The Snakeskin Gourami (Trichopodus pectoralis) Tends to Exhibit XX/XY Sex Determination
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens and DNA Extraction
2.2. Development of DArTseq Arrays
2.3. Comparison of Potential Sex-Linked Loci
2.4. Homology Searching
2.5. Chromosome Preparations and Mapping the Chromosomal Locations of Microsatellite Repeat Motifs, Telomeric (TTAGGG)n Sequences, and Major Ribosomal RNA Genes with Fluorescence In Situ Hybridization
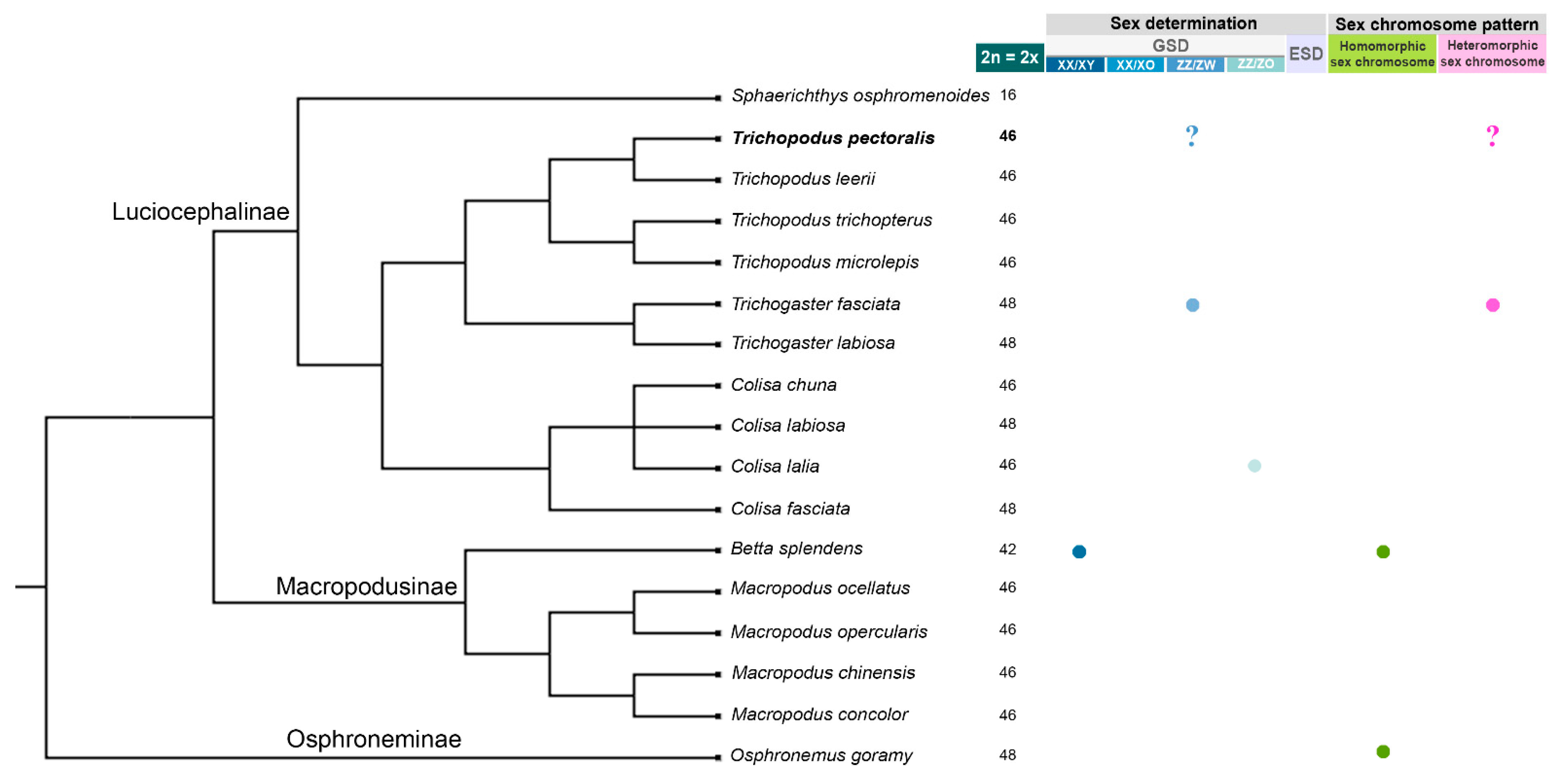
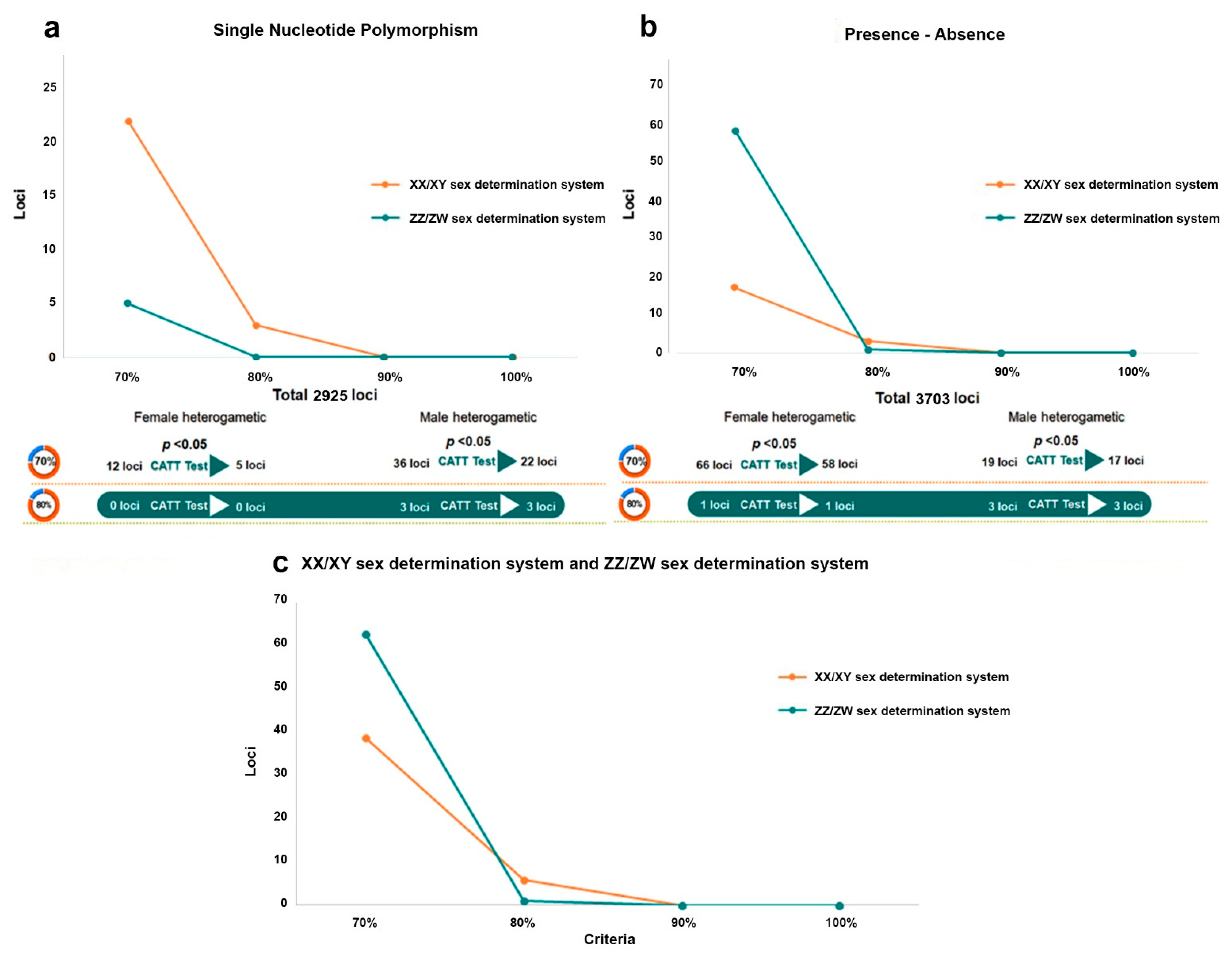
3. Results
3.1. Identification of the Sex Determination System and Sex-Linked Loci in the Snakeskin Gourami
3.2. Random Sex-Linkage Estimation
3.3. Homology of Putative Sex-Linked Loci
3.4. Karyotype and Meiotic Configuration
3.5. Chromosomal Locations of the 18S–28S rRNA Genes, Telomeric (TTAGGG)n Sequences, and Microsatellite Repeat Motifs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Regan, C.T. The Asiatic fishes of the family Anabantidae. J. Zool. 1910, 1909, 767–787. [Google Scholar]
- Morioka, S.; Vongvichith, B.; Phommachan, P.; Chantasone, P. Growth and morphological development of laboratory-reared larval and juvenile giant gourami Osphronemus goramy (Perciformes: Osphronemidae). Ichthyol. Res. 2013, 60, 209–217. [Google Scholar] [CrossRef]
- Froese, R. Cube law, condition factor and weight–length relationships: History, meta-analysis and recommendations. J. Appl. Ichthyol. 2006, 22, 241–253. [Google Scholar] [CrossRef]
- Tacon, A.G.; Hasan, M.R.; Metian, M. Demand and Supply of Feed Ingredients for Farmed Fish and Crustaceans: Trends and Prospects. 2011. Available online: https://www.proquest.com/docview/1328361199?pq-origsite=gscholar&fromopenview=true (accessed on 21 August 2020).
- Minh, N.P.; Mai, P.X.; Van Linh, N.T. Several aspects influencing to production of dry-salted snakeskin gourami (Trichogaster pectoralis). Orient J. Chem. 2019, 35, 773–777. [Google Scholar] [CrossRef]
- Needham, S.; Funge-Smith, S.J. The consumption of fish and fish products in the Asia-Pacific region based on household surveys. Bangk. FAO Reg. Off. Asia Pac. 2015, 12, 87. [Google Scholar]
- Budd, A.M.; Banh, Q.Q.; Domingos, J.A.; Jerry, D.R. Sex control in fish: Approaches, challenges and opportunities for aquaculture. J. Mar. Sci. Eng. 2015, 3, 329–355. [Google Scholar] [CrossRef]
- Kohn, Y.Y.; Lokman, P.M.; Kilimnik, A.; Symonds, J.E. Sex identification in captive hapuku (Polyprion oxygeneios) using ultrasound imagery and plasma levels of vitellogenin and sex steroids. Aquaculture 2013, 384, 87–93. [Google Scholar] [CrossRef]
- Ramadhani, D.G.; Utomo, S.B.; Indriyanti, N.Y. Content analysis of 13 dimensions to support student teachers’ PCK in the environmental chemistry textbooks. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2018; p. 012077. [Google Scholar]
- Hu, Q.; Chang, C.; Wang, Q.; Tian, H.; Qiao, Z.; Wang, L.; Meng, Y.; Xu, C.; Xiao, H. Genome-wide RAD sequencing to identify a sex-specific marker in Chinese giant salamander Andrias davidianus. BMC Genom. 2019, 20, 415. [Google Scholar] [CrossRef]
- Heule, C.; Salzburger, W.; Böhne, A. Genetics of sexual development: An evolutionary playground for fish. Genetics 2014, 196, 579–591. [Google Scholar] [CrossRef]
- Luckenbach, J.A.; Borski, R.J.; Daniels, H.V.; Godwin, J. Sex determination in flatfishes: Mechanisms and environmental influences. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2009; pp. 256–263. [Google Scholar]
- Penman, D.J.; Piferrer, F. Fish gonadogenesis. Part I: Genetic and environmental mechanisms of sex determination. Rev. Fish. Sci. 2008, 16, 16–34. [Google Scholar] [CrossRef]
- Arai, R. Fish Karyotypes: A Check List; Springer: Tokyo, Japan, 2011. [Google Scholar]
- Kottler, V.A.; Schartl, M. The colorful sex chromosomes of teleost fish. Genes 2018, 9, 233. [Google Scholar] [CrossRef]
- Trukhina, A.V.; Lukina, N.A.; Wackerow-Kouzova, N.D.; Smirnov, A.F. The variety of vertebrate mechanisms of sex determination. Biomed. Res. Int. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vidthayanon, C. Thailand Red Data: Fishes; Office of Natural Resources and Environmental Policy and Planning: Bangkok, Thailand, 2005. [Google Scholar]
- Ross, J.A.; Urton, J.R.; Boland, J.; Shapiro, M.D.; Peichel, C.L. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 2009, 5, e1000391. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.M.; Panthum, T.; Ponjarat, J.; Laopichiengpong, N.; Kraichak, E.; Singchat, W.; Muangmai, N.; Peyachoknagul, S.; Na-Nakorn, U.; Srikulnath, K. An investigation of ZZ/ZW and XX/XY sex determination systems in North African catfish (Clarias gariepinus, Burchell 1822). Front. Genet. 2021, 11, 1719. [Google Scholar] [CrossRef]
- Nguyen, D.H.M.; Ponjarat, J.; Laopichienpong, N.; Kraichak, E.; Panthum, T.; Singchat, W.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; et al. Genome-wide SNP analysis suggests male heterogamety in bighead catfish (Clarias macrocephalus). Aquaculture 2021, 545, 737005. [Google Scholar] [CrossRef]
- Cioffi, M.D.B.; Yano, C.F.; Sember, A.; Bertollo, L.A.C. Chromosomal evolution in lower vertebrates: Sex chromosomes in neotropical fishes. Genes 2017, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Sember, A.; Bertollo, L.; Ráb, P.; Yano, C.F.; Hatanaka, T.; de Oliveira, E.A.; Cioffi, M.B. Sex chromosome evolution and genomic divergence in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Front. Genet. 2018, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Shams, F.; Dyer, F.; Thompson, R.; Duncan, R.P.; Thiem, J.D.; Majtánová, Z.; Ezaz, T. Karyotypes and sex chromosomes in two Australian native freshwater fishes, golden perch (Macquaria ambigua) and murray cod (Maccullochella peelii) (Percichthyidae). Int. J. Mol. Sci. 2019, 20, 4244. [Google Scholar] [CrossRef]
- Singchat, W.; O’Connor, R.E.; Tawichasri, P.; Suntronpong, A.; Sillapaprayoon, S.; Suntrarachun, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genom. 2018, 19, 939. [Google Scholar] [CrossRef]
- Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Indananda, C.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”? Chromosome Res. 2020, 28, 209–228. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Panthum, T.; Srikulnath, K. Impact of repetitive DNA elements on snake genome biology and evolution. Cells 2021, 10, 1707. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Nishida, C.; Olsson, M.; Matsuda, Y. Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 2014, 123, 563–575. [Google Scholar] [CrossRef]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Ota, H.; Matsuda, Y. Karyotype reorganization in the Hokou gecko (Gekko hokouensis, Gekkonidae): The process of microchromosome disappearance in Gekkota. PLoS ONE 2015, 10, e0134829. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Azad, B.; Singchat, W.; Ezaz, T. Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS ONE 2019, 14, e0212683. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerfulfacilitators of chromosomal rearrangements for diversity and evolution (running title: Genomic impact of repeats on chromosomal dynamics inreptiles). Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Singchat, W.; Ahmad, S.F.; Sillapaprayoon, S.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Partial amniote sex chromosomal linkage homologies shared on snake W sex chromosomes support the ancestral super-sex chromosome evolution in Amniotes. Front. Genet. 2020, 11, 948. [Google Scholar] [CrossRef] [PubMed]
- Pennell, M.W.; Mank, J.E.; Peichel, C.L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2018, 27, 3950–3963. [Google Scholar] [CrossRef]
- Pokorná, M.; Kratochvíl, L.; Kejnovský, E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 2011, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Ponjarat, J.; Singchat, W.; Monkheang, P.; Suntronpong, A.; Tawichasri, P.; Sillapaprayoon, S.; Ogawa, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; et al. Evidence of dramatic sterility in F1 male hybrid catfish [male Clarias gariepinus (Burchell, 1822) × female C. macrocephalus (Günther, 1864)] resulting from the failure of homologous chromosome pairing in meiosis I. Aquaculture 2019, 505, 84–91. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Steinberg, K.M.; Larson, D.E.; Wilson, R.K.; Mardis, E.R. The next-generation sequencing revolution and its impact on genomics. Cell 2013, 155, 27–38. [Google Scholar] [CrossRef]
- Yáñez, J.M.; Naswa, S.; López, M.E.; Bassini, L.; Correa, K.; Gilbey, J.; Bernatchez, L.; Norris, A.; Neira, R.; Lhorente, J.P.; et al. Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): Validation in wild and farmed American and European populations. Mol. Ecol. Resour. 2016, 16, 1002–1011. [Google Scholar] [CrossRef]
- Koomgun, T.; Laopichienpong, N.; Singchat, W.; Panthum, T.; Phatcharakullawarawat, R.; Kraichak, E.; Sillapaprayoon, S.; Ahmad, S.F.; Muangmai, N.; Peyachoknagul, S.; et al. Genome complexity reduction high-throughput genome sequencing of green iguana (Iguana iguana) reveal a paradigm shift in understanding sex-chromosomal linkages on homomorphic X and Y sex chromosomes. Front. Genet. 2020, 11, 1217. [Google Scholar] [CrossRef]
- Rüber, L.; Britz, R.; Zardoya, R. Molecular phylogenetics and evolutionary diversification of labyrinth fishes (Perciformes: Anabantoidei). Syst. Biol. 2006, 55, 374–397. [Google Scholar] [CrossRef] [PubMed]
- Koref-Santibanez, S.; Paepke, H. Karyotypes of the Trichogasterinae Liem (Teleostei, Anabantoidei). In Abstract of the VIII Congress of the Society of European Ichthyologists; Society of European Ichthyologists: Oviedo, Spain, 1994; p. 55. [Google Scholar]
- Manna, G.K.; Prasad, R. Chromosome analysis of five species of freshwater fishes. Nucleus 1977, 20, 264–271. [Google Scholar]
- Grazyna, F.S.; Fopp-Bayat, D.; Jankun, M.; Krejszeff, S.; Mamcarz, A. Note on the karyotype and NOR location of Siamese fighting fish Betta splendens (Perciformes, Osphronemidae). Caryologia 2008, 61, 349–353. [Google Scholar] [CrossRef]
- Koref-Santibanez, S.; Paepke, H.J.; Tjio, H.J. Karyotypen der arten der gattung macropodus lac. (Teleostei Anabantoidei). Zool. Anz. 1991, 227, 271–278. [Google Scholar]
- Jiaxun, C.; Xiuhai, R.; Qixing, Y. Nuclear DNA content variation in fishes. Cytologia 1991, 56, 425–429. [Google Scholar] [CrossRef][Green Version]
- Elhag, A.I.; Rahmah, S.; Sheriff, S.M.; Tan, W.C.; Jong, K.F.; Ambak, M.A.; Liew, H.J. Sexual characteristic differences between male and female of jade perch Scortum barcoo. Int. J. Fish. Aquat. Stud. 2019, 7, 258–264. [Google Scholar]
- Kitano, J.; Mori, S.; Peichel, C.L. Sexual dimorphism in the external morphology of the threespine stickleback (Gasterosteus aculeatus). Copeia 2007, 2007, 336–349. [Google Scholar] [CrossRef]
- Wyneken, J.; Epperly, S.P.; Crowder, L.B.; Vaughan, J.; Blair Esper, K. Determining sex in posthatchling loggerhead sea turtles using multiple gonadal and accessory duct characteristics. Herpetologica 2007, 63, 19–30. [Google Scholar] [CrossRef]
- Hubert, N.; Hanner, R.; Holm, E.; Mandrak, N.E.; Taylor, E.; Burridge, M.; Watkinson, D.; Dumont, P.; Curry, A.; Bentzen, P.; et al. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE 2008, 3, e2490. [Google Scholar] [CrossRef]
- Rüber, L.; Britz, R.; Kullander, S.O.; Zardoya, R. Evolutionary and biogeographic patterns of the Badidae (Teleostei: Perciformes) inferred from mitochondrial and nuclear DNA sequence data. Mol. Phylogenet. Evol. 2004, 32, 1010–1022. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015, 14, 13981–13997. [Google Scholar] [CrossRef] [PubMed]
- Laopichienpong, N.; Kraichak, E.; Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; Ezaz, T.; et al. Genome-wide SNP analysis of Siamese cobra (Naja kaouthia) reveals the molecular basis of transitions between Z and W sex chromosomes and supports the presence of an ancestral super-sex chromosome in amniotes. Genomics 2021, 113, 624–636. [Google Scholar] [CrossRef]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef] [PubMed]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics; Humana Press: Totowa, NJ, USA, 2012; pp. 67–89. [Google Scholar]
- Ren, R.; Ray, R.; Li, P.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A.; Yang, X. Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genom. 2015, 290, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Sven, K.; Klaus, R. HapEstXXR: Multi-locus stepwise regression. 2019. Available online: https://rdrr.io/cran/HapEstXXR/man/coding.baseline.allele.html (accessed on 21 August 2020).
- Gruber, B.; Georges, A. dartR: Importing and Analysing SNP and Silico Dart Data Generated by Genome-Wide Restriction Fragment Analysis, R Package Version 1111; 2019. Available online: https://cran.r-project.org/web/packages/dartR/index.html (accessed on 21 August 2020).
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Broughton, R.E.; Milam, J.E.; Roe, B.A. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001, 11, 1958–1967. [Google Scholar] [CrossRef] [PubMed]
- Elmerot, C.; Arnason, U.; Gojobori, T.; Janke, A. The mitochondrial genome of the pufferfish, Fugu rubripes, and ordinal teleostean relationships. Gene 2002, 295, 163–172. [Google Scholar] [CrossRef]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Suntronpong, A.; Thapana, W.; Twilprawat, P.; Prakhongcheep, O.; Somyong, S.; Muangmai, N.; Peyachoknagul, S.; Srikulnath, K. Karyological characterization and identification of four repetitive element groups (the 18S–28S rRNA gene, telomeric sequences, microsatellite repeat motifs, Rex retroelements) of the Asian swamp eel (Monopterus albus). Comp. Cytogenet. 2017, 11, 435. [Google Scholar] [CrossRef]
- Sumner, M.A.; Bradley, T.R.; Hodgson, G.S.; Cline, M.J.; Fry, P.A.; Sutherland, L. The growth of bone marrow cells in liquid culture. Br. J. Haematol. 1972, 23, 221–234. [Google Scholar] [CrossRef]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y.; Nishida, C. Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet. Genome Res. 2009, 125, 213–223. [Google Scholar] [CrossRef]
- Imai, H.T.; Matsuda, Y.; Shiroishi, T.; Moriwaki, K. High frequency fo X-Y chromosome dissociation in primary spermatocytes of F1 hybrids between Japanese wild mice (Mus musculus molossinus) and inbred laboratory mice. Cytogenet. Cell Genet. 1981, 29, 166–175. [Google Scholar] [CrossRef]
- Matsuda, Y.; Chapman, V.M. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 1995, 16, 261–272. [Google Scholar] [CrossRef] [PubMed]
- National Bureau of Agricultural Commodity and Food Standards Ministry of Agriculture and Cooperatives. Thai agricultural standard: Organic snakeskin gourami. Publ. R. Gaz. 2010, 127, 150. [Google Scholar]
- Mei, J.; Gui, J.F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Skelly, D.K.; Ezaz, T. Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genom. 2016, 17, 844. [Google Scholar] [CrossRef]
- Gamble, T.; Coryell, J.; Ezaz, T.; Lynch, J.; Scantlebury, D.P.; Zarkower, D. Restriction site-associated DNA sequences (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 2015, 32, 1296–1309. [Google Scholar] [CrossRef]
- Samonte-Padilla, I.E.; Eizaguirre, C.; Scharsack, P.J.; Lenz, T.L.; Milinski, M. Induction of diploid gynogenesis in an evolutionary model organism, the three-spined stickleback (Gasterosteus aculeatus). BMC Dev. Biol. 2011, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Boonanuntanasarn, S.; Jangprai, A.; Na-Nakorn, U. Transcriptomic analysis of female and male gonads in juvenile snakeskin gourami (Trichopodus pectoralis). Sci. Rep. 2020, 10, 1–13. [Google Scholar]
- Martínez, P.; Viñas, A.M.; Sánchez, L.; Díaz, N.; Ribas, L.; Piferrer, F. Genetic architecture of sex determination in fish: Applications to sex ratio control in aquaculture. Front. Genet. 2014, 5, 340. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F. Endocrine sex control strategies for the feminization of teleost fish. Aquaculture 2001, 197, 229–281. [Google Scholar] [CrossRef]
- Saenjundaeng, P.; de Bello Cioffi, M.; de Oliveira, E.A.; Tanomtong, A.; Supiwong, W.; Phimphan, S.; Collares-Pereira, M.J.; Sember, A.; Bertollo, L.A.C.; Liehr, T.; et al. Chromosomes of Asian cyprinid fishes: Cytogenetic analysis of two representatives of small paleotetraploid tribe Probarbini. Mol. Cytogenet. 2018, 11, 1–9. [Google Scholar] [CrossRef]
- Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Akberdina, E.A.; Kizilova, E.A.; Romanenko, M.V.; Shnaider, E.P.; Scherbakova, M.M.; Korobitsyn, I.G.; Rubtsov, N.B.; et al. Germline-restricted chromosome (GRC) in the sand martin and the pale martin (Hirundinidae, Aves): Synapsis, recombination and copy number variation. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kawai, A.; Nishida-Umehara, C.; Ishijima, J.; Tsuda, Y.; Ota, H.; Matsuda, Y. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 2007, 117, 92–102. [Google Scholar] [CrossRef]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef]
- Mackiewicz, D.; Posacki, P.; Burdukiewicz, M.; Błażej, P. Role of recombination and faithfulness to partner in sex chromosome degeneration. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Manolakou, P.; Lavranos, G.; Angelopoulou, R. Molecular patterns of sex determination in the animal kingdom: A comparative study of the biology of reproduction. Reprod. Biol. Endocrinol. 2006, 4, 59. [Google Scholar] [CrossRef][Green Version]
- Boehne, A.; Schultheis, C.; Galiana-Arnoux, D.; Froschauer, A.; Zhou, Q.; Schmidt, C.; Selz, Y.; Ozouf-Costaz, C.; Dettai, A.; Segurens, B.; et al. Molecular analysis of the sex chromosomes of the platyfish Xiphophorus maculatus: Towards the identification of a new type of master sexual regulator in vertebrates. Integr. Zool. 2009, 4, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D. The guppy sex chromosome system and the sexually antagonistic polymorphism hypothesis for Y chromosome recombination suppression. Genes 2018, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Myosho, T.; Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. Turnover of sex chromosomes in celebensis group medaka fishes. G3 Genes Genomes Genet. 2015, 5, 2685–2691. [Google Scholar] [CrossRef] [PubMed]
- Volff, J.N.; Schartl, M. Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus. Cytogenet. Genome Res. 2002, 99, 170–177. [Google Scholar] [CrossRef]
- Charlesworth, D. Evolution of recombination rates between sex chromosomes. Phil. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160456. [Google Scholar] [CrossRef]
- Mank, J.E.; Avise, J.C. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex. Dev. 2009, 3, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Liew, W.C.; Orbán, L. Zebrafish sex: A complicated affair. Brief. Funct. Genom. 2014, 13, 172–187. [Google Scholar] [CrossRef]
- Vandeputte, M.; Dupont-Nivet, M.; Chavanne, H.; Chatain, B. A polygenic hypothesis for sex determination in the European sea bass Dicentrarchus labrax. Genetics 2007, 176, 1049–1057. [Google Scholar] [CrossRef]
- Moore, E.C.; Roberts, R.B. Polygenic sex determination. Curr. Biol. 2013, 23, R510–R512. [Google Scholar] [CrossRef]
- Casey, J.; Yue, X.; Nguyen, T.D.; Acun, A.; Zellmer, V.R.; Zhang, S.; Zorlutana, P. 3D hydrogel-based microwell arrays as a tumor microenvironment model to study breast cancer growth. Biomed. Mater. 2017, 12, 025009. [Google Scholar] [CrossRef]
- Livernois, A.M.; Waters, S.A.; Deakin, J.E.; Graves, J.A.M.; Waters, P.D. Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals. PLoS Genet. 2013, 9, e1003635. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; O’Brien, P.C.M.; Grützner, F.; Clarke, O.; Graphodatskaya, D.; Tsend-Ayush, E.; Trifonov, V.A.; Skelton, H.; Wallis, M.C.; Johnston, S.; et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007, 8, R243. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.L.; Glor, R.E.; Turelli, M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- Chalopin, D.; Volff, J.N.; Galiana, D.; Anderson, J.L.; Schartl, M. Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 2015, 23, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Dechaud, C.; Volff, J.; Schartl, M.; Naville, M. Sex and the TEs: Transposable elements in sexual development and function in animals. Mob. DNA 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Nishida, C.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y. Karyotypic evolution in squamate reptiles: Comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res. 2009, 17, 975. [Google Scholar] [CrossRef]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Matsuda, Y. Karyotype evolution in monitor lizards: Cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 2013, 21, 805–819. [Google Scholar] [CrossRef]
- Ezaz, T.; Srikulnath, K.; Graves, J.A.M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements? J. Hered. 2017, 108, 94–105. [Google Scholar] [CrossRef]
- Matsubara, K.; O’Meally, D.; Sarre, S.D.; Georges, A.; Srikulnath, K.; Ezaz, T. ZW sex chromosomes in Australian dragon lizards (Agamidae) originated from a combination of duplication and translocation in the nucleolar organising region. Genes 2019, 10, 861. [Google Scholar] [CrossRef]
- Gamble, T.; Geneva, A.J.; Glor, R.E.; Zarkower, D. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 2014, 68, 1027–1041. [Google Scholar] [CrossRef]
- Liu, S.; Aagaard, A.; Bechsgaard, J.; Bilde, T. DNA methylation patterns in the social spider, Stegodyphus dumicola. Genes 2019, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Ma, X.; Li, T.; Zhu, C.; You, X. Developing single nucleotide polymorphisms for identification of cod products by RAD-Seq. Animals 2020, 10, 423. [Google Scholar] [CrossRef]
- Gamble, T. Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Mol. Ecol. 2016, 25, 2114–2116. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Liu, T.; Yu, C.H.; Chiang, T.Y.; Hwang, C.C. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS ONE 2013, 8, e62856. [Google Scholar] [CrossRef]
- Gamble, T.; Zarkower, D. Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol. Ecol. Resour. 2014, 14, 902–913. [Google Scholar] [CrossRef]
- Phua, A.C.Y.; Abdullah, R.B.; Mohamed, Z. A PCR-based sex determination method for possible application in caprine gender selection by simultaneous amplification of the Sry and Aml-X genes. J. Reprod. Dev. 2003, 49, 307–311. [Google Scholar] [CrossRef] [PubMed][Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panthum, T.; Laopichienpong, N.; Kraichak, E.; Singchat, W.; Ho My Nguyen, D.; Ariyaraphong, N.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; et al. The Snakeskin Gourami (Trichopodus pectoralis) Tends to Exhibit XX/XY Sex Determination. Fishes 2021, 6, 43. https://doi.org/10.3390/fishes6040043
Panthum T, Laopichienpong N, Kraichak E, Singchat W, Ho My Nguyen D, Ariyaraphong N, Ahmad SF, Muangmai N, Duengkae P, Peyachoknagul S, et al. The Snakeskin Gourami (Trichopodus pectoralis) Tends to Exhibit XX/XY Sex Determination. Fishes. 2021; 6(4):43. https://doi.org/10.3390/fishes6040043
Chicago/Turabian StylePanthum, Thitipong, Nararat Laopichienpong, Ekaphan Kraichak, Worapong Singchat, Dung Ho My Nguyen, Nattakan Ariyaraphong, Syed Farhan Ahmad, Narongrit Muangmai, Prateep Duengkae, Surin Peyachoknagul, and et al. 2021. "The Snakeskin Gourami (Trichopodus pectoralis) Tends to Exhibit XX/XY Sex Determination" Fishes 6, no. 4: 43. https://doi.org/10.3390/fishes6040043
APA StylePanthum, T., Laopichienpong, N., Kraichak, E., Singchat, W., Ho My Nguyen, D., Ariyaraphong, N., Ahmad, S. F., Muangmai, N., Duengkae, P., Peyachoknagul, S., Ezaz, T., & Srikulnath, K. (2021). The Snakeskin Gourami (Trichopodus pectoralis) Tends to Exhibit XX/XY Sex Determination. Fishes, 6(4), 43. https://doi.org/10.3390/fishes6040043










