Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi
Abstract
1. Introduction
2. Results
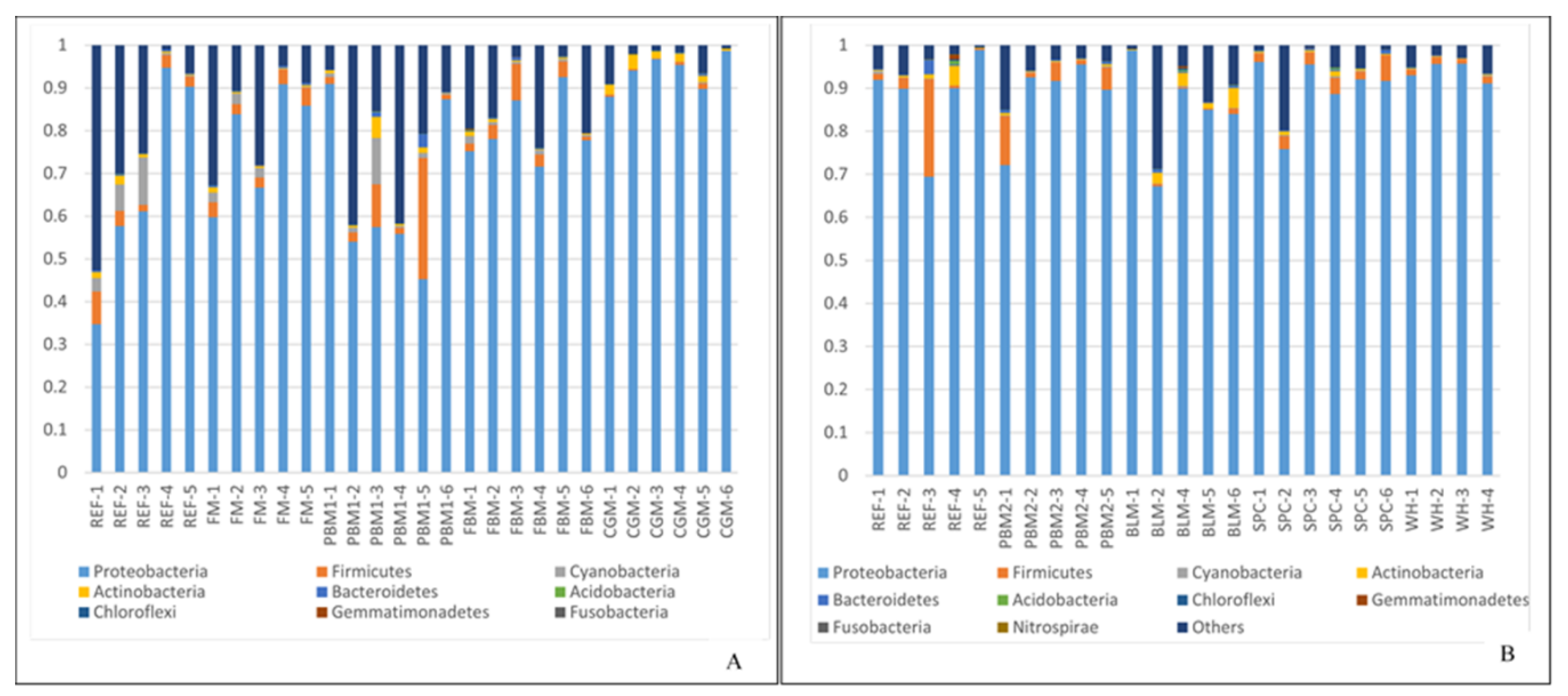
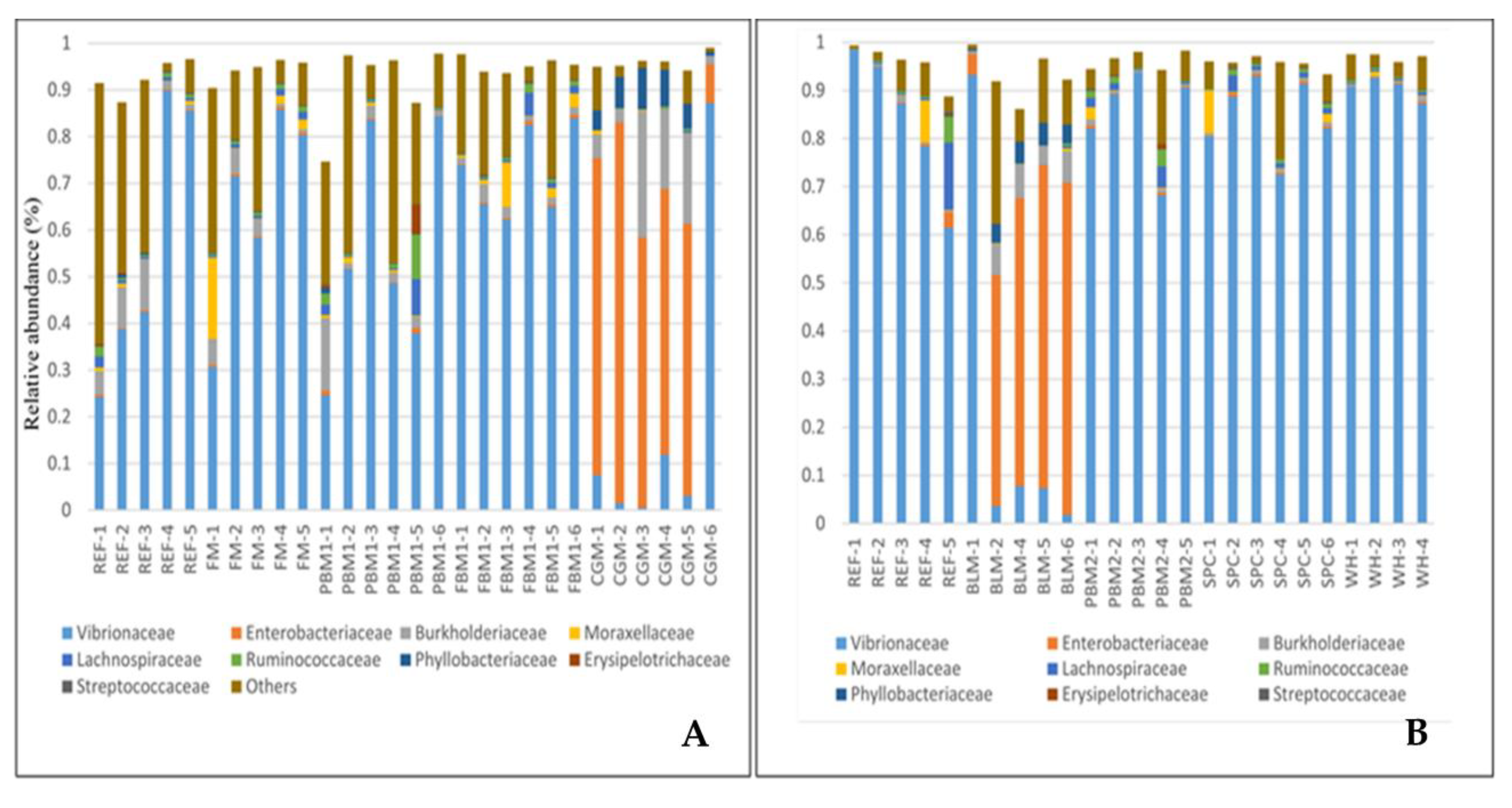
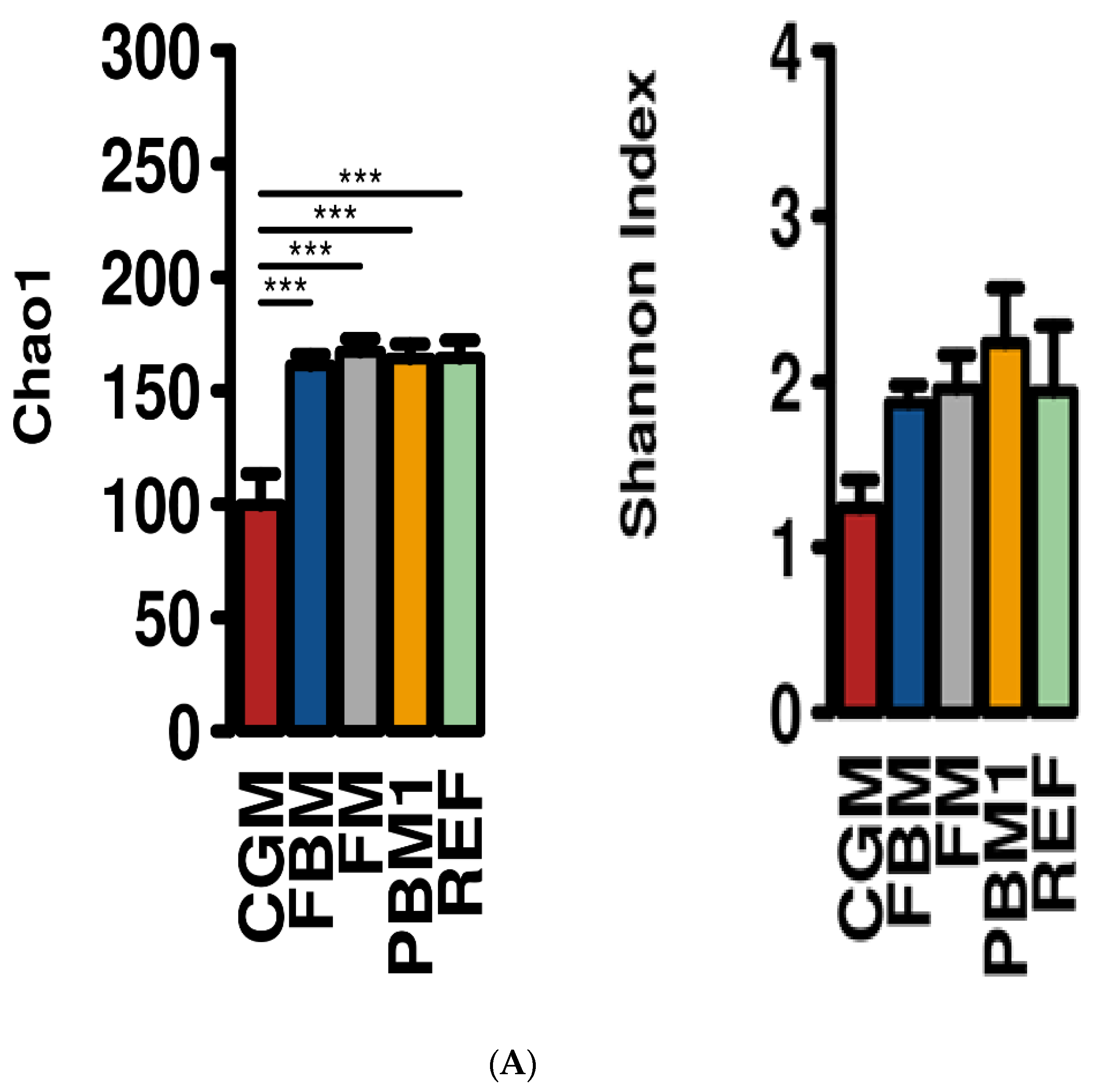
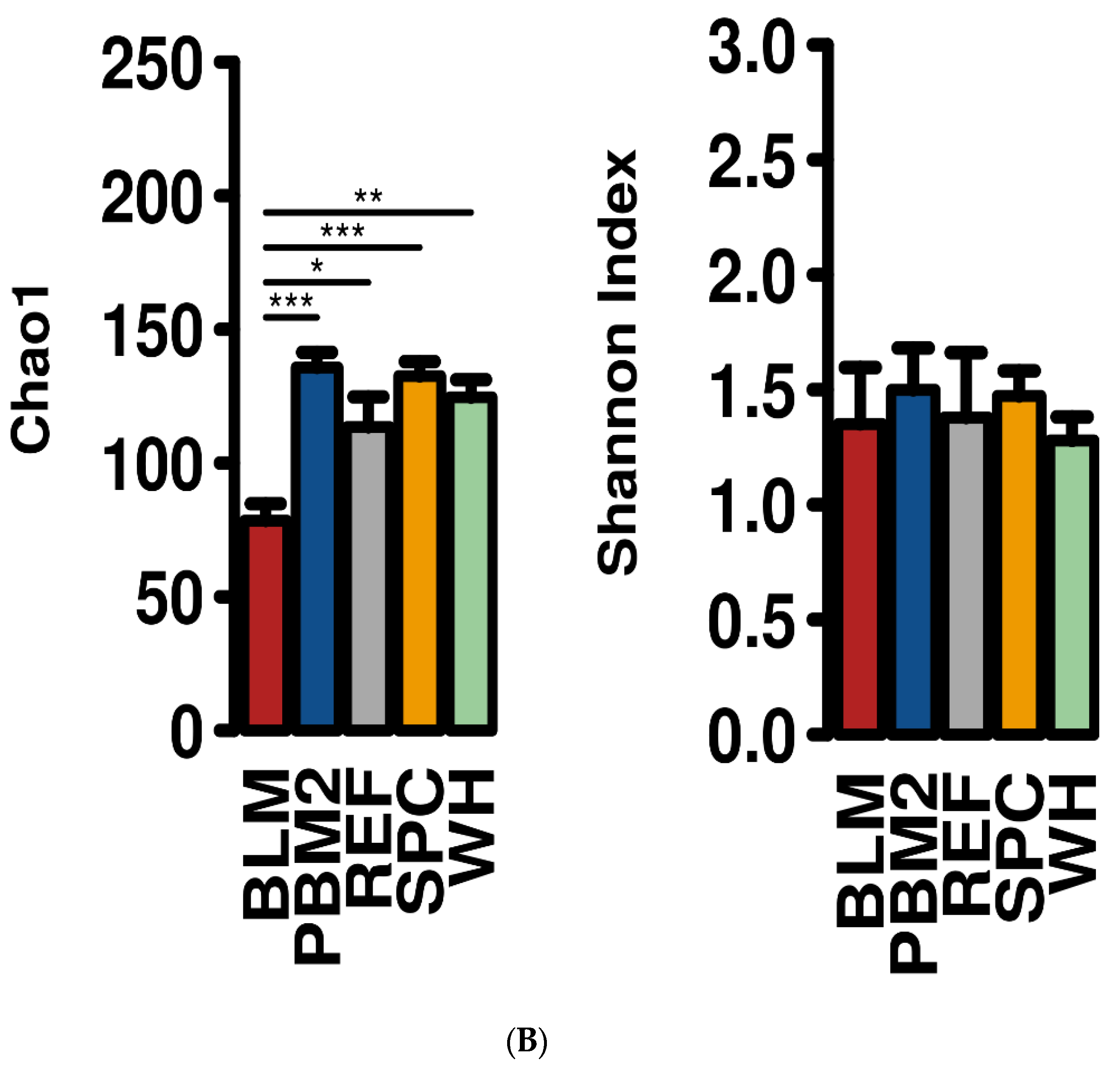
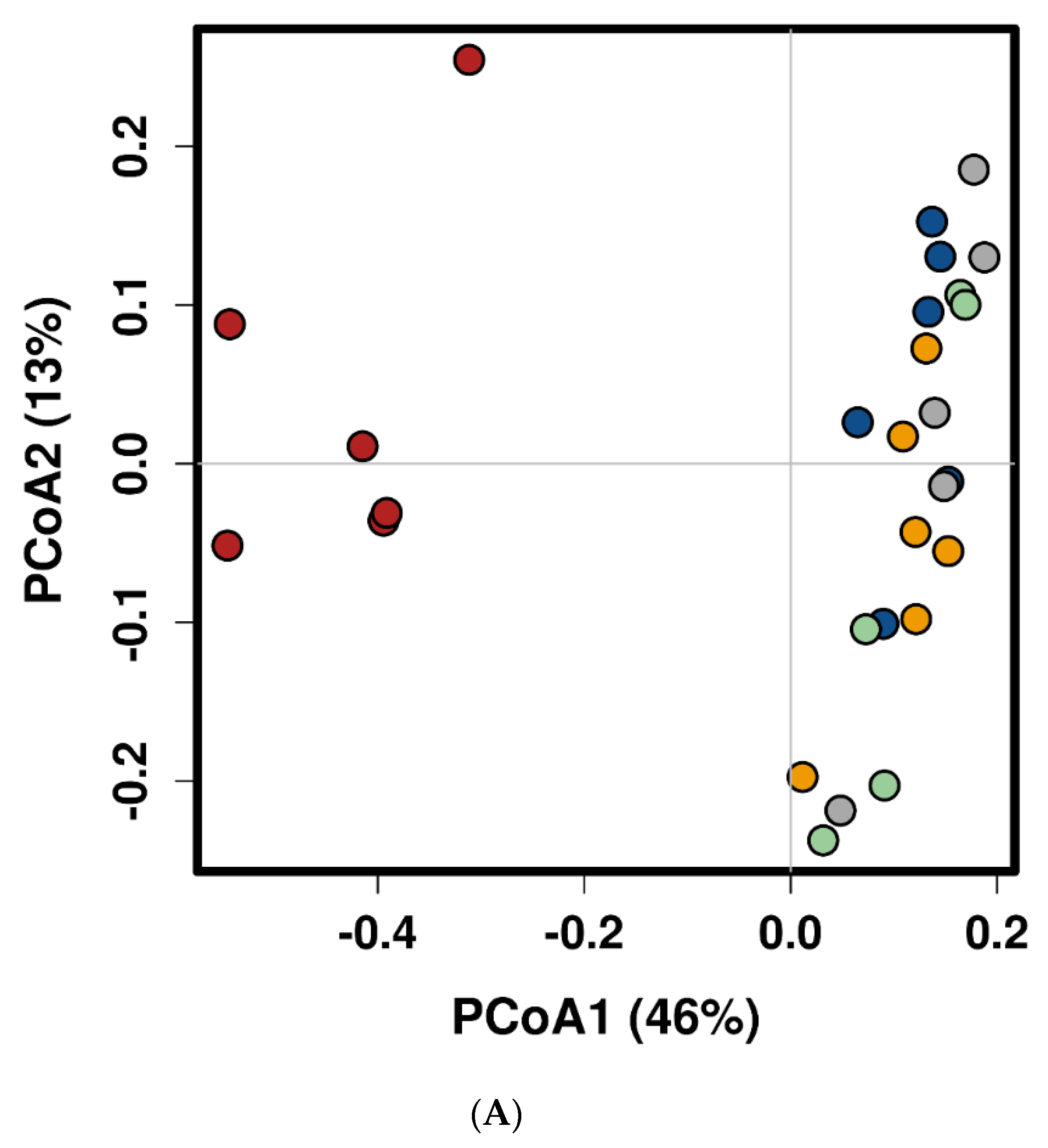
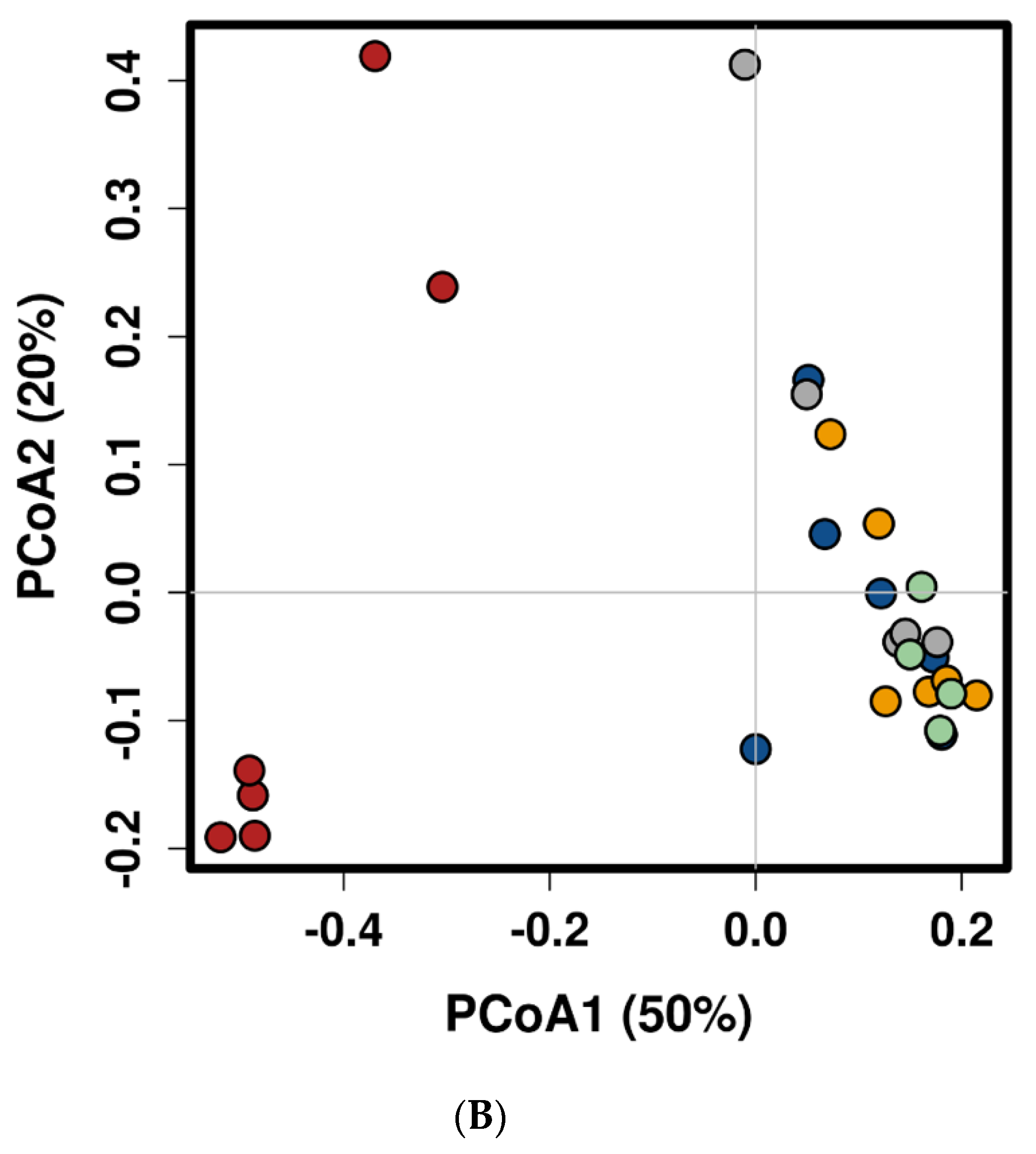
2.1. Feed Raw Materials Change the Intestinal Bacterial Community Composition
2.2. Feed Raw Materials Modulate Predicted Intestinal Microbiome Function
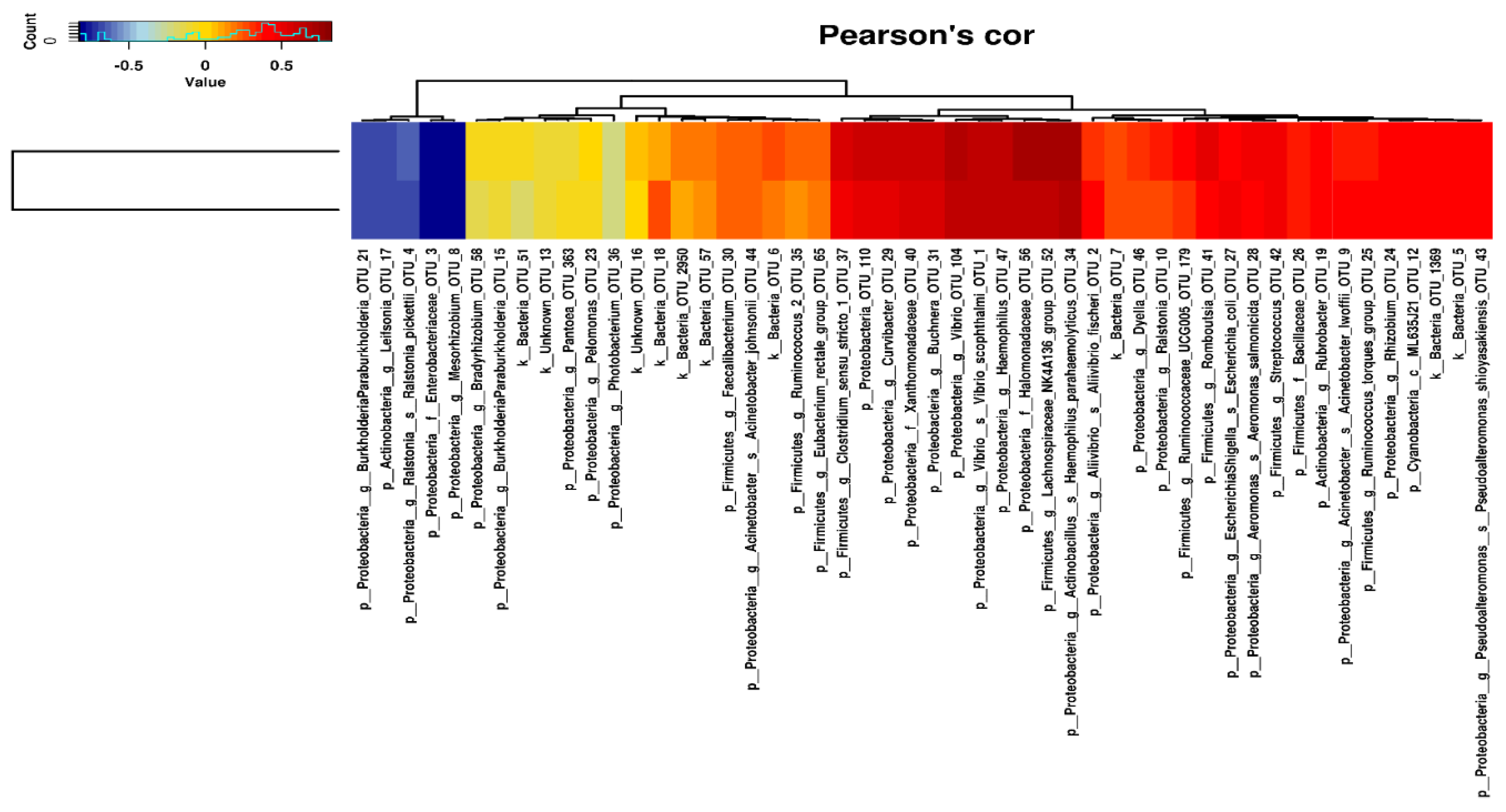
2.3. Intestinal Microbiota Correlates with Apparent Nutrient Digestibility
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experiment Design
4.3. Sampling
4.4. DNA Extraction and High-Throughput Sequencing of Bacterial 16S rRNA Genes
4.5. Sequencing Data Processing
4.6. Operational Taxonomic Units (OTUs) Cluster and Species Annotation
4.7. Predicting Function and Metagenome Contribution
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture (SOFIA); FAO: Rome, Italy, 2018; p. 227. [Google Scholar]
- Bureau, D.; Harris, A.; Cho, C. Apparent digestibility of rendered animal protein ingredients for rainbow trout (Oncorhynchus mykiss). Aquaculture 1999, 180, 345–358. [Google Scholar] [CrossRef]
- Glencross, B.D.; Carter, C.G.; Duijster, N.; Evans, D.R.; Dods, K.; McCafferty, P.; Hawkins, W.E.; Maas, R.; Sipsas, S. A comparison of the digestibility of a range of lupin and soybean protein products when fed to either Atlantic salmon (Salmo salar) or rainbow trout (Oncorhynchus mykiss). Aquaculture 2004, 237, 333–346. [Google Scholar] [CrossRef]
- Gatlin, D.M., III; Barrows, F.T.; Brown, P.; Dabrowski, K.; Gaylord, T.G.; Hardy, R.W.; Herman, E.; Hu, G.; Krogdahl, Å.; Nelson, R. Expanding the utilization of sustainable plant products in aquafeeds: A review. Aquaculture 2007, 38, 551–579. [Google Scholar] [CrossRef]
- Hardy, R.W. Utilization of plant proteins in fish diets: Effects of global demand and supplies of fishmeal. Aquac. Res. 2010, 41, 770–776. [Google Scholar] [CrossRef]
- Booth, M.A.; Allan, G.L.; Frances, J.; Parkinson, S. Replacement of fish meal in diets for Australian silver perch, Bidyanus bidyanus: IV. Effects of dehulling and protein concentration on digestibility of grain legumes. Aquaculture 2001, 196, 67–85. [Google Scholar] [CrossRef]
- Francis, G.; Makkar, H.P.; Becker, K.J. Antinutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture 2001, 199, 197–227. [Google Scholar] [CrossRef]
- Hartviksen, M.; Bakke, A.M.; Vecino, J.G.; Ringø, E.; Krogdahl, Å. Evaluation of the effect of commercially available plant and animal protein sources in diets for Atlantic salmon (Salmo salar L.): Digestive and metabolic investigations. Fish Physiol. Boichem. 2014, 40, 1621–1637. [Google Scholar] [CrossRef]
- Rimoldi, S.; Terova, G.; Ascione, C.; Giannico, R.; Brambilla, F. Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PLoS ONE 2018, 13, e0193652. [Google Scholar] [CrossRef]
- Cheli, F.; Battaglia, D.; Gallo, R.; Dell’Orto, V. EU legislation on cereal safety: An update with a focus on mycotoxins. Food Control 2014, 37, 315–325. [Google Scholar] [CrossRef]
- Bates, J.M.; Mittge, E.; Kuhlman, J.; Baden, K.N.; Cheesman, S.E.; Guillemin, K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006, 297, 374–386. [Google Scholar] [CrossRef]
- Fraune, S.; Bosch, T.C. Why bacteria matter in animal development and evolution. Bioessays 2010, 32, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Stephens, W.Z.; Burns, A.R.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. The composition of the zebrafish intestinal microbial community varies across development. Isme J. 2016, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Givens, C.E.; Ransom, B.; Bano, N.; Hollibaugh, J.T. Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 2015, 518, 209–223. [Google Scholar] [CrossRef]
- Ramírez, C.; Romero, J. The microbiome of Seriola lalandi of wild and aquaculture origin reveals differences in composition and potential function. Front. Microbiol. 2017, 8, 1844. [Google Scholar] [CrossRef] [PubMed]
- Wilkes Walburn, J.; Wemheuer, B.; Thomas, T.; Copeland, E.; O’Connor, W.; Booth, M.; Fielder, S.; Egan, S. Diet and diet-associated bacteria shape early microbiome development in Yellowtail Kingfish (Seriola lalandi). Microb. Biotechnol. 2019, 12, 275–288. [Google Scholar] [CrossRef]
- Schmidt, V.; Amaral-Zettler, L.; Davidson, J.; Summerfelt, S.; Good, C. Influence of fishmeal-free diets on microbial communities in Atlantic salmon (Salmo salar) recirculation aquaculture systems. Appl. Environ. Microbiol. 2016, 82, 4470–4481. [Google Scholar] [CrossRef]
- Gajardo, K.; Jaramillo-Torres, A.; Kortner, T.M.; Merrifield, D.L.; Tinsley, J.; Bakke, A.M.; Krogdahl, Å. Alternative protein sources in the diet modulate microbiota and functionality in the distal intestine of Atlantic salmon (Salmo salar). Appl. Environ. Microbiol. 2017, 83, e02615–e02616. [Google Scholar] [CrossRef]
- Cahill, M.M. Bacterial flora of fishes: A review. Microb. Ecol. 1990, 19, 21–41. [Google Scholar] [CrossRef]
- Ringø, E.; Strøm, E.; Tabachek, J.A. Intestinal microflora of salmonids: A review. Aquac. Res. 1995, 26, 773–789. [Google Scholar] [CrossRef]
- Fowler, A.J.; Ham, J.M.; Jennings, P.R. Discriminating between Cultured and Wild Yellowtail Kingfish (Seriola Lalandi) in South Australia; SARDI Aquatic Sciences Publication No. RD03/0159: Adelaide, Australia, 2003. [Google Scholar]
- Green, T.J.; Smullen, R.; Barnes, A.C. Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet. Microbiol. 2013, 166, 286–292. [Google Scholar] [CrossRef]
- Booman, M.; Forster, I.; Vederas, J.C.; Groman, D.B.; Jones, S.R. Soybean meal-induced enteritis in Atlantic salmon (Salmo salar) and Chinook salmon (Oncorhynchus tshawytscha) but not in pink salmon (O. gorbuscha). Aquaculture 2018, 483, 238–243. [Google Scholar] [CrossRef]
- Parma, L.; Candela, M.; Soverini, M.; Turroni, S.; Consolandi, C.; Brigidi, P.; Mandrioli, L.; Sirri, R.; Fontanillas, R.; Gatta, P.P. Next-generation sequencing characterization of the gut bacterial community of gilthead sea bream (Sparus aurata, L.) fed low fishmeal based diets with increasing soybean meal levels. Anim. Feed Sci. Techonol. 2016, 222, 204–216. [Google Scholar] [CrossRef]
- Desai, A.R.; Links, M.G.; Collins, S.A.; Mansfield, G.S.; Drew, M.D.; Van Kessel, A.G.; Hill, J.E. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 2012, 350, 134–142. [Google Scholar] [CrossRef]
- Huyben, D.; Vidaković, A.; Hallgren, S.W.; Langeland, M. High-throughput sequencing of gut microbiota in rainbow trout (Oncorhynchus mykiss) fed larval and pre-pupae stages of black soldier fly (Hermetia illucens). Aquaculture 2019, 500, 485–491. [Google Scholar] [CrossRef]
- Hutson, K.; Ernst, I.; Whittington, I. Risk assessment for metazoan parasites of yellowtail kingfish Seriola lalandi (Perciformes: Carangidae) in South Australian sea-cage aquaculture. Aquaculture 2007, 271, 85–99. [Google Scholar] [CrossRef]
- Moran, D.; Pether, S.J.; Lee, P.S. Growth, feed conversion and faecal discharge of yellowtail kingfish (Seriola lalandi) fed three commercial diets. N. Z. J. Mar. Freshw. Res. 2009, 43, 917–927. [Google Scholar] [CrossRef]
- Miegel, R.; Pain, S.; Van Wettere, W.; Howarth, G.; Stone, D. Effect of water temperature on gut transit time, digestive enzyme activity and nutrient digestibility in yellowtail kingfish (Seriola lalandi). Aquaculture 2010, 308, 145–151. [Google Scholar] [CrossRef]
- Bowyer, J.N.; Booth, M.A.; Qin, J.G.; D’Antignana, T.; Thomson, M.J.; Stone, D.A. Temperature and dissolved oxygen influence growth and digestive enzyme activities of yellowtail kingfish S eriola lalandi (V alenciennes, 1833). Aquaculture 2014, 45, 2010–2020. [Google Scholar] [CrossRef]
- Stone, D.A.; Bellgrove, E.J.; Forder, R.E.; Howarth, G.S.; Bansemer, M.S. Inducing subacute enteritis in Yellowtail Kingfish Seriola lalandi: The effect of dietary inclusion of soybean meal and grape seed extract on hindgut morphology and inflammation. N. Am. J. Aquac. 2018, 80, 59–68. [Google Scholar] [CrossRef]
- Sicuro, B.; Luzzana, U. The state of Seriola spp. other than yellowtail (S. quinqueradiata) farming in the world. Rev. Fish. Sci. Aquac. 2016, 24, 314–325. [Google Scholar] [CrossRef]
- Poortenaar, C.; Hooker, S.; Sharp, N. Assessment of yellowtail kingfish (Seriola lalandi lalandi) reproductive physiology, as a basis for aquaculture development. Aquaculture 2001, 201, 271–286. [Google Scholar] [CrossRef]
- Nakada, M. Yellowtail culture development and solutions for the future. Rev. Fish. Sci. 2002, 10, 559–575. [Google Scholar] [CrossRef]
- Booth, M.A.; Moses, M.; Allan, G.L. Utilisation of carbohydrate by yellowtail kingfish Seriola lalandi. Aquaculture 2013, 376–379, 151–161. [Google Scholar] [CrossRef]
- Booth, M.A.; Allan, G.L.; Pirozzi, I. Estimation of digestible protein and energy requirements of yellowtail kingfish Seriola lalandi using a factorial approach. Aquaculture 2010, 307, 247–259. [Google Scholar] [CrossRef]
- Stone, D.A.J.; Bellgrove, E. A literature review: The current status of knowledge of the nutritional requirements of yellowtail kingfish. In Sustainable Feeds and Feed Management for Yellowtail Kingfish (Seriola lalandi), ASCRC Project No. 2009/728; Vol. SARDI Publication No. F2013/000200-1. SARDI Research Report Series No. 751; Stone, D.A.J., Bowyer, J., Eds.; South Australian Research and Development Institute (Aquatic Sciences): Adelaide, Australia, 2013; pp. 92–121. [Google Scholar]
- Booth, M.; Pirozzi, I. Making sense of nonsense: Using regression analysis to deal with highly variable data collected from a yellowtail kingfish (Seriola lalandi) digestibility experiment. Aquaculture 2018, 485, 39–48. [Google Scholar] [CrossRef]
- Bansemer, M.S.; Stone, D.A.; D’Antignana, T.; Skordas, P.; Kuerschner, L.; Currie, K.L. Optimizing Feeding Strategies for Yellowtail Jack at Winter Water Temperatures. N. Am. J. Aquac. 2018, 80, 128–140. [Google Scholar] [CrossRef]
- Dam, C.T.; Elizur, A.; Ventura, T.; Salini, M.; Smullen, R.; Pirozzi, I.; Booth, M. Apparent digestibility of raw materials by yellowtail kingfish (Seriola lalandi). Aquaculture 2019, 734233. [Google Scholar] [CrossRef]
- Legrand, T.P.; Catalano, S.R.; Wos-Oxley, M.L.; Stephens, F.; Landos, M.; Bansemer, M.S.; Stone, D.A.; Qin, J.G.; Oxley, A. The inner workings of the outer surface: Skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 2018, 8, 2664. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef]
- Allan, G.L.; Parkinson, S.; Booth, M.A.; Stone, D.A.; Rowland, S.J.; Frances, J.; Warner-Smith, R. Replacement of fish meal in diets for Australian silver perch, Bidyanus bidyanus: I. Digestibility of alternative ingredients. Aquaculture 2000, 186, 293–310. [Google Scholar] [CrossRef]
- Booth, M.; Allan, G.; Smullen, R. Digestibility of common feed ingredients by juvenile mulloway Argyrosomus japonicus. Aquaculture 2013, 414, 140–148. [Google Scholar] [CrossRef]
- Blyth, D.; Tabrett, S.; Bourne, N.; Glencross, B. Comparison of faecal collection methods and diet acclimation times for the measurement of digestibility coefficients in barramundi (Lates calcarifer). Aquac. Nutr. 2015, 21, 248–255. [Google Scholar] [CrossRef]
- Che, J.; Su, B.; Tang, B.; Bu, X.; Li, J.; Lin, Y.; Yang, Y.; Ge, X. Apparent digestibility coefficients of animal and plant feed ingredients for juvenile Pseudobagrus ussuriensis. Aquac. Nutr. 2017, 23, 1128–1135. [Google Scholar] [CrossRef]
- Aguilera, E.; Yany, G.; Romero, J. Cultivable intestinal microbiota of yellowtail juveniles (Seriola lalandi) in an aquaculture system. Lat. Am. J. Aquat. Res. 2013, 41, 395–403. [Google Scholar]
- Hovda, M.B.; Lunestad, B.T.; Fontanillas, R.; Rosnes, J.T. Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquaculture 2007, 272, 581–588. [Google Scholar] [CrossRef]
- Villasante, A.; Ramírez, C.; Catalán, N.; Opazo, R.; Dantagnan, P.; Romero, J. Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar). Animals 2019, 9, 89. [Google Scholar] [CrossRef]
- Estruch, G.; Collado, M.; Peñaranda, D.; Vidal, A.T.; Cerdá, M.J.; Martínez, G.P.; Martinez-Llorens, S. Impact of fishmeal replacement in diets for gilthead sea bream (Sparus aurata) on the gastrointestinal microbiota determined by pyrosequencing the 16S rRNA gene. PLoS ONE 2015, 10, e0136389. [Google Scholar] [CrossRef]
- Ringø, E.; Sperstad, S.; Myklebust, R.; Refstie, S.; Krogdahl, Å. Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.): The effect of fish meal, standard soybean meal and a bioprocessed soybean meal. Aquaculture 2006, 261, 829–841. [Google Scholar]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Calduch-Giner, J.A.; Fouz, B.; Estensoro, I.; Simó-Mirabet, P.; Puyalto, M.; Karalazos, V.; Palenzuela, O.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. Under control: How a dietary additive can restore the gut microbiome and proteomic profile, and improve disease resilience in a marine teleostean fish fed vegetable diets. Microbiome 2017, 5, 164. [Google Scholar] [CrossRef] [PubMed]
- Gatesoupe, F.-J.; Huelvan, C.; Le Bayon, N.; Le Delliou, H.; Madec, L.; Mouchel, O.; Quazuguel, P.; Mazurais, D.; Zambonino-Infante, J.L. The highly variable microbiota associated to intestinal mucosa correlates with growth and hypoxia resistance of sea bass, Dicentrarchus labrax, submitted to different nutritional histories. BMC Microbiol. 2016, 16, 266. [Google Scholar] [CrossRef]
- Silva, F.C.d.P.; Nicoli, J.R.; Zambonino-Infante, J.L.; Kaushik, S.; Gatesoupe, F.J. Influence of the diet on the microbial diversity of faecal and gastrointestinal contents in gilthead sea bream (Sparus aurata) and intestinal contents in goldfish (Carassius auratus). FEMS Microbiol. 2011, 78, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Star, B.; Haverkamp, T.H.; Jentoft, S.; Jakobsen, K.S. Next generation sequencing shows high variation of the intestinal microbial species composition in Atlantic cod caught at a single location. BMC Microbiol. 2013, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.L.; Steven, B.; Penn, K.; Methé, B.A.; Detrich, W.H. Characterization of the intestinal microbiota of two Antarctic notothenioid fish species. Extremophiles 2009, 13, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, H.; Ling, Z.; Chang, J.; Ye, J. Gut microbiota of fast and slow growing grouper Epinephelus coioides. Afr. J. Microbiol. Res. 2009, 3, 637–640. [Google Scholar]
- Smriga, S.; Sandin, S.A.; Azam, F. Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol. Ecol. 2010, 73, 31–42. [Google Scholar] [CrossRef]
- Vandenberghe, J.; Thompson, F.L.; Gomez-Gil, B.; Swings, J. Phenotypic diversity amongst Vibrio isolates from marine aquaculture systems. Aquaculture 2003, 219, 9–20. [Google Scholar] [CrossRef]
- Gatesoupe, F.-J.; Infante, J.-L.Z.; Cahu, C.; Quazuguel, P. Early weaning of seabass larvae, Dicentrarchus labrax: The effect on microbiota, with particular attention to iron supply and exoenzymes. Aquaculture 1997, 158, 117–127. [Google Scholar] [CrossRef]
- Henderson, R.J.; Millar, R.-M. Characterization of lipolytic activity associated with a Vibrio species of bacterium isolated from fish intestines. J. Mar. Biotechnol. 1998, 6, 168–173. [Google Scholar]
- Itoi, S.; Okamura, T.; Koyama, Y.; Sugita, H. Chitinolytic bacteria in the intestinal tract of Japanese coastal fishes. Can. J. Microbiol. 2006, 52, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Ghosh, K.; Ringø, E. Enzyme-producing bacteria isolated from fish gut: A review. Aquac. Nutr. 2012, 18, 465–492. [Google Scholar] [CrossRef]
- Austin, B.; Stuckey, L.; Robertson, P.; Effendi, I.; Griffith, D. A probiotic strain of Vibrio alginolyticus effective in reducing diseases caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. Fish Dis. 1995, 18, 93–96. [Google Scholar] [CrossRef]
- Sugita, H.; Matsuo, N.; Hirose, Y.; Iwato, M.; Deguchi, Y. Vibrio sp. strain NM 10, isolated from the intestine of a Japanese coastal fish, has an inhibitory effect against Pasteurella piscicida. Appl. Environ. Microbiol. 1997, 63, 4986–4989. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gao, T.; Zheng, Y.; Wang, W.; Cheng, Y.; Wang, G. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2010, 303, 1–7. [Google Scholar] [CrossRef]
- Kim, D.H.; Brunt, J.; Austin, B.J. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol. 2007, 102, 1654–1664. [Google Scholar] [CrossRef]
- Carda-Diéguez, M.; Mira, A.; Fouz, B. Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. Fems Microbiol. Ecol. 2014, 87, 451–459. [Google Scholar] [CrossRef]
- Wamala, S.P.; Mugimba, K.K.; Mutoloki, S.; Evensen, Ø.; Mdegela, R.; Byarugaba, D.K.; Sørum, H. Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fish. Aquatttic Sci. 2018, 21, 6. [Google Scholar] [CrossRef]
- Apper, E.; Weissman, D.; Respondek, F.; Guyonvarch, A.; Baron, F.; Boisot, P.; Rodiles, A.; Merrifield, D. Hydrolysed wheat gluten as part of a diet based on animal and plant proteins supports good growth performance of Asian seabass (Lates calcarifer), without impairing intestinal morphology or microbiota. Aquaculture 2016, 453, 40–48. [Google Scholar] [CrossRef]
- Cerezuela, R.; Fumanal, M.; Tapia-Paniagua, S.T.; Meseguer, J.; Moriñigo, M.Á.; Esteban, M.Á. Changes in intestinal morphology and microbiota caused by dietary administration of inulin and Bacillus subtilis in gilthead sea bream (Sparus aurata L.) specimens. Fish Shellfish Immunol. 2013, 34, 1063–1070. [Google Scholar] [CrossRef]
- Nyman, A.; Huyben, D.; Lundh, T.; Dicksved, J. Effects of microbe-and mussel-based diets on the gut microbiota in Arctic charr (Salvelinus alpinus). Aquaculture 2017, 5, 34–40. [Google Scholar] [CrossRef]
- Ringø, E.; Løvmo, L.; Kristiansen, M.; Bakken, Y.; Salinas, I.; Myklebust, R.; Olsen, R.E.; Mayhew, T.M. Lactic acid bacteria vs. pathogens in the gastrointestinal tract of fish: A review. Aquaculture 2010, 41, 451–467. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef] [PubMed]
- Bai, N.; Gu, M.; Liu, M.; Jia, Q.; Pan, S.; Zhang, Z. Corn gluten meal induces enteritis and decreases intestinal immunity and antioxidant capacity in turbot (Scophthalmus maximus) at high supplementation levels. PLoS ONE 2019, 14, e0213867. [Google Scholar] [CrossRef] [PubMed]
- Le Sciellour, M.; Labussière, E.; Zemb, O.; Renaudeau, D. Effect of dietary fiber content on nutrient digestibility and fecal microbiota composition in growing-finishing pigs. PLoS ONE 2018, 13, e0206159. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Ma, X.; Gao, S.; He, L.; Wu, W. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015, 5, 9938. [Google Scholar] [CrossRef] [PubMed]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Ntrition 2011, 94, 58–65. [Google Scholar] [CrossRef]
- Ramirez, R.F.; Dixon, B.A. Enzyme production by obligate intestinal anaerobic bacteria isolated from oscars (Astronotus ocellatus), angelfish (Pterophyllum scalare) and southern flounder (Paralichthys lethostigma). Aquaculture 2003, 227, 417–426. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dam, C.T.M.; Booth, M.; Pirozzi, I.; Salini, M.; Smullen, R.; Ventura, T.; Elizur, A. Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi. Fishes 2020, 5, 14. https://doi.org/10.3390/fishes5020014
Dam CTM, Booth M, Pirozzi I, Salini M, Smullen R, Ventura T, Elizur A. Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi. Fishes. 2020; 5(2):14. https://doi.org/10.3390/fishes5020014
Chicago/Turabian StyleDam, Chinh Thi My, Mark Booth, Igor Pirozzi, Michael Salini, Richard Smullen, Tomer Ventura, and Abigail Elizur. 2020. "Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi" Fishes 5, no. 2: 14. https://doi.org/10.3390/fishes5020014
APA StyleDam, C. T. M., Booth, M., Pirozzi, I., Salini, M., Smullen, R., Ventura, T., & Elizur, A. (2020). Alternative Feed Raw Materials Modulate Intestinal Microbiota and Its Relationship with Digestibility in Yellowtail Kingfish Seriola lalandi. Fishes, 5(2), 14. https://doi.org/10.3390/fishes5020014






