The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles
Abstract
1. Introduction
2. Material and Methods
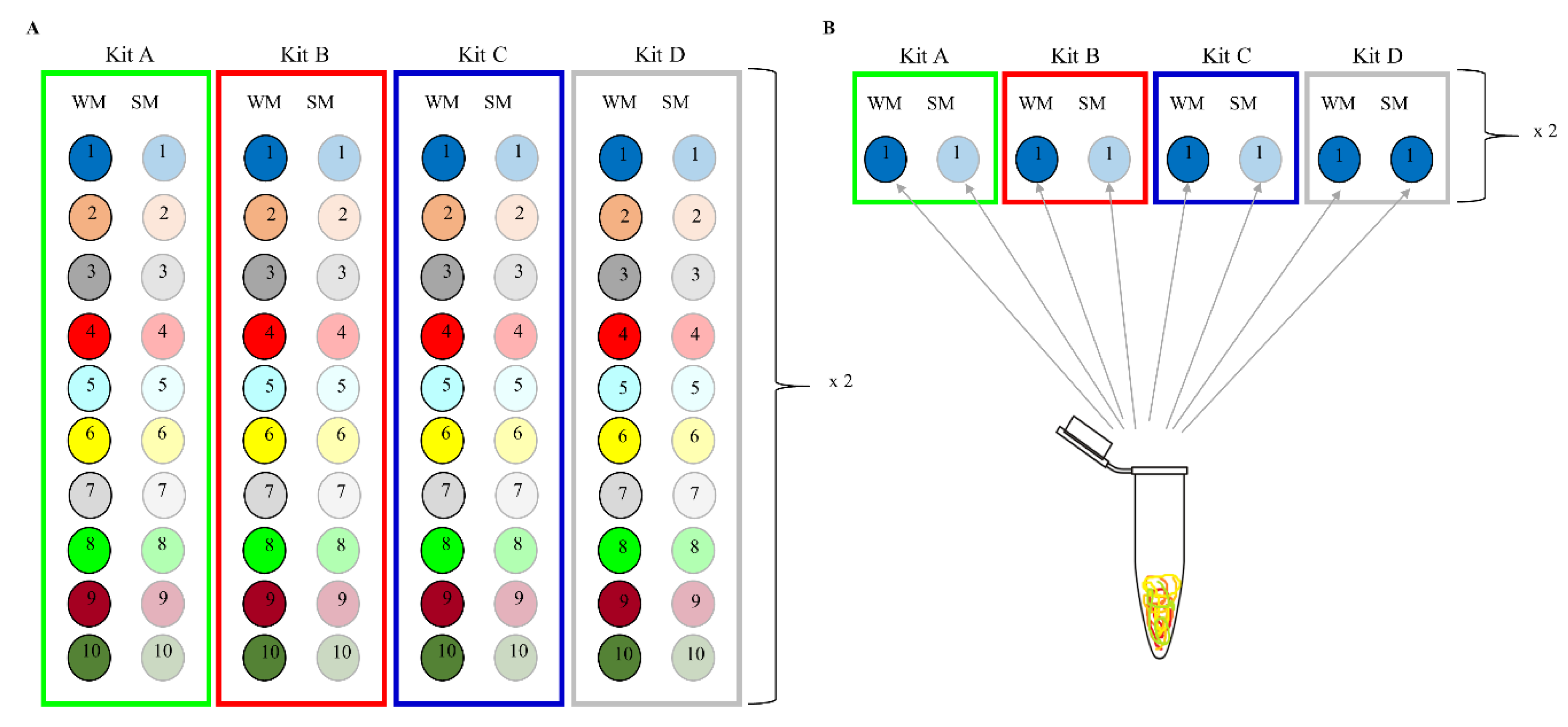
2.1. Subjects and Sample Collection
2.2. Methods of DNA Extraction
2.3. DNA Quantification
2.4. Extraction and Sequencing Controls
2.5. 16S Ribosomal Ribonucleic Acid (rRNA) Amplicon Library Preparation
2.6. 16S Ribosomal Ribonucleic Acid (rRNA) Gene Sequencing
2.7. Bioinformatics Workflow
2.8. Data Analysis
3. Results
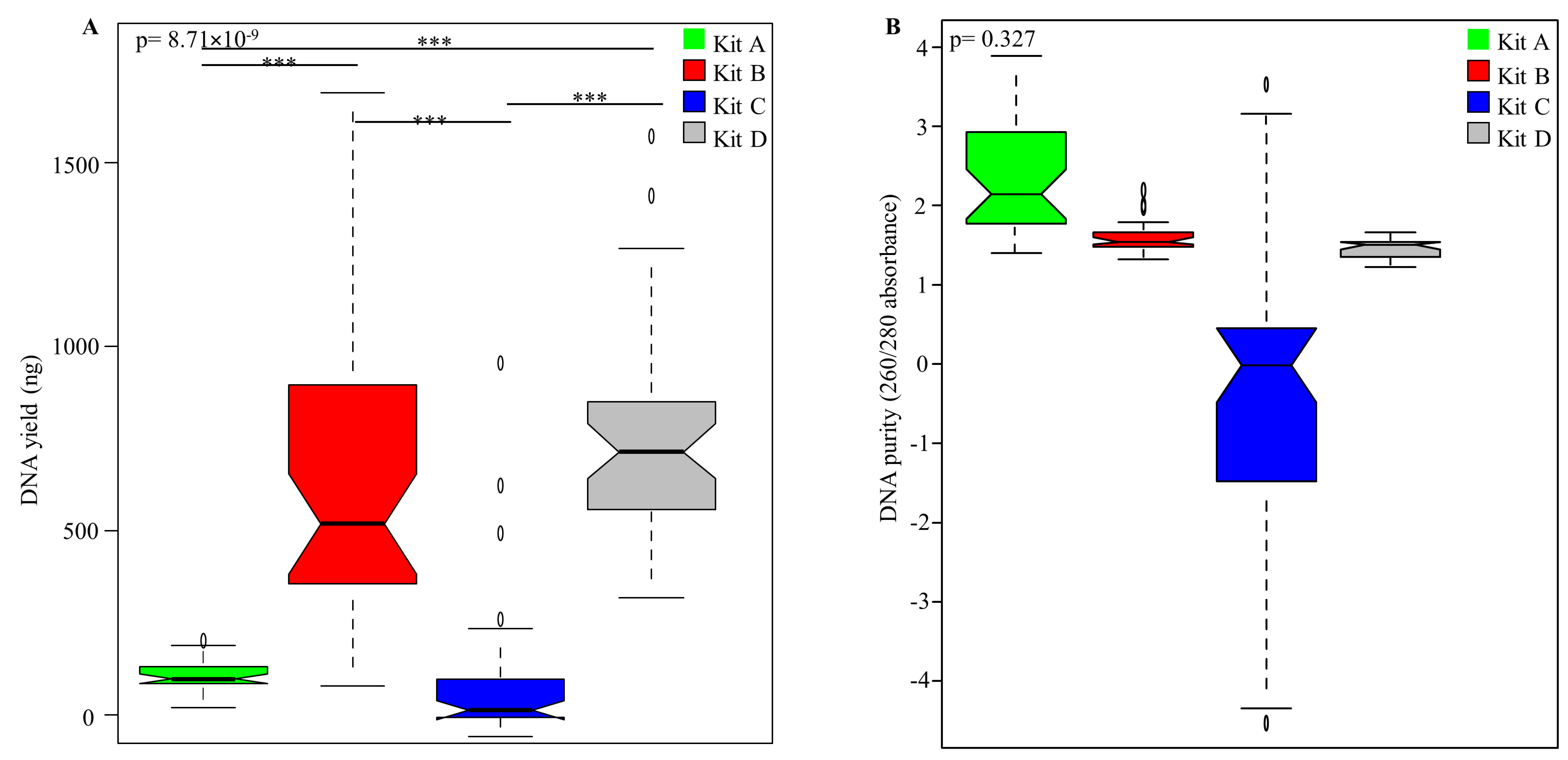
3.1. Influence of DNA Extraction Kits and Lipid Removal on Yield and Quality of DNA Extracted from Un-Spiked Human Milk Samples
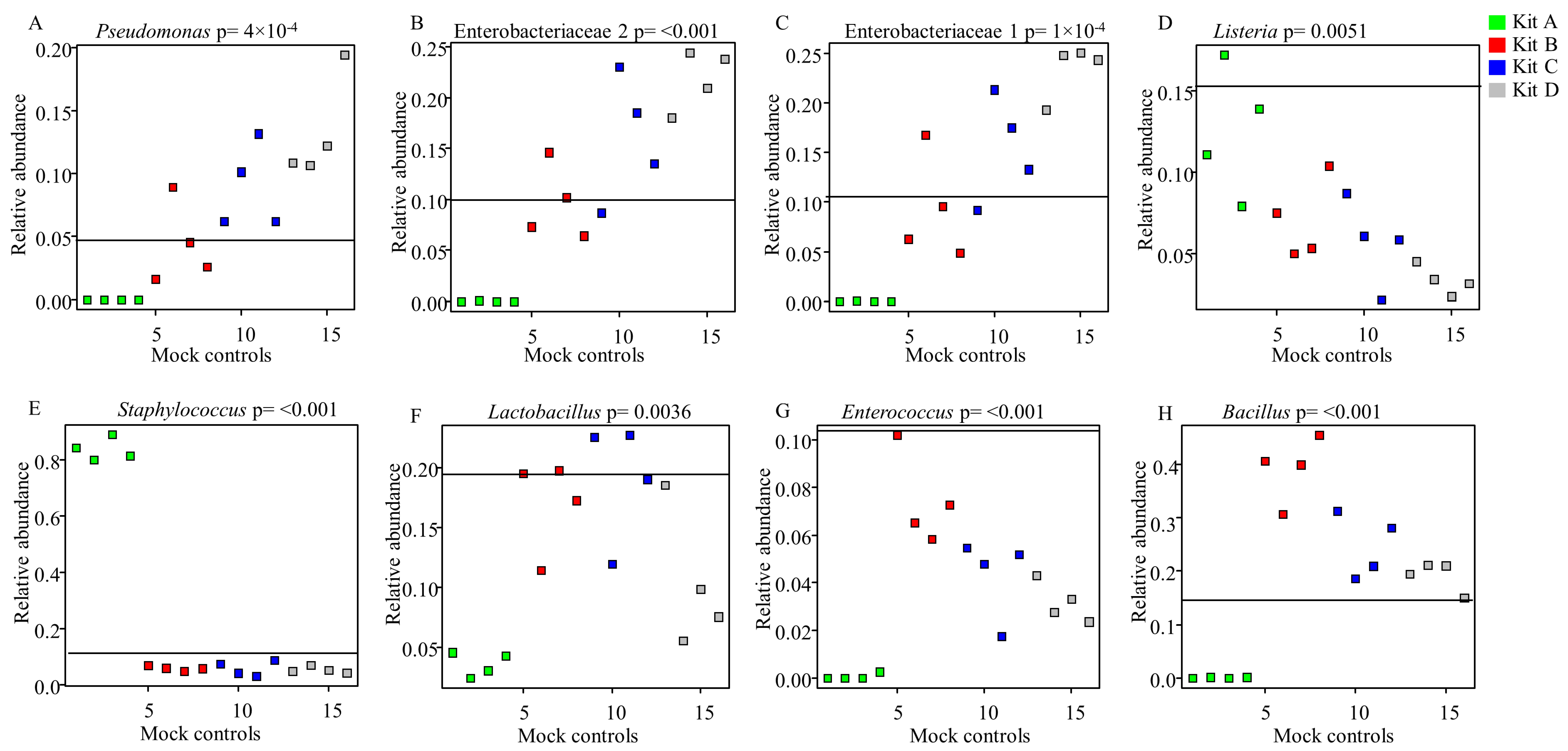
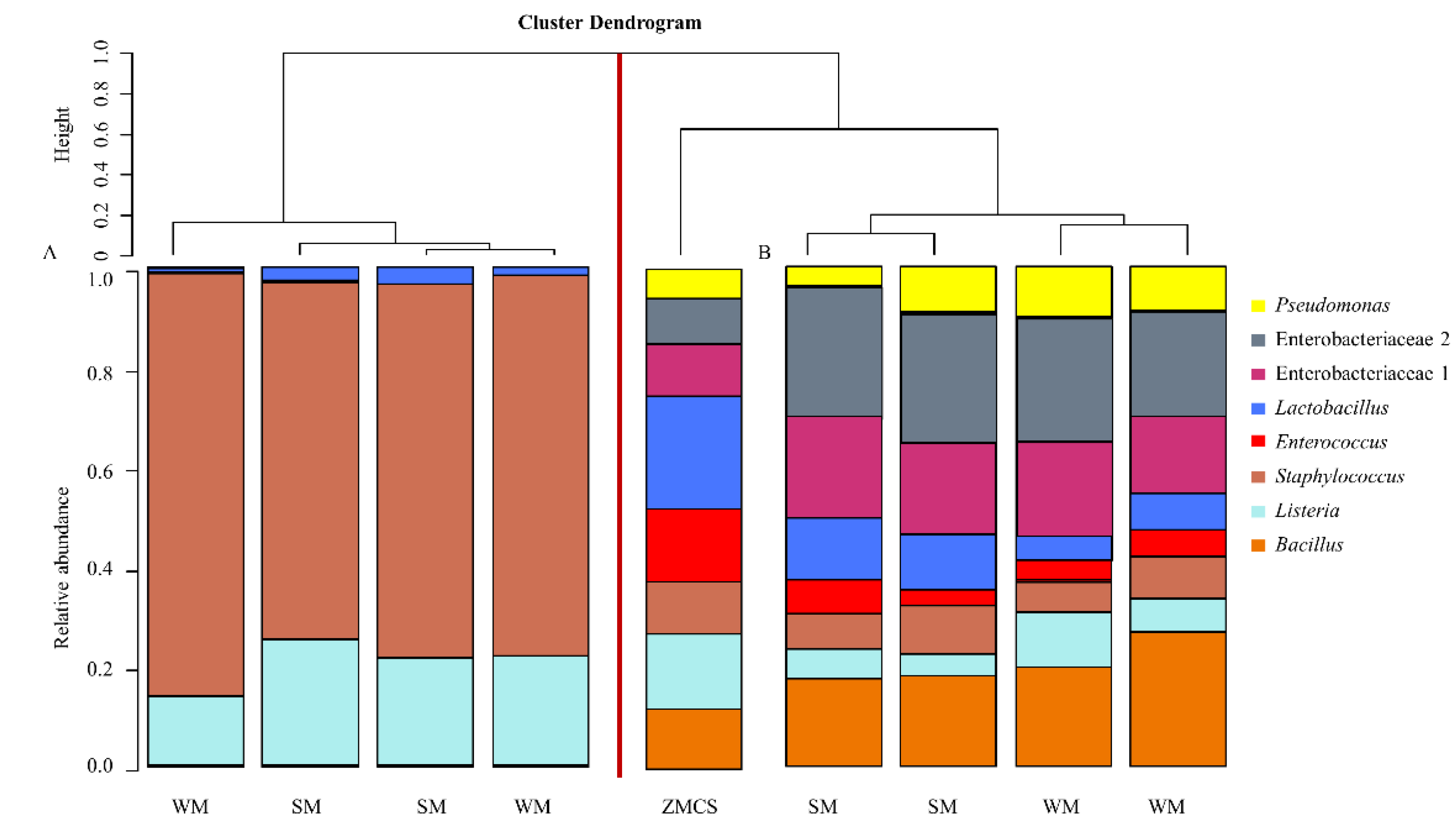
3.2. Influence of DNA Extraction Kits and Lipid Removal on Bacterial Profiles Obtained from Mock Microbial Community Standard Spiked into Human Milk
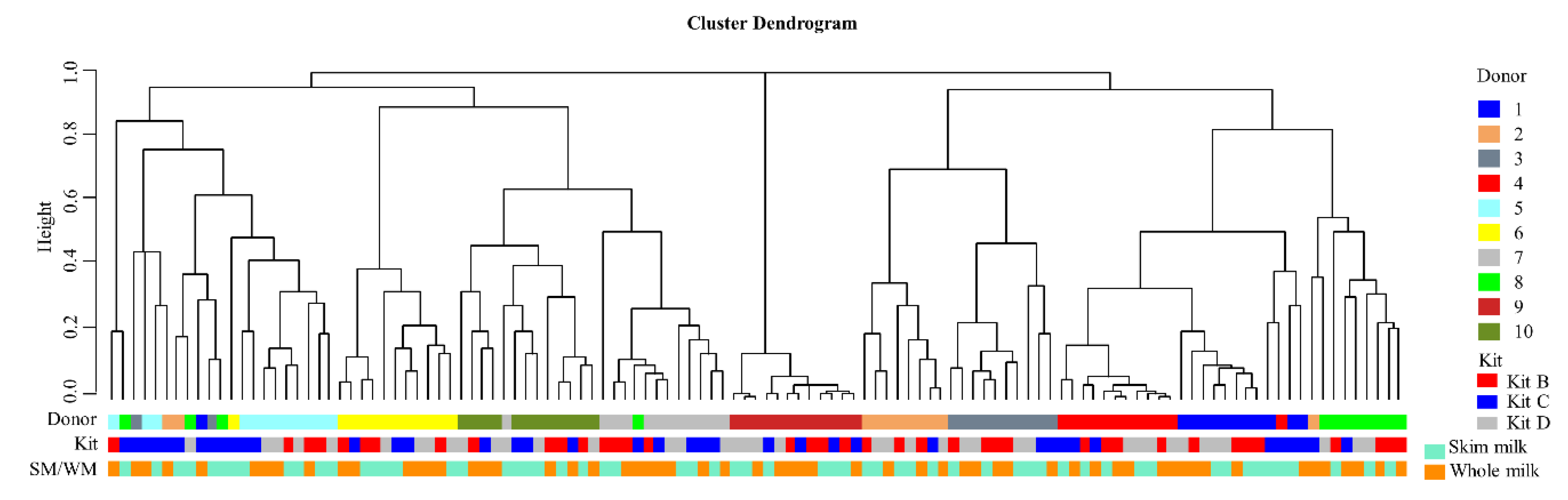
3.3. Influence of DNA Extraction Kit and Milk Type on Bacterial Diversity in Un-Spiked HM Samples
3.4. Repeatability of Extractions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferretti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Civardi, E.; Garofoli, F.; Tzialla, C.; Paolillo, P.; Bollani, L.; Stronati, M. Microorganisms in human milk: Lights and shadows. J. Matern. Fetal Neonatal Med. 2013, 26 (Suppl. S2), 30–34. [Google Scholar] [CrossRef] [PubMed]
- Jimenéz, E.; Fernández, L.; Maldonado, A.; Martin, R.; Olivares, M.; Xaus, J.; Rodriguez, J.M. Oral administration of lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Environ. Microbiol. 2008, 74, 4650–4655. [Google Scholar] [CrossRef]
- Fernández, L.; Cárdenas, N.; Arroyo, R.; Manzano, S.; Jiménez, E.; Martín, V.; Rodríguez, J.M. Prevention of infectious mastitis by oral administration of lactobacillus salivarius ps2 during late pregnancy. Clin. Infect. Dis. 2016, 62, 568–573. [Google Scholar] [CrossRef]
- Yuan, S.; Cohen, D.B.; Ravel, J.; Abdo, Z.; Forney, L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 2012, 7, e33865. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Ross, R.P.; Fitzgerald, G.; Stanton, C.; Cotter, P.D. Composition of the early intestinal microbiota:Knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes 2012, 3, 203–220. [Google Scholar] [CrossRef]
- Ojo-Okunola, A.; Nicol, M.; du Toit, E. Human breast milk bacteriome in health and disease. Nutrients 2018, 10, 1643. [Google Scholar] [CrossRef]
- Claesson, M.J.; Wang, Q.; O’sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16s rrna gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Salipante, S.J.; Sengupta, D.J.; Rosenthal, C.; Costa, G.; Spangler, J.; Sims, E.H.; Jacobs, M.A.; Miller, S.I.; Hoogestraat, D.R.; Cookson, B.T.; et al. Rapid 16s rrna next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS ONE 2013, 8, e65226. [Google Scholar] [CrossRef]
- Ariefdjohan, M.W.; Savaiano, D.A.; Nakatsu, C.H. Comparison of DNA extraction kits for pcr-dgge analysis of human intestinal microbial communities from fecal specimens. Nutr. J. 2010, 9, 23. [Google Scholar] [CrossRef]
- Claassen, S.; du Toit, E.; Kaba, M.; Moodley, C.; Zar, H.J.; Nicol, M.P. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J. Microbiol. Methods 2013, 94, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Rangasamy, M.; Tan, S.Y.; Wang, H.; Siegfried, B.D. Evaluation of five methods for total DNA extraction from western corn rootworm beetles. PLoS ONE 2010, 5, e11963. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, V.; Gaia, N.; Girard, M.; Francois, P.; Schrenzel, J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. PLoS ONE 2013, 8, e67699. [Google Scholar] [CrossRef] [PubMed]
- Ó Cuív, P.; Aguirre de Cárcer, D.; Jones, M.; Klaassens, E.S.; Worthley, D.L.; Whitehall, V.L.J.; Kang, S.; McSweeney, C.S.; Leggett, B.A.; Morrison, M. The effects from DNA extraction methods on the evaluation of microbial diversity associated with human colonic tissue. Microb. Ecol. 2011, 61, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Gill, C.; van de Wijgert, J.H.; Blow, F.; Darby, A.C. Evaluation of lysis methods for the extraction of bacterial DNA for analysis of the vaginal microbiota. PLoS ONE 2016, 11, e0163148. [Google Scholar] [CrossRef]
- Usman, T.; Yu, Y.; Liu, C.; Fan, Z.; Wang, Y. Comparison of methods for high quantity and quality genomic DNA extraction from raw cow milk. Genet. Mol. Res. 2014, 13, 3319–3328. [Google Scholar] [CrossRef]
- Douglas, C.A.; Ivey, K.L.; Papanicolas, L.E.; Best, K.P.; Muhlhausler, B.S.; Rogers, G.B. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci. Rep. 2020, 10, 123. [Google Scholar] [CrossRef]
- Wagner Mackenzie, B.; Waite, D.W.; Taylor, M.W. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front. Microbiol. 2015, 6, 130. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schutte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Macherey-Nagel. Genomic DNA from Lipid-Rich Tissue: User Manual [Internet]. Available online: https://www.mn-net.com/Portals/8/attachments/Redakteure_Bio/Protocols/Genomic%20DNA/UM_gDNALipidTissue.pdf (accessed on 10 May 2019).
- Lucey, J.A.; Tamehana, M.; Singh, H.; Munro, P.A. Effect of interactions between denatured whey proteins and casein micelles on the formation and rheological properties of acid skim milk gels. J. Dairy Res. 1998, 65, 555–567. [Google Scholar] [CrossRef]
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef]
- Wu, L.; Wen, C.; Qin, Y.; Yin, H.; Tu, Q.; Van Nostrand, J.D.; Yuan, T.; Yuan, M.; Deng, Y.; Zhou, J. Phasing amplicon sequencing on illumina miseq for robust environmental microbial community analysis. BMC Microbiol. 2015, 15, 125. [Google Scholar] [CrossRef]
- Claassen-Weitz, S.; Gardner-Lubbe, S.; Nicol, P.; Botha, G.; Mounaud, S.; Shankar, J.; Nierman, W.C.; Mulder, N.; Budree, S.; Zar, H.J. Hiv-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a south african birth cohort. Sci. Rep. 2018, 8, 5078. [Google Scholar] [CrossRef]
- Ojo-Okunola, A.; Claassen-Weitz, S.; Mwaikono, K.S.; Gardner-Lubbe, S.; Stein, D.J.; Zar, H.J.; Nicol, M.P.; du Toit, E. Influence of socio-economic and psychosocial profiles on the human breast milk bacteriome of south african women. Nutrients 2019, 11, 1390. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16s rrna diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Illumina, P. Miseq® System User Guide; Illumina: San Diego, CA, USA, 2014; pp. 1–94. [Google Scholar]
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data; 2010; Available online: Http://www.Bioinformatics.Babraham.Ac.Uk/projects/fastqc (accessed on 30 October 2018).
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R foundation for statistical computing: Vienna, Austria, 2017. [Google Scholar]
- Hartigan, J.A. Clustering Algorithms; Pages113129; John Willey and Sons Inc.: New York, NY, USA, 1975. [Google Scholar]
- Murtagh, F. Multidimensional clustering algorithms. In Compstat Lectures; Physika Verlag: Vienna, Austria, 1985. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.4-4, Available online: https://www.researchgate.net/publication/323265820_vegan_Community_Ecology_Package_R_package_version_24-4_http_s (accessed on 31 January 2017).
- Hill, M.O. Diversity and evenness: A unifying notation and its consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef]
- Chambers, J.M.; Hastie, T.J. Statistical Models in s; Wadsworth & Brooks/Cole Advanced Books & Software: Pacific Grove, CA, USA, 1992; Volume 251. [Google Scholar]
- Greenacre, M.J. Biplots in Practice; Fundacion BBVA: Bilbao, Spain; Fundacion, Dominica, 2010. [Google Scholar]
- Gower, J.C.; Lubbe, S.G.; Le Roux, N.J. Understanding Biplots; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing rna-seq, 16s rrna gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Devroye, L. Sample-based non-uniform random variate generation. In Proceedings of the 18th Conference on Winter Simulation, Washington, DC, USA, 8–10 December 1986; ACM: New York, NY, USA, 1986; pp. 260–265. [Google Scholar]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models; CRC Press: Boca Raton, FL, USA, 1989; Volume 37. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yandell, B.S. Practical Data Analysis for Designed Experiments; CRC Press: Boca Raton, FL, USA, 1997; Volume 39. [Google Scholar]
- Chambers, J.M.; Cleveland, W.S.; Kleiner, B.; Tukey, P.A. Graphical Methods for Data Analysis; Wadsworth & Brooks/Cole Publishing Company: Pacific Grove, CA, USA, 1983. [Google Scholar]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 2013, 110, 1253–1262. [Google Scholar] [CrossRef]
- Mathay, C.; Hamot, G.; Henry, E.; Georges, L.; Bellora, C.; Lebrun, L.; de Witt, B.; Ammerlaan, W.; Buschart, A.; Wilmes, P.; et al. Method optimization for fecal sample collection and fecal DNA extraction. Biopreserv. Biobank. 2015, 13, 79–93. [Google Scholar] [CrossRef]
- Abusleme, L.; Hong, B.Y.; Dupuy, A.K.; Strausbaugh, L.D.; Diaz, P.I. Influence of DNA extraction on oral microbial profiles obtained via 16s rrna gene sequencing. J. Oral Microbiol. 2014, 6. [Google Scholar] [CrossRef]
- Terrat, S.; Christen, R.; Dequiedt, S.; Lelievre, M.; Nowak, V.; Regnier, T.; Bachar, D.; Plassart, P.; Wincker, P.; Jolivet, C.; et al. Molecular biomass and metataxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. Microb. Biotechnol. 2012, 5, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Castiglioni, B.; Malferrari, G.; Biunno, I.; Vimercati, C.; Moroni, P.; Morandi, S.; Luzzana, M. Technical note: Improved method for rapid DNA extraction of mastitis pathogens directly from milk. J. Dairy Sci. 2006, 89, 163–169. [Google Scholar] [CrossRef]
- Jervis-Bardy, J.; Leong, L.E.X.; Marri, S.; Smith, R.J.; Choo, J.M.; Smith-Vaughan, H.C.; Nosworthy, E.; Morris, P.S.; O’Leary, S.; Rogers, G.B.; et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of illumina miseq data. Microbiome 2015, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Glassing, A.; Dowd, S.E.; Galandiuk, S.; Davis, B.; Chiodini, R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Willner, D.; Daly, J.; Whiley, D.; Grimwood, K.; Wainwright, C.E.; Hugenholtz, P. Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples. PLoS ONE 2012, 7, e34605. [Google Scholar] [CrossRef]
- Wen, Y.; Xiao, F.; Wang, C.; Wang, Z. The impact of different methods of DNA extraction on microbial community measures of balf samples based on metagenomic data. Am. J. Transl. Res. 2016, 8, 1412–1425. [Google Scholar]
- Nelson, M.T.; Pope, C.E.; Marsh, R.L.; Wolter, D.J.; Weiss, E.J.; Hager, K.R.; Vo, A.T.; Brittnacher, M.J.; Radey, M.C.; Hayden, H.S. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep. 2019, 26, 2227–2240. [Google Scholar] [CrossRef]
- Wesolowska-Andersen, A.; Bahl, M.I.; Carvalho, V.; Kristiansen, K.; Sicheritz-Pontén, T.; Gupta, R.; Licht, T.R. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014, 2, 19. [Google Scholar] [CrossRef]
- Kennedy, N.A.; Walker, A.W.; Berry, S.H.; Duncan, S.H.; Farquarson, F.M.; Louis, P.; Thomson, J.M. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16s rrna gene sequencing. PLoS ONE 2014, 9, e88982. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojo-Okunola, A.; Claassen-Weitz, S.; Mwaikono, K.S.; Gardner-Lubbe, S.; Zar, H.J.; Nicol, M.P.; du Toit, E. The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles. Methods Protoc. 2020, 3, 39. https://doi.org/10.3390/mps3020039
Ojo-Okunola A, Claassen-Weitz S, Mwaikono KS, Gardner-Lubbe S, Zar HJ, Nicol MP, du Toit E. The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles. Methods and Protocols. 2020; 3(2):39. https://doi.org/10.3390/mps3020039
Chicago/Turabian StyleOjo-Okunola, Anna, Shantelle Claassen-Weitz, Kilaza S. Mwaikono, Sugnet Gardner-Lubbe, Heather J. Zar, Mark P. Nicol, and Elloise du Toit. 2020. "The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles" Methods and Protocols 3, no. 2: 39. https://doi.org/10.3390/mps3020039
APA StyleOjo-Okunola, A., Claassen-Weitz, S., Mwaikono, K. S., Gardner-Lubbe, S., Zar, H. J., Nicol, M. P., & du Toit, E. (2020). The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles. Methods and Protocols, 3(2), 39. https://doi.org/10.3390/mps3020039




