Custom Multiphoton/Raman Microscopy Setup for Imaging and Characterization of Biological Samples
Abstract
:1. Introduction
2. Materials and Methods
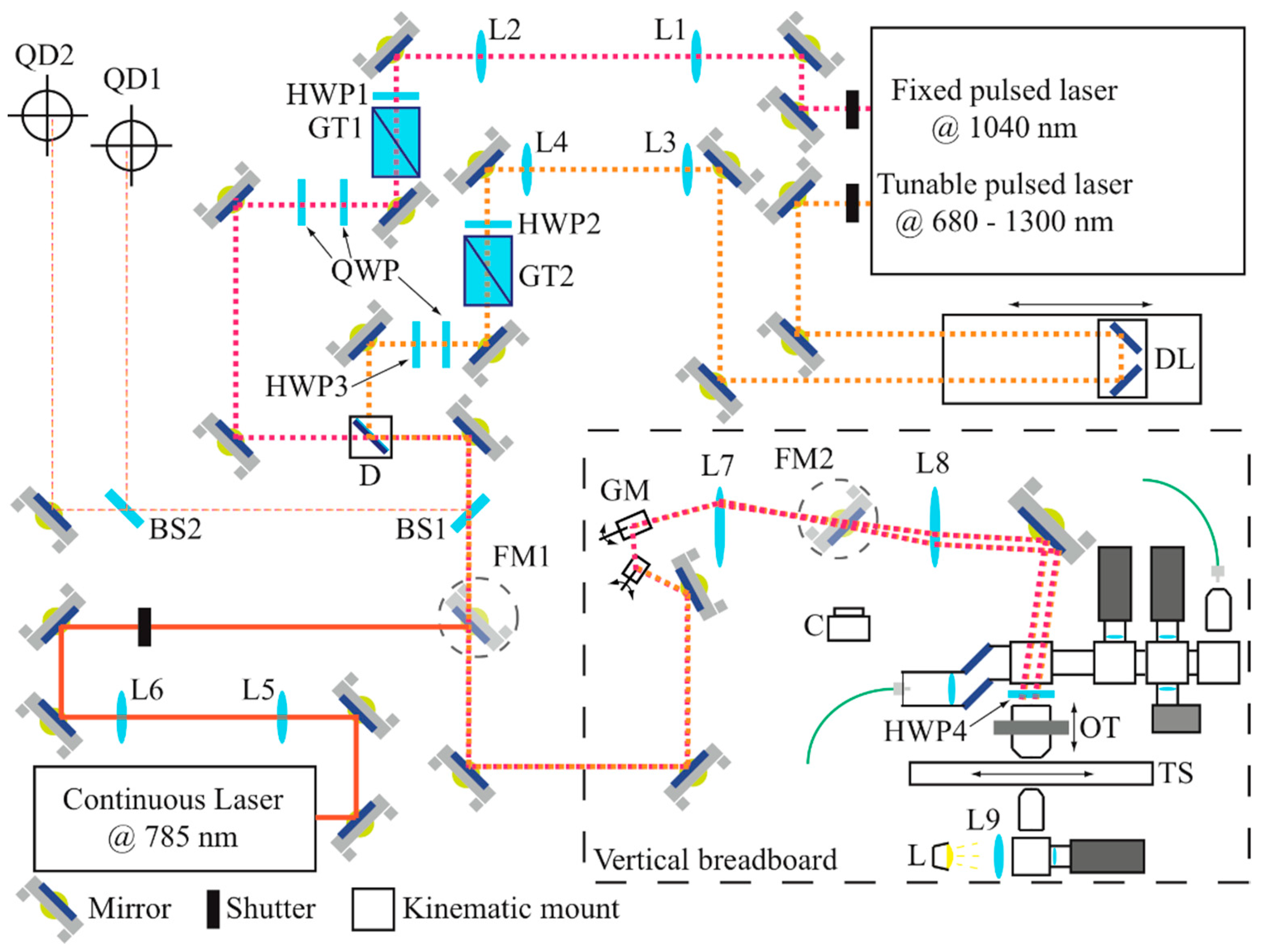
2.1. The Opto-Mechanical System
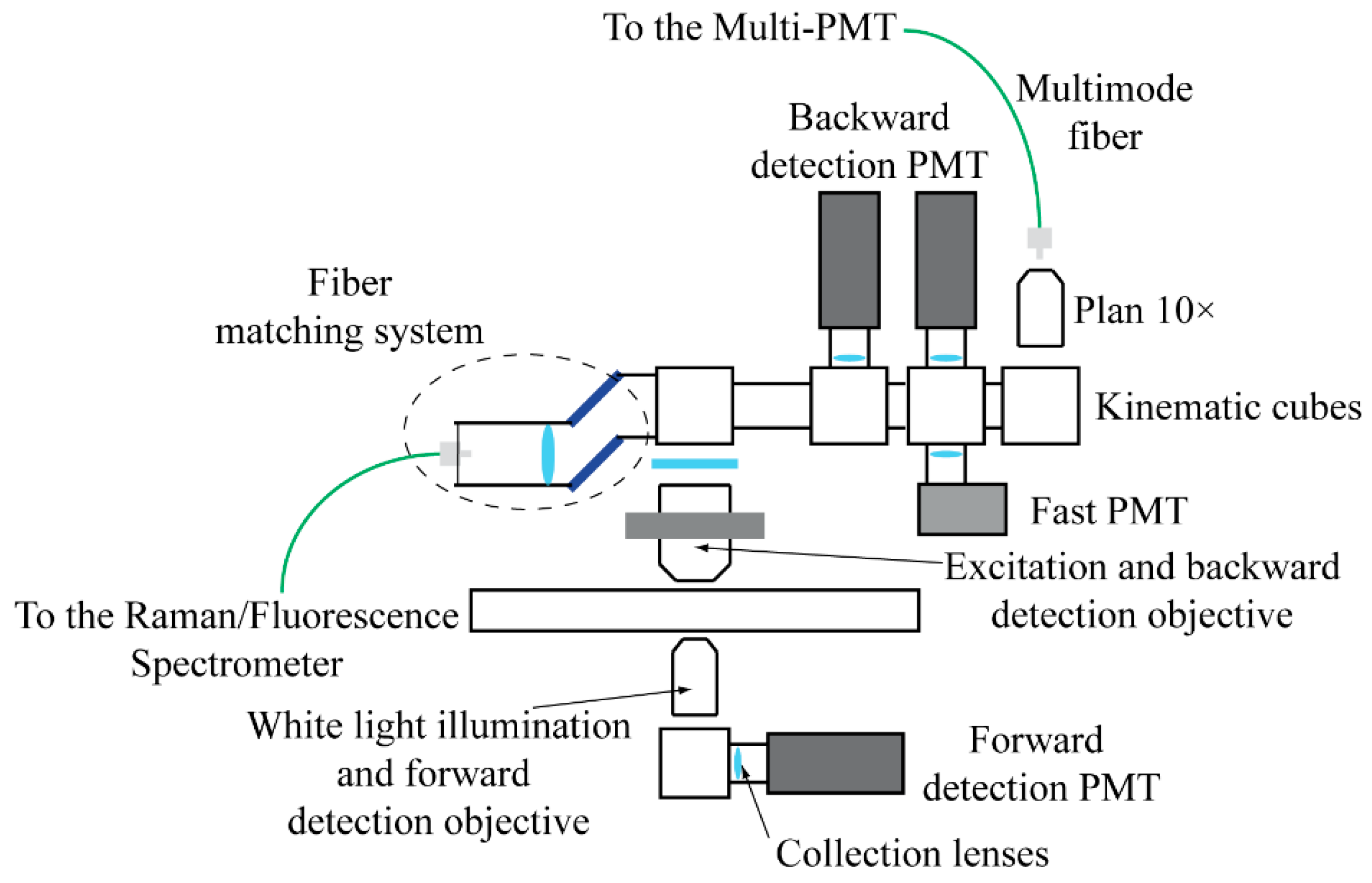
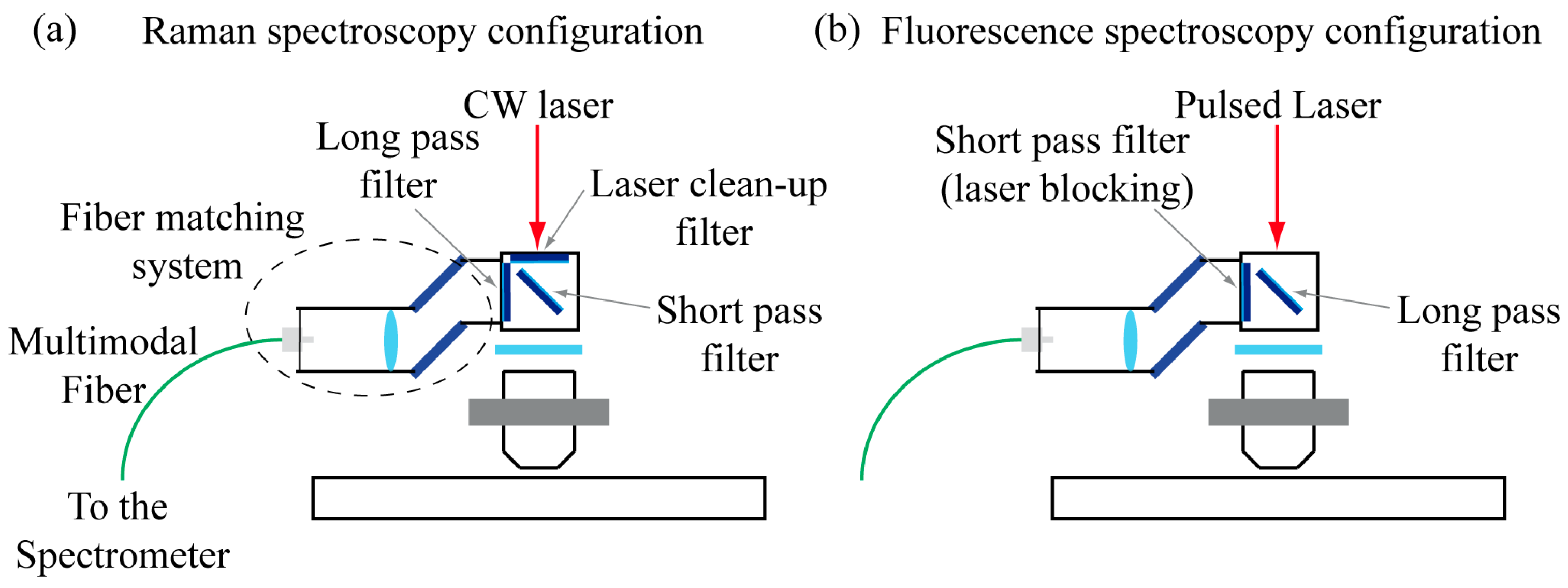
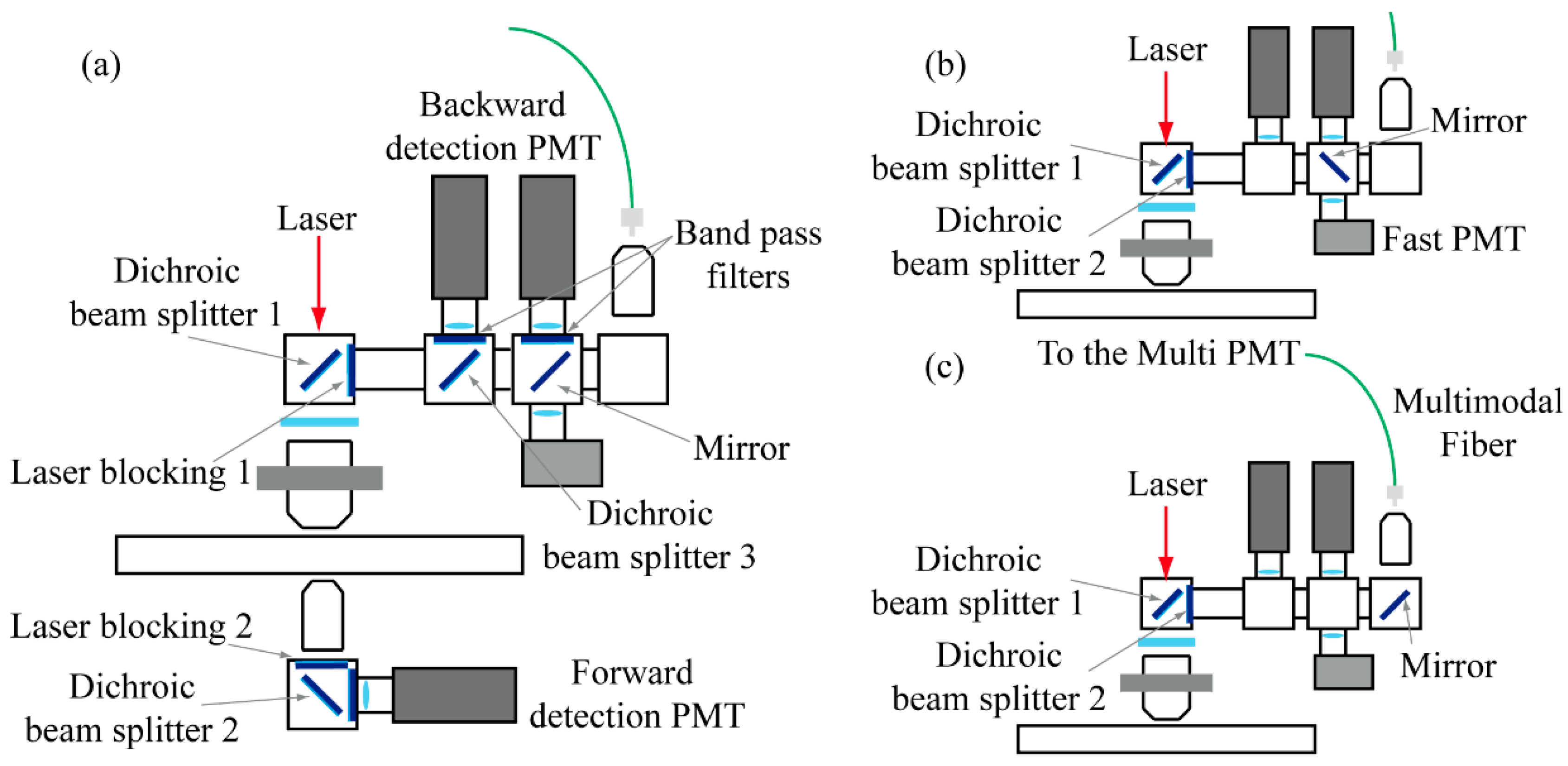
2.2. The Detection System
2.3. The Control and Acquisition System
2.4. Sample Description and Preparation
3. Results and Discussion
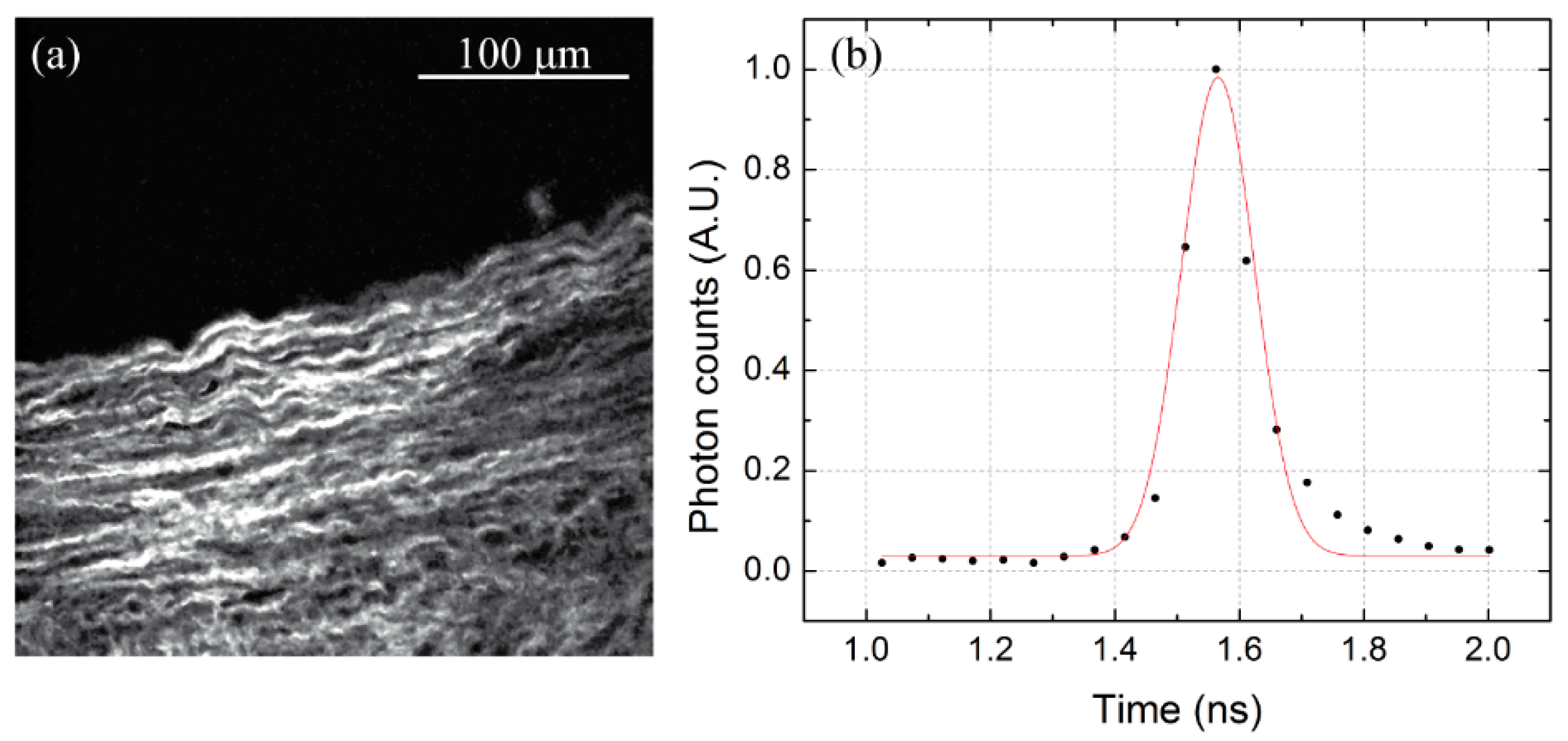
3.1. Microscope Calibration: Spatial and Temporal Resolution
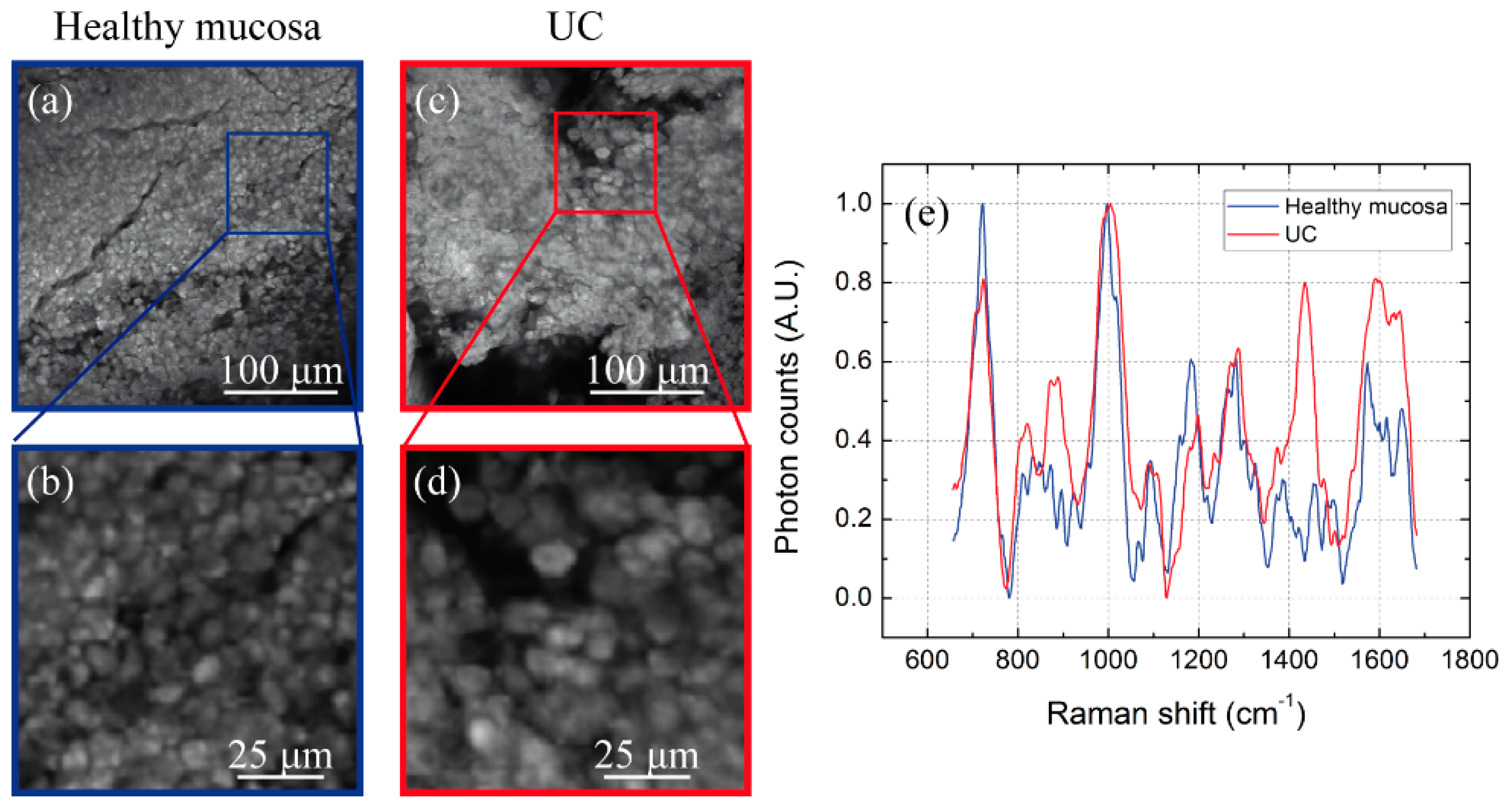
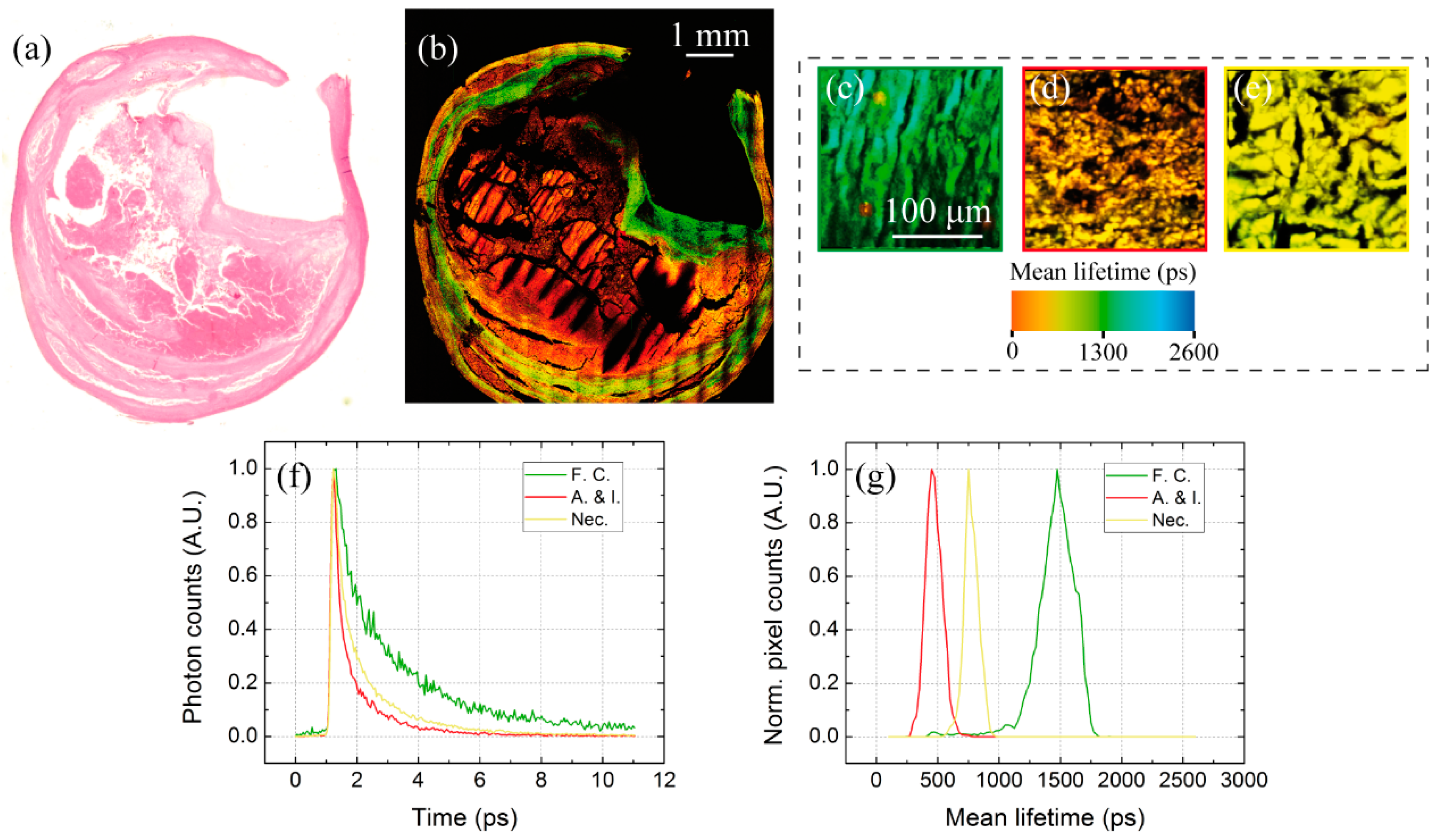
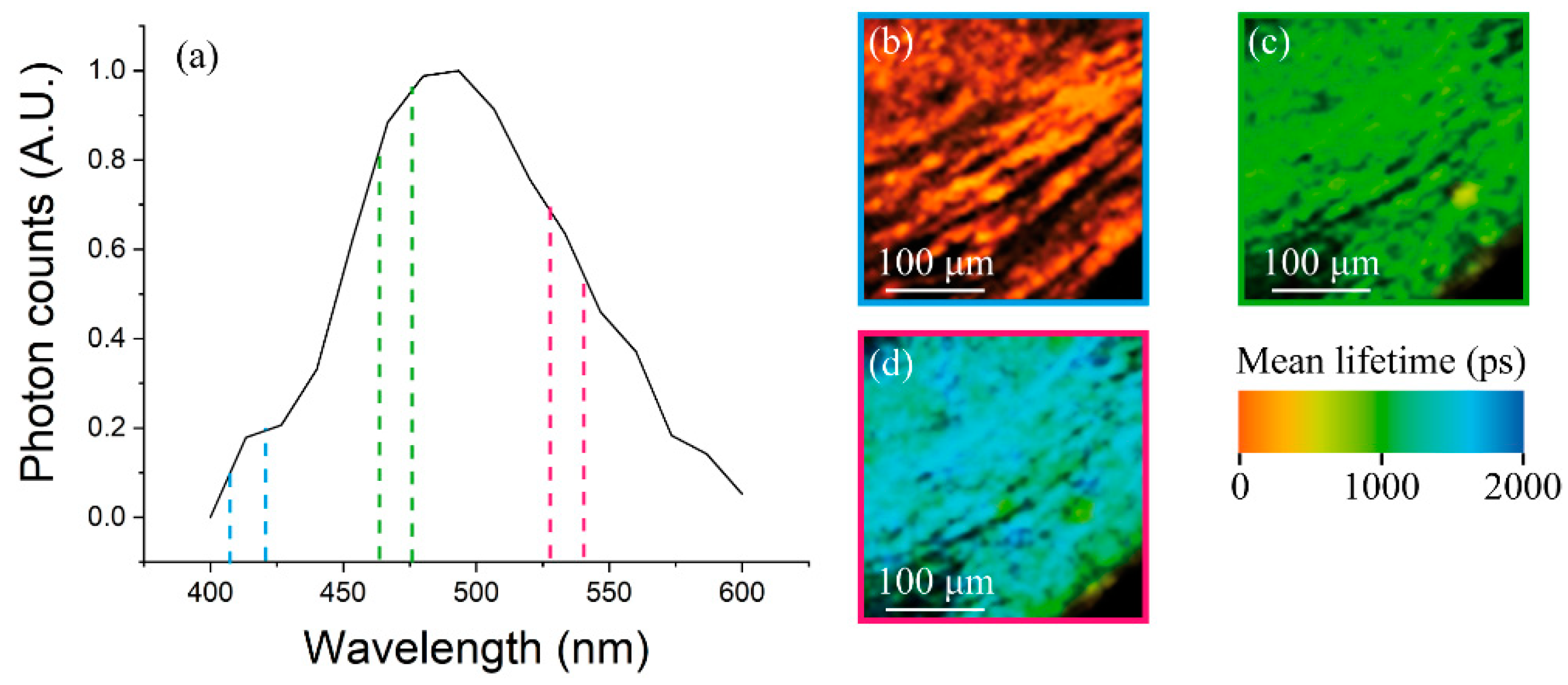
3.2. Multimodal Imaging of Human Ex Vivo Tissue Samples
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alturkistani, H.A.; Tashkandi, F.M.; Mohammedsaleh, Z.M. Histological Stains: A Literature Review and Case Study. Glob. J. Health Sci. 2016, 8, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K. The wonderful colors of the hematoxylin-eosin stain in diagnostic surgical pathology. Int. J. Surg. Pathol. 2014, 22, 12–32. [Google Scholar] [CrossRef] [PubMed]
- De Matos, L.L.; Trufelli, D.C.; De Matos MG, L.; da Silva Pinhal, M.A. Immunohistochemistry as an Important Tool in Biomarkers Detection and Clinical Practice. Biomark. Insights 2010, 5, 9–20. [Google Scholar] [CrossRef]
- Zipfel, W.R.; Williams, R.M.; Webb, W.W. Nonlinear magic: Multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Zoumi, A.; Yeh, A.; Tromberg, B.J. Imaging cells and extracellular matrix in vivo by using second harmonic generation and two-photon excited fluorescence. Proc. Natl. Acad. Sci. USA 2002, 99, 11014–11019. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-X.; Xie, X.S. Coherent Anti-Stokes Raman Scattering Microscopy: Instrumentation, Theory, and Applications. J. Phys. Chem. B 2004, 108, 827–840. [Google Scholar] [CrossRef]
- Puppels, G.J.; de Mul, F.F.M.; Otto, C.; Greve, J.; Robert-Nicoud, M.; Arndt-Jovin, D.J.; Jovin, T.M. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature 1990, 347, 301–303. [Google Scholar] [CrossRef]
- Scarcelli, G.; Yun, S.H. Confocal Brillouin microscopy for three-dimensional mechanical imaging. Nat. Photonics 2008, 2, 39–43. [Google Scholar] [CrossRef]
- Zysk, A.M.; Nguyen, F.T.; Oldenburg, A.L.; Marks, D.L.; Boppart, S.A. Optical coherence tomography: A review of clinical development from bench to bedside. J. Biomed. Opt. 2007, 12, 051403. [Google Scholar] [CrossRef]
- Lim, C.S.; Cho, B.R. Two-photon probes for biomedical applications. BMB Rep. 2013, 46, 188–194. [Google Scholar] [CrossRef] [Green Version]
- Eibl, M.; Karpf, S.; Hakert, H.; Weng, D.; Huber, R. Two-photon-excited fluorescence (TPEF) and fluorescence lifetime imaging (FLIM) with sub-nanosecond pulses and a high analog bandwidth signal detection. Proc. SPIE 2017. [Google Scholar] [CrossRef]
- Huang, S.; Heikal, A.A.; Webb, W.W. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys. J. 2002, 82, 2811–2825. [Google Scholar] [CrossRef]
- Zipfel, W.R.; Williams, R.M.; Christie, R.; Nikitin, A.Y.; Hyman, B.T.; Webb, W.W. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. USA 2003, 100, 7075–7080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadrous, P.J. Methods for imaging the structure and function of living tissues and cells: 2. Fluorescence lifetime imaging. J. Pathol. 2000, 191, 229–234. [Google Scholar] [CrossRef]
- Palero, J.A.; de Bruijn, H.S.; van der Ploeg van den Heuvel, A.; Sterenborg, H.J.; Gerritsen, H.C. Spectrally resolved multiphoton imaging of in vivo and excised mouse skin tissues. Biophys. J. 2007, 93, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Campagnola, P.J.; Millard, A.C.; Terasaki, M.; Hoppe, P.E.; Malone, C.J.; Mohler, W.A. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys. J. 2002, 82, 493–508. [Google Scholar] [CrossRef]
- Suhalim, J.L.; Chung, C.Y.; Lilledahl, M.B.; Lim, R.S.; Levi, M.; Tromberg, B.J.; Potma, E.O. Characterization of cholesterol crystals in atherosclerotic plaques using stimulated Raman scattering and second-harmonic generation microscopy. Biophys. J. 2012, 102, 1988–1995. [Google Scholar] [CrossRef]
- Cicchi, R.; Vogler, N.; Kapsokalyvas, D.; Dietzek, B.; Popp, J.; Pavone, F.S. From molecular structure to tissue architecture: Collagen organization probed by SHG microscopy. J. Biophotonics 2013, 6, 129–142. [Google Scholar] [CrossRef]
- Stoller, P.; Reiser, K.M.; Celliers, P.M.; Rubenchik, A.M. Polarization-modulated second harmonic generation in collagen. Biophys. J. 2002, 82, 3330–3342. [Google Scholar] [CrossRef]
- Amat-Roldan, I.; Psilodimitrakopoulos, S.; Loza-Alvarez, P.; Artigas, D. Fast image analysis in polarization SHG microscopy. Opt. Express 2010, 18, 17209–17219. [Google Scholar] [CrossRef]
- Choo-Smith, L.P.; Edwards, H.G.; Endtz, H.P.; Kros, J.M.; Heule, F.; Barr, H.; Robinson, J.S., Jr.; Bruining, H.A.; Puppels, G.J. Medical applications of Raman spectroscopy: From proof of principle to clinical implementation. Biopolymers 2002, 67, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kuhar, N.; Sil, S.; Verma, T.; Umapathy, S. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 2018, 8, 25888–25908. [Google Scholar] [CrossRef] [Green Version]
- Tolles, W.M.; Nibler, J.V.; McDonald, J.R.; Harvey, A.B. A Review of the Theory and Application of Coherent Anti-Stokes Raman Spectroscopy (CARS). Appl. Spectrosc. 1977, 31, 253–271. [Google Scholar] [CrossRef]
- Rodriguez, L.G.; Lockett, S.J.; Holtom, G.R. Coherent anti-stokes Raman scattering microscopy: A biological review. Cytometry Part A 2006, 69, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Book, L.D.; Xie, X.S. Polarization coherent anti-Stokes Raman scattering microscopy. Opt. Lett. 2001, 26, 1341–1343. [Google Scholar] [CrossRef] [PubMed]
- Cicchi, R.; Baria, E.; Matthäus, C.; Lange, M.; Lattermann, A.; Brehm, B.R.; Popp, J.; Pavone, F.S. Non-linear imaging and characterization of atherosclerotic arterial tissue using combined SHG and FLIM microscopy. J. Biophotonics 2015, 8, 347–356. [Google Scholar] [CrossRef]
- Chen, H.; Wang, H.; Slipchenko, M.N.; Jung, Y.K.; Shi, Y.; Zhu, J.; Buhman, K.K.; Cheng, J.-X. A multimodal platform for nonlinear optical microscopy and microspectroscopy. Opt. Express 2009, 17, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, V.; Massi, D.; Sestini, S.; Cicchi, R.; Pavone, F.S.; Lotti, T. Combined non-linear laser imaging (two-photon excitation fluorescence microscopy, fluorescence lifetime imaging microscopy, multispectral multiphoton microscopy) in cutaneous tumours: First experiences. J. Eur. Acad. Dermatol. Venereol. 2009, 23, 314–316. [Google Scholar] [CrossRef]
- Theer, P.; Mongis, C.; Knop, M. PSFj: Know your fluorescence microscope. Nat. Methods 2014, 11, 981–982. [Google Scholar] [CrossRef]
- Dietrich, B.; Srinivas, S. Urothelial carcinoma: The evolving landscape of immunotherapy for patients with advanced disease. Res. Rep. Urol. 2018, 26, 7–16. [Google Scholar] [CrossRef]
- Baba, A.I.; Câtoi, C. Chapter 3: Tumor cell morphology. In Comparative Oncology; The Publishing House of the Romanian Academy: Bucharest, Romania, 2007. [Google Scholar]
- Bandeira Cartaxo, S.; Dunshee de Abranches Oliveira Santos, I.; Bitar, R.; Fernandes Oliveira, A.; Masako Ferreira, L.; Silva Martinho, H.; Martin, A.A. FT-Raman spectroscopy for the differentiation between cutaneous melanoma and pigmented nevus. Acta Cir. Bras. 2010, 25, 351–356. [Google Scholar] [CrossRef]
- Feng, S.; Huang, S.; Lin, D.; Chen, G.; Xu, Y.; Li, Y.; Huang, Z.; Pan, J.; Chen, R.; Zeng, H. Surface-enhanced Raman spectroscopy of saliva proteins for the noninvasive differentiation of benign and malignant breast tumors. Int. J. Nanomed. 2015, 12, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Sahu, D.; Lotan, Y.; Wittmann, B.; Neri, B.; Hansel, D.E. Metabolomics analysis reveals distinct profiles of nonmuscle-invasive and muscle-invasive bladder cancer. Cancer Med. 2017, 6, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Sheena Mary, Y.; Ushakumari, L.; Harikumar, B.; Tresa Varghese, H.; Yohannan Panicker, C. FT-IR, FT-Raman and SERS Spectra of L-Proline. J. Iran. Chem. Soc. 2009, 6, 138–144. [Google Scholar] [CrossRef]
- Pierna, J.A.F.; Abbas, O.; Dardenne, P.; Baeten, V. Discrimination of Corsican honey by FT-Raman spectroscopy and chemometrics. Biotechnol. Agron. Soc. Environ. 2011, 15, 75–84. [Google Scholar]
- US Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/heartdisease/facts.htm (accessed on 19 June 2019).
- US American Heart Association. Available online: https://www.heart.org/en/health-topics/cholesterol/about-cholesterol/atherosclerosis (accessed on 19 June 2019).
- Steinberg, D.; Gotto, A.M. Preventing coronary artery disease by lowering cholesterol levels. Fifty years from bench to bedside. JAMA 1999, 282, 2043–2050. [Google Scholar] [CrossRef]
- Finn, A.V.; Nakano, M.; Narula, J.; Kolodgie, F.D.; Virmani, R. Concept of Vulnerable/Unstable Plaque. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1282–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baria, E.; Nesi, G.; Santi, R.; Maio, V.; Massi, D.; Pratesi, C.; Cicchi, R.; Pavone, F.S. Improved label-free diagnostics and pathological assessment of atherosclerotic plaques through nonlinear microscopy. J. Biophotonics 2018, 11, e201800106. [Google Scholar] [CrossRef] [Green Version]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchetti, M.; Baria, E.; Cicchi, R.; Pavone, F.S. Custom Multiphoton/Raman Microscopy Setup for Imaging and Characterization of Biological Samples. Methods Protoc. 2019, 2, 51. https://doi.org/10.3390/mps2020051
Marchetti M, Baria E, Cicchi R, Pavone FS. Custom Multiphoton/Raman Microscopy Setup for Imaging and Characterization of Biological Samples. Methods and Protocols. 2019; 2(2):51. https://doi.org/10.3390/mps2020051
Chicago/Turabian StyleMarchetti, Marco, Enrico Baria, Riccardo Cicchi, and Francesco Saverio Pavone. 2019. "Custom Multiphoton/Raman Microscopy Setup for Imaging and Characterization of Biological Samples" Methods and Protocols 2, no. 2: 51. https://doi.org/10.3390/mps2020051




