giacomelli, S.; Oreste, U.; Motta, C.M. Investigation of immunoglobulins in skin of the Antarctic teleost Trematomus bernacchii. Fish Shellfish Immunol. 2014, 39, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.Y.; Kong, W.; Yin, Y.X.; Dong, F.; Huang, Z.Y.; Yin, G.M.; Dong, S.; Salinas, I.; Zhang, Y.A.; Xu, Z. Mucosal immunoglobulins protect the olfactory organ of teleost fish against parasitic infection. PLoS Pathog. 2018, 14, e1007251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Yu, Y.; Zhang, X.; Xu, Z. Immune responses of fish to Ichthyophthirius multifiliis (Ich): A model for understanding immunity against protozoan parasites. Dev. Comp. Immunol. 2019, 93, 93–102. [Google Scholar] [CrossRef] [PubMed]

| Species | IgT Heavy Chain (Identity, %) | NCBI Accession Number |
|---|---|---|
| Entire Sequence | ||
| Larimichthys crocea | 100 | MW450786 |
| Scophthalmus maximus | 58.73 | AMQ49170.1 |
| Sparus aurata | 56.10 | ASK39430.1 |
| Paralichthys olivaceus | 55.88 | ANS12795.1 |
| Dicentrarchus labrax | 55.04 | AKK32388.1 |
| Epinephelus coioides | 53.96 | ACZ54909.1 |
| Oncorhynchus mykiss | 44.14 | AAW66978.1 |
| Oreochromis niloticus | 36.84 | AUV64181.1 |
| Danio rerio | 32.14 | ABF19723.1 |
| Ctenopharyngodon idella | 28.24 | AAT67444.1 |
| Species | IgT Heavy Chain (Identity, %) | |||
|---|---|---|---|---|
| VH | CH1 | CH2 | CH4 | |
| Larimichthys crocea | 100 | 100 | 100 | 100 |
| Scophthalmus maximus | 72.50 | 61.29 | 52.05 | 63.33 |
| Oreochromis niloticus | 41.05 | 67.61 | 61.11 | 63.74 |
| Paralichthys olivaceus | 56.10 | 61.29 | 52.05 | 63.33 |
| Epinephelus coioides | 43.97 | 66.20 | 56.94 | 63.74 |
| Sparus aurata | 47.47 | 62.90 | 61.11 | 60.24 |
| Dicentrarchus labrax | 51.90 | 47.89 | 56.94 | 58.43 |
| Oncorhynchus mykiss | 44.21 | 50.00 | 40.85 | 47.73 |
| Ctenopharyngodon idella | 45.30 | 35.14 | 35.14 | 25.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, Y.; Fu, Q.; Tan, Y.; Ding, Y.; Ding, Y.; Chen, X. Identification and Characterization of Immunoglobulin T Heavy Chain in Large Yellow Croaker (Larimichthys crocea). Fishes 2022, 7, 29. https://doi.org/10.3390/fishes7010029
Teng Y, Fu Q, Tan Y, Ding Y, Ding Y, Chen X. Identification and Characterization of Immunoglobulin T Heavy Chain in Large Yellow Croaker (Larimichthys crocea). Fishes. 2022; 7(1):29. https://doi.org/10.3390/fishes7010029
Chicago/Turabian StyleTeng, Yan, Qiuling Fu, Yuanzhen Tan, Yangyang Ding, Yang Ding, and Xinhua Chen. 2022. "Identification and Characterization of Immunoglobulin T Heavy Chain in Large Yellow Croaker (Larimichthys crocea)" Fishes 7, no. 1: 29. https://doi.org/10.3390/fishes7010029
APA StyleTeng, Y., Fu, Q., Tan, Y., Ding, Y., Ding, Y., & Chen, X. (2022). Identification and Characterization of Immunoglobulin T Heavy Chain in Large Yellow Croaker (Larimichthys crocea). Fishes, 7(1), 29. https://doi.org/10.3390/fishes7010029





