Analysis of the Level of Plasmid-Derived mRNA in the Presence of Residual Plasmid DNA by Two-Step Quantitative RT-PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Transfection of the MC3T3-E1 Cell Line
2.3. RNA Isolation, RT Reaction and Quantitative PCR Analysis
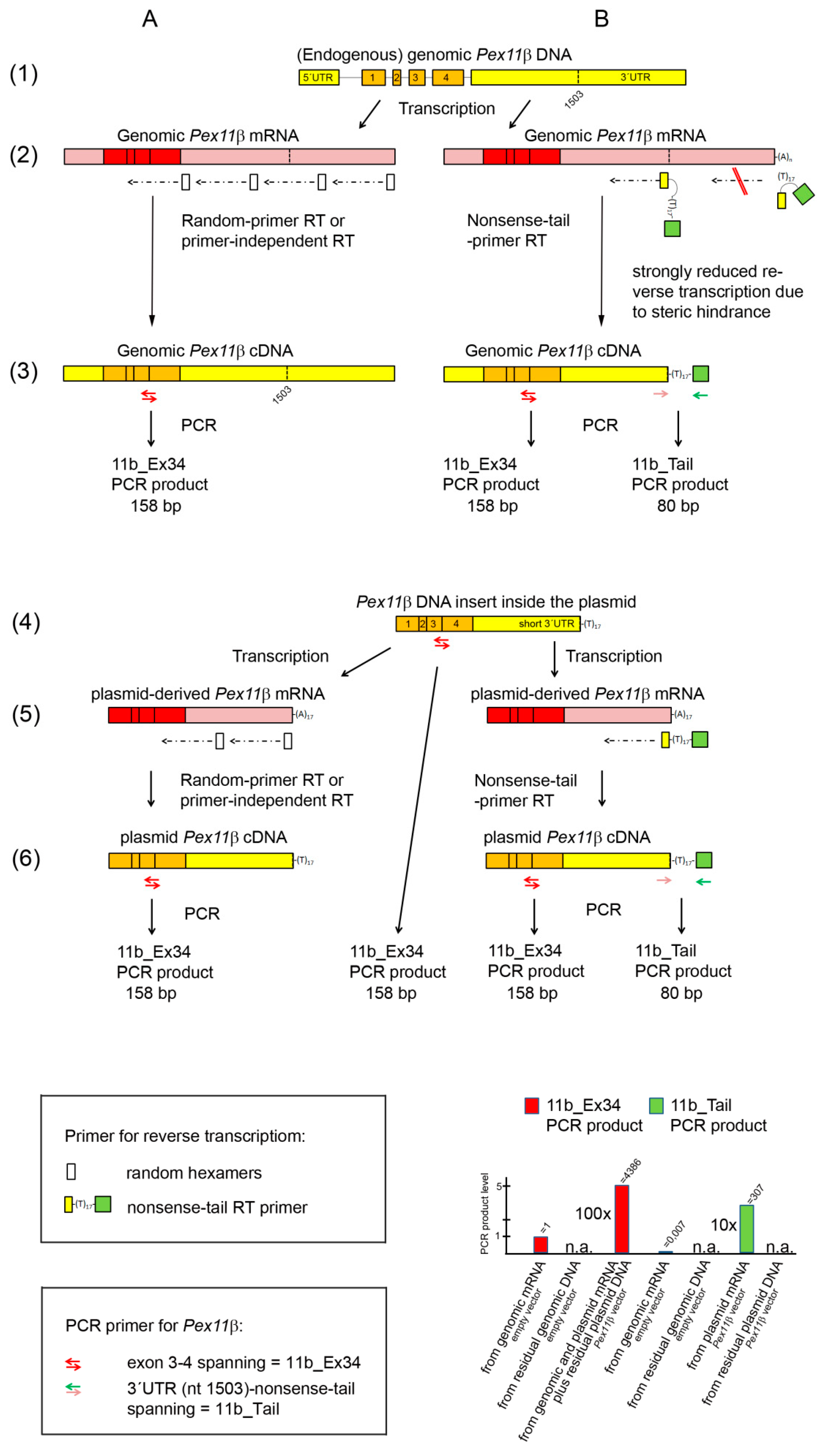
3. Results and Discussion
3.1. Detection of Contaminating Plasmid DNA in Transfected MC3T3-E1 cells
3.2. A Simple RT-qPCR Protocol to Distinguish between Plasmid-Derived mRNA and Plasmid DNA/Endogenous Genome-Derived mRNA
3.3. High Amounts of Intracellular Plasmid DNA Disturbed the Removal of (Plasmid and Genomic) DNA during Standard RNA Isolation
3.4. Necessity for a Selective Detection of Plasmid cDNA
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomic data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Laurell, H.; Iacovoni, J.C.; Abot, A.; Svec, D.; Maoret, J.J.; Arnal, J.F.; Kubista, M. Correction of RT-qPCR data for genomic DNA-derived signals with ValidPrime. Nucleic Acids Res. 2012, 40, e51. [Google Scholar] [CrossRef] [PubMed]
- Padhi, B.K.; Singh, M.; Huang, N.; Pelletier, G.A. PCR-based approach to assess genomic DNA contamination in RNA: Application to rat RNA samples. Anal. Biochem. 2016, 494, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Hashemipetroudi, S.H.; Nematzadeh, G.; Ahmadian, G.; Yamchi, A.; Kuhlmann, M. Assessment of DNA contamination in RNA samples based on ribosomal DNA. J. Vis. Exp. 2018, 131. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Luo, D.; Liao, D.J. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS ONE 2012, 7, e41659. [Google Scholar] [CrossRef]
- Shuldiner, A.R.; Nirula, A.; Roth, J. RNA template-specific polymerase chain reaction (RS-PCR): A novel strategy to reduce dramatically false positives. Gene 1990, 91, 139–142. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Spivack, S.D. mRNA-specific reverse transcription-polymerase chain reaction from human tissue extracts. Anal. Biochem. 2002, 307, 304–315. [Google Scholar] [CrossRef]
- Smith, R.D.; Ogden, C.W.; Penny, M.A. Exclusive amplification of cDNA template (EXACT) RT-PCR to avoid amplifying contaminating genomic pseudogenes. Biotechniques 2001, 31, 776–778. [Google Scholar] [CrossRef]
- Sudo, H.; Kodama, H.A.; Amagai, Y.; Yamamoto, S.; Kasai, S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 1983, 96, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Baumgart, E.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, D.; Gould, S.J. PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 2002, 22, 4358–4365. [Google Scholar] [CrossRef] [PubMed]
- Ahlemeyer, B.; Gottwald, M.; Baumgart-Vogt, E. Deletion of a single allele of the Pex11β gene is sufficient to cause oxidative stress, delayed differentiation and neuronal death in mouse brain. Dis. Model. Mech. 2012, 5, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Fan, W.; Ahlemeyer, B.; Karnati, S.; Baumgart-Vogt, E. Peroxisomes in different skeletal cell types during intramembranous and endochondral ossification and their regulation during osteoblast differentiation by distinct peroxisome proliferator-activated receptors. PLoS ONE 2015, 10, e0143439. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.J.; Zhang, M.; Champoux, J.J. Specific cleavages by RNase H facilitate initiation of plus-strand RNA synthesis by Moloney murine leukemia virus. J. Virol. 2003, 77, 5275–5285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Huang, Z.; Fasco, M.J.; Kaminsky, L.S. Optimization of DNase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Biotechniques 1996, 20, 1012–1020. [Google Scholar] [CrossRef]
- Ivarsson, K.; Weijdegård, B. Evaluation of the effects of DNase treatment on signal specificity in RT-PCR and in situ RT-PCR. Biotechniques 1998, 25, 630–632. [Google Scholar] [CrossRef]
- Jalkanen, A.L.; Coleman, S.J.; Wilusz, J. Determinants and implications of mRNA poly(A) tail size--does this protein make my tail look big? Semin. Cell Dev. Biol. 2014, 34, 24–32. [Google Scholar] [CrossRef]
- Feng, L.; Lintula, S.; Ho, T.H.; Anastasina, M.; Paju, A.; Haglund, C.; Stenman, U.H.; Hotakainen, K.; Orpana, A.; Kainov, D.; et al. Technique for strand-specific gene-expression analysis and monitoring of primer-independent cDNA synthesis in reverse transcription. Biotechniques 2012, 52, 263–270. [Google Scholar] [CrossRef]
- Peretti, S.W.; Bailey, J.E.; Lee, J.J. Transcription from plasmid genes, macromolecular stability, and cell-specific productivity in Escherichia coli carrying copy number mutant plasmids. Biotechnol. Bioeng. 1989, 34, 902–908. [Google Scholar] [CrossRef]
- Wood, T.K.; Peretti, S.W. Depression of protein synthetic capacity due to cloned-gene expression in E. coli. Biotechnol. Bioeng. 1990, 36, 865–878. [Google Scholar] [CrossRef]
- Nowakowski, A.; Andrzejewska, A.; Boltze, J.; Nitzsche, F.; Cui, L.L.; Jolkkonen, J.; Walczak, P.; Lukomska, B.; Janowski, M. Translation, but not transfection limits clinically relevant, exogenous mRNA based induction of alpha-4 integrin expression on human mesenchymal stem cells. Sci. Rep. 2017, 7, 1103. [Google Scholar] [CrossRef] [PubMed]
- Leahy, P.; Carmichael, G.G.; Rossomando, E.F. Transcription from plasmid expression vectors is increased up to 14-fold when plasmids are transfected as concatemers. Nucleic Acids Res. 1997, 25, 449–450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, C.C.; Aylott, M.C.; Fisher, K.J.; Lynch, A.M.; Gooderham, N.J. DNA damage responses after exposure to DNA-based products. J. Gene Med. 2006, 8, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Emerson, M.; Renwick, L.; Tate, S.; Rhind, S.; Milne, E.; Painter, H.A.; Boyd, A.C.; McLachlan, G.; Griesenbach, U.; Cheng, S.H.; et al. Transfection efficiency and toxicity following delivery of naked plasmid DNA and cationic lipid-DNA complexes to ovine lung segments. Mol. Ther. 2003, 8, 646–653. [Google Scholar] [CrossRef]
- Yew, N.S.; Zhao, H.; Wu, I.H.; Song, A.; Tousignant, J.D.; Przybylska, M.; Cheng, S.H. Reduced inflammatory response to plasmid DNA vectors by elimination and inhibition of immunostimulatory CpG motifs. Mol. Ther. 2000, 1, 255–262. [Google Scholar] [CrossRef]
- Yenofsky, R.; Bergmann, I.; Brawerman, G. Cloned complementary deoxyribonucleic acid probes for untranslated messenger ribonucleic acid components of mouse sarcoma ascites cells. Biochemistry 1982, 21, 3909–3913. [Google Scholar] [CrossRef]
- Fliedl, L.; Kast, F.; Grillari, J.; Wieser, M.; Grillari-Voglauer, R. Optimization of a quantitative PCR based method for plasmid copy number determination in human cell lines. New Biotechnol. 2015, 32, 716–719. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 536–537. [Google Scholar]
- Chomczynski, P.; Sacci, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Hart, S.L.; Mayall, E.; Stern, M.; Munkonge, F.M.; Frost, A.; Huang, L.; Vasilliou, M.; Williamson, R.; Alton, E.W.; Coutelle, C. The introduction of two silent mutations into a CFTR cDNA construct allows improved detection of exogenous mRNA in gene transfer experiments. Hum. Mol. Genet. 1995, 4, 1597–1602. [Google Scholar] [CrossRef]
- Mayall, E.S.; Coutelle, C. RT-PCR method specific for the detection of transgenic CFTR mRNA in the presence of transgene plasmid DNA and endogenous CFTR mRNA. Gene Ther. 1997, 4, 875–878. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fairman, J.; Roche, L.; Pieslak, I.; Lay, M.; Corson, S.; Fox, E.; Luong, C.; Koe, G.; Lemos, B.; Grove, R.; et al. Quantitative RT-PCR to evaluate in vivo expression of multiple transgenes using a common intron. Biotechniques 1999, 27, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Buchman, A.R.; Berg, P. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 1988, 8, 4395–4405. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.L.; Chung, M.; Matyas, R.J. Persistent DNA contamination in competitive RT-PCR using cRNA internal standards: Identity, quantity, and control. Biotechniques 2002, 32, 1412–1417. [Google Scholar] [CrossRef]
| Gene name NCBI Accession Number | PCR Primer Sequence (5´-3´end) | Position Ms Transcript | Position Plasmid DNA | Am-pli-con size (bp) | Effi-cien-cy | Abbre-viation |
|---|---|---|---|---|---|---|
| Pex11b, NM_011069.3 | ||||||
| Aaccgagccttgtactttgc aggcgaatctcataagcatca | 532-551 689-669 | 281-300 418-438 | 158 | 1.99 | 11b_Ex34 | |
| cgcctattgatggaacaagagact tccaggtcccacagtttctactc | 685-708 780-758 | 434-457 507-529 | 96 | 1.99 | 11b_Ex4 | |
| ctggtccttgcccacagagc ccgatcgcgatttcgataaaa | 1470-1484 Ø | Ø 1721-1740 | 80 | 1.99 | 11b_Tail | |
| Hprt1 NM_013556.2 | ||||||
| agtcccagcgtcgtgattag tttccaaatcctcggcataatga | 159-178 246-224 | Ø Ø | 88 | 1.86 | Hprt_Ex12 | |
| RT primer sequence (5´-3´end) | ||||||
| Nonsense-tail RT primer | Ggctagcgctaaagctattttttttttttttttt gagcaaactgat | 1770-1782 | 1518-1530 |
| High-capacity cDNA reverse transcription kit; Appl. Biosystems, Cat. No. 4368881 | ||||
| Reaction | Amount | Volume (µL) | defined as | |
| Reverse Transcription | RNA template | 2 µg | 10.0 | |
| + | 10 x RT buffer | 2.0 | ||
| + | 25 x dNTP Mix | 100 mM | 0.8 | |
| either | Multiscribe™ Reverse Transcriptase | 50 U/µL | 1.0 | |
| or | Water, nuclease-free | 1.0 | no RT | |
| either | 10 x random hexamers | 1.0 | Random RT | |
| or | Nonsense tail RT primer | 1pmol/µL | 1.0 | Nonsense-tail RT |
| or | Water, nuclease-free | 1.0 | no RT primer RT | |
| + | Water, nuclease-free | to 20.0 | ||
| 10 min, 25°C; 120 min, 37°C; 5 min, 85 °C, ∞ 4°C | ||||
| Maxima SYBR Green/Fluorescein qPCR Master Mix (2x); Thermo Fisher Scientific, #K0243 | ||||
| Reaction | Amount | Volume (µL) | Defined as | |
| PCR | DNA template | cDNA from 0.2 µg total RNA | 2.0 | |
| + | Forward and reverse primer | 2.5 pmol/µL | 1.0 each | |
| 11b_Ex34 | 11b_Ex34 PCR | |||
| 11b_Ex4 | 11b_Ex4 PCR | |||
| 11b_Tail | 11b_Tail PCR | |||
| Hprt_Ex12 | Hprt_Ex12 PCR | |||
| + | 2 x SYBR Green qPCR Master Mix | |||
| + | Water, nuclease-free | 10.0 | ||
| 10 min, 95°C; 40 cycles: 15 s, 95°C; 60 s, 60°C | ||||
| 1 | 2 | 3 | 4 | ||
|---|---|---|---|---|---|
| Experimental Condition | Primer Pex11β | Primer Hprt | |||
| 11b_Ex34 | Hprt_Ex12 | normalized to Hprt | set to1 | ||
| ct values | ct values | ||||
| 1 | 6 h, empty vector | 17.10 | 19.30 | 1.13 | 1.00 |
| 2 | 6 h, Pex11β vector | 9.00 | 20.55 | 674.87 | 595.98 |
| 3 | 12 h, empty vector | 22.80 | 25.80 | 1.23 | 1.00 |
| 4 | 12 h, Pex11β vector | 12.85 | 28.20 | 5395.47 | 4386.11 |
| 5 | 12 h, Pex11β vector, no RT | 18.38 | N/A | ||
| 6 | 12 h, Pex11β vector, DNase | 14.85 | 26.50 | 469.68 | 381.81 |
| 1 | 2 | 3 | ||
|---|---|---|---|---|
| Experimental Condition | Primer Pex11β | Primer Hprt | Primer Pex11β | |
| 11b_Tail | Hprt_Ex12 | 11b_Ex34 | ||
| 6 h, Nonsense-Tail RT | ||||
| 1 | empty vector | 24.60 | 26.10 | 17.45 |
| 2 | Pex11β vector | 13.20 | 25.90 | 9.30 |
| 6 h, Random RT | ||||
| 3 | empty vector | N/A | 22.65 | 18.50 |
| 4 | Pex11β vector | N/A | 22.90 | 11.30 |
| 5 | 6 h, No Transfection | |||
| 6 | random RT | N/A | 21.44 | 21.02 |
| 7 | nonsense-tail RT | 26.83 | 25.7 | 20.17 |
| 8 | no RT-primer RT | N/A | 28.2 | 20.74 |
| 9 | no RT | N/A | N/A | N/A |
| Experimental Condition | Primer Pex11β | |
|---|---|---|
| 6 h, No Transfection | 11b_Ex4 | |
| 1 | no RT, DNase | 30.20 |
| 2 | no RT, no DNase | 30.50 |
| 3 | random RT, DNase | 21.50 |
| 4 | random RT, no DNase | 22.00 |
| 6 h, Empty Vector | ||
| 5 | no RT, DNase | 29.80 |
| 6 | no RT, no DNase | 24.60 |
| 7 | random RT, DNase | 25.30 |
| 8 | random RT, no DNase | 24.50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahlemeyer, B.; Colasante, C.; Baumgart-Vogt, E. Analysis of the Level of Plasmid-Derived mRNA in the Presence of Residual Plasmid DNA by Two-Step Quantitative RT-PCR. Methods Protoc. 2020, 3, 40. https://doi.org/10.3390/mps3020040
Ahlemeyer B, Colasante C, Baumgart-Vogt E. Analysis of the Level of Plasmid-Derived mRNA in the Presence of Residual Plasmid DNA by Two-Step Quantitative RT-PCR. Methods and Protocols. 2020; 3(2):40. https://doi.org/10.3390/mps3020040
Chicago/Turabian StyleAhlemeyer, Barbara, Claudia Colasante, and Eveline Baumgart-Vogt. 2020. "Analysis of the Level of Plasmid-Derived mRNA in the Presence of Residual Plasmid DNA by Two-Step Quantitative RT-PCR" Methods and Protocols 3, no. 2: 40. https://doi.org/10.3390/mps3020040
APA StyleAhlemeyer, B., Colasante, C., & Baumgart-Vogt, E. (2020). Analysis of the Level of Plasmid-Derived mRNA in the Presence of Residual Plasmid DNA by Two-Step Quantitative RT-PCR. Methods and Protocols, 3(2), 40. https://doi.org/10.3390/mps3020040





