The DIVER Microscope for Imaging in Scattering Media
Abstract
:1. Introduction
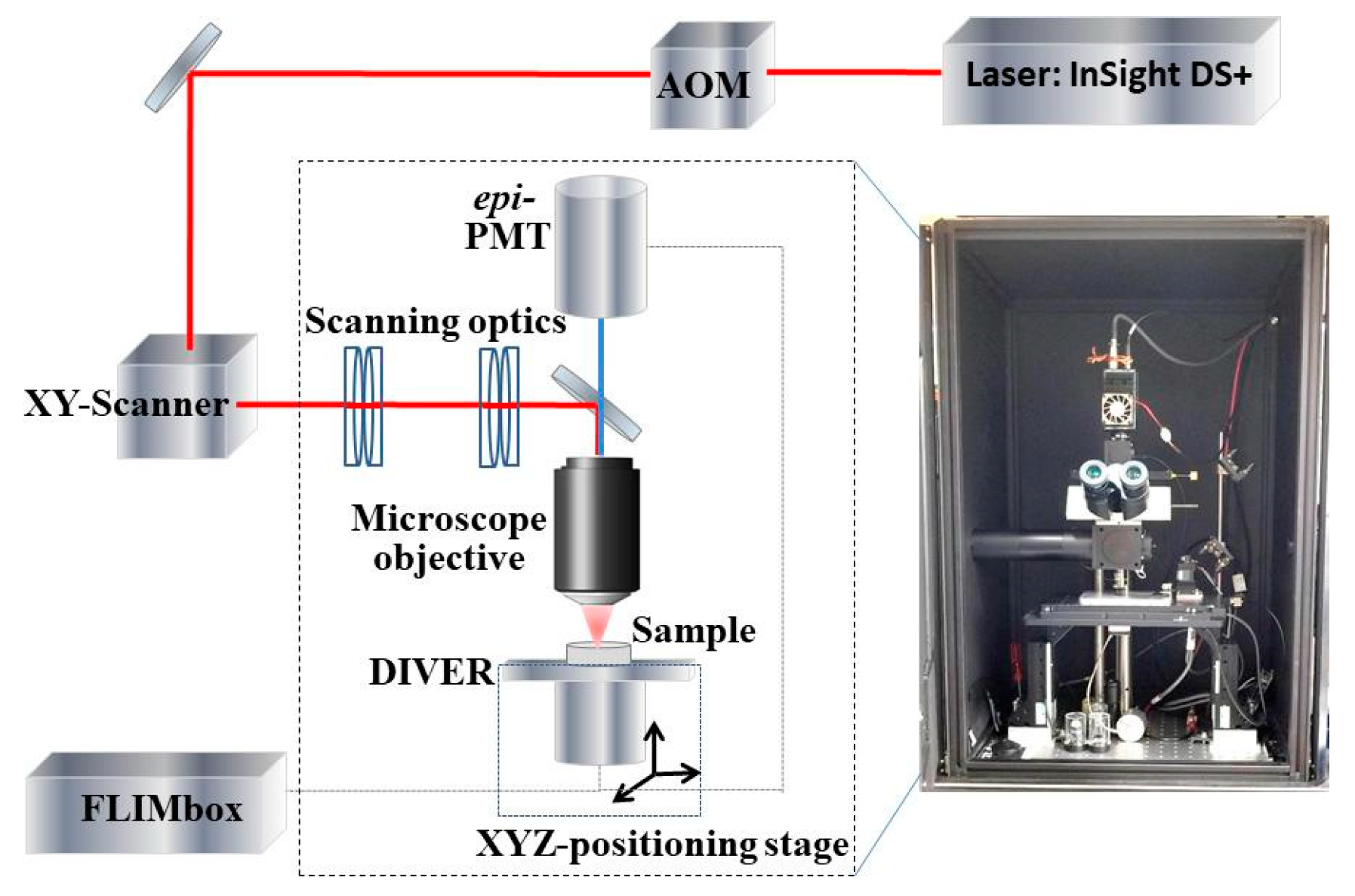
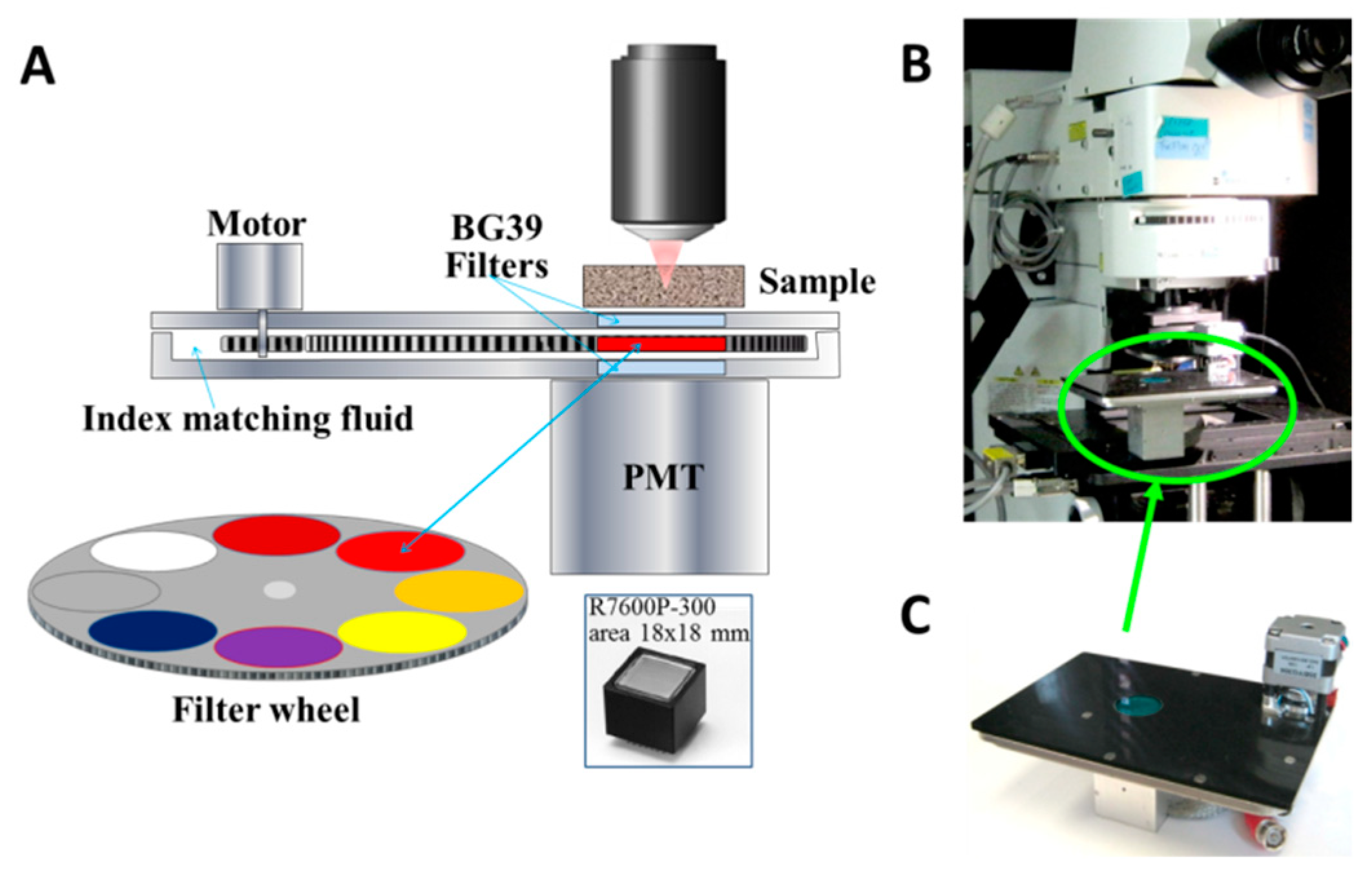
2. Materials and Methods
3. Results
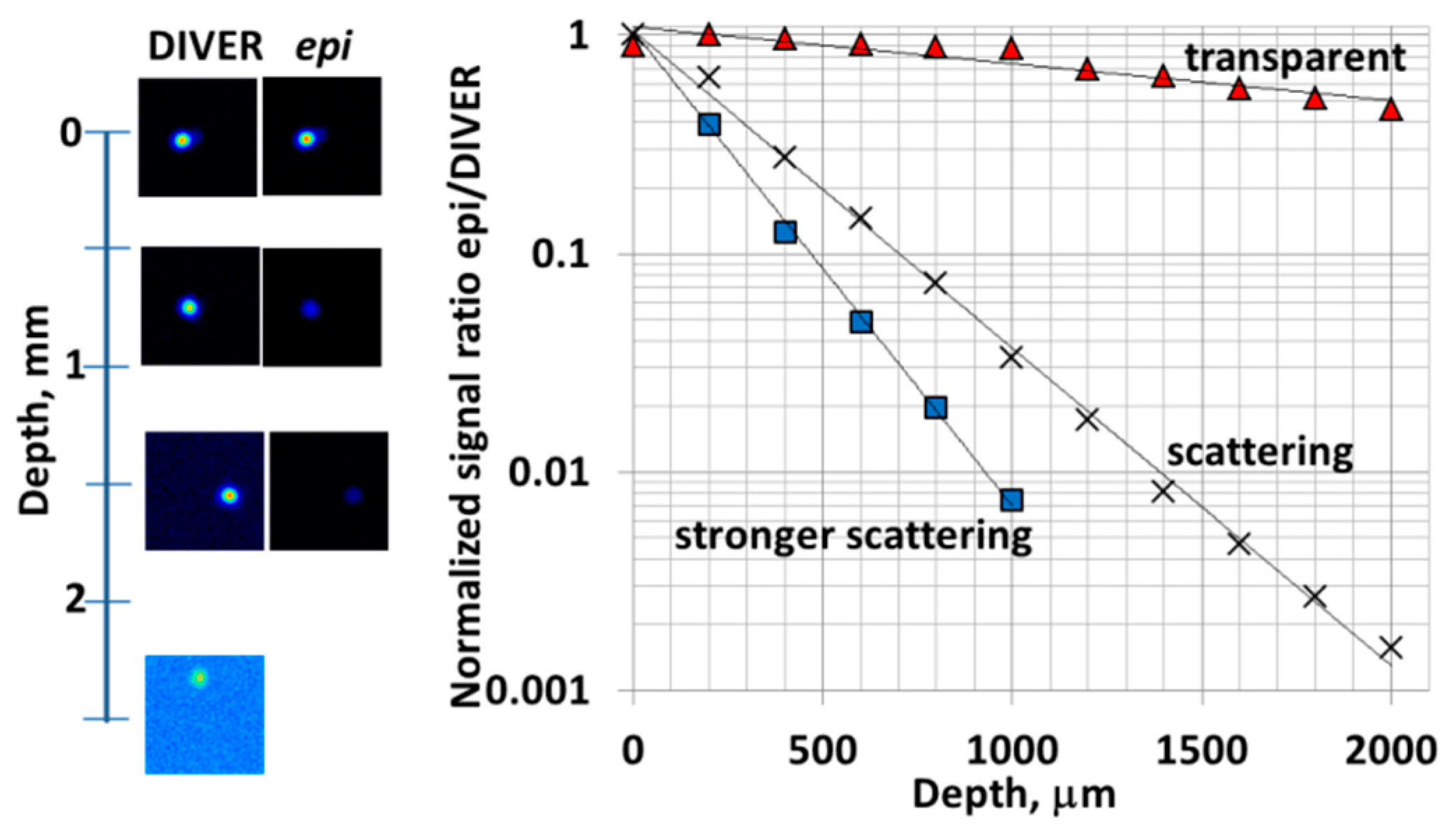
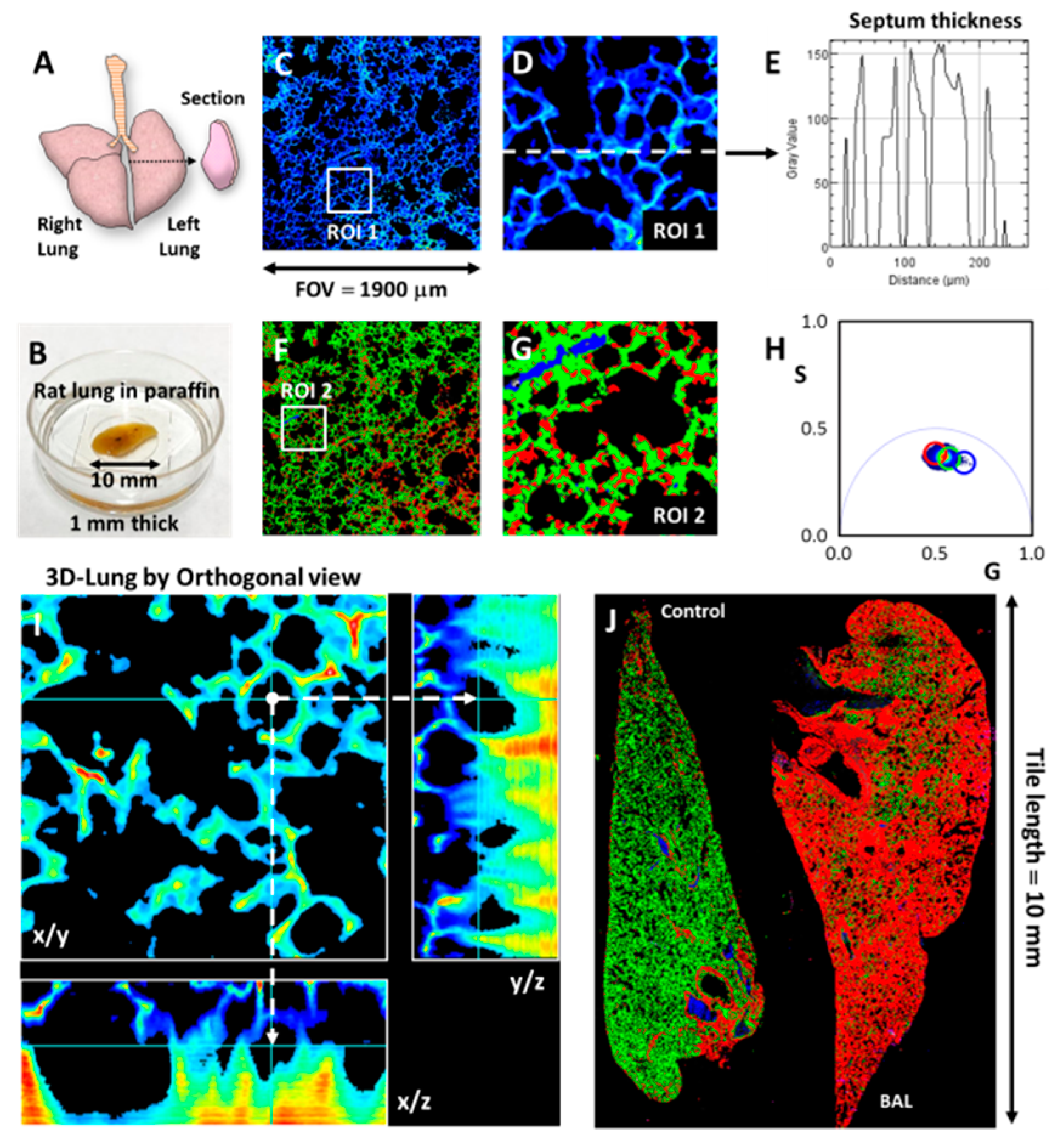
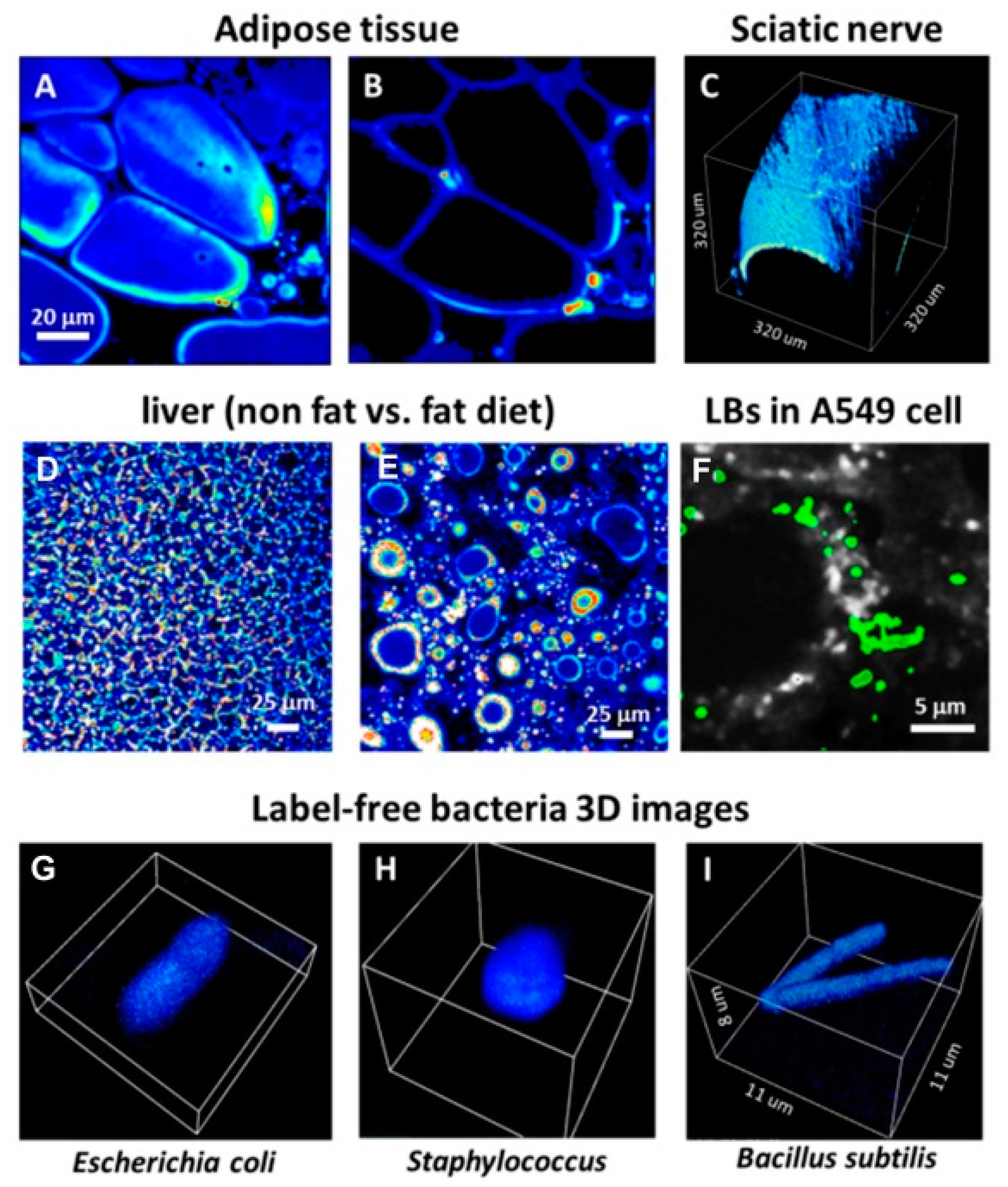
3.1. Imaging in Scattering Media: the DIVER Versus Epi-Detector
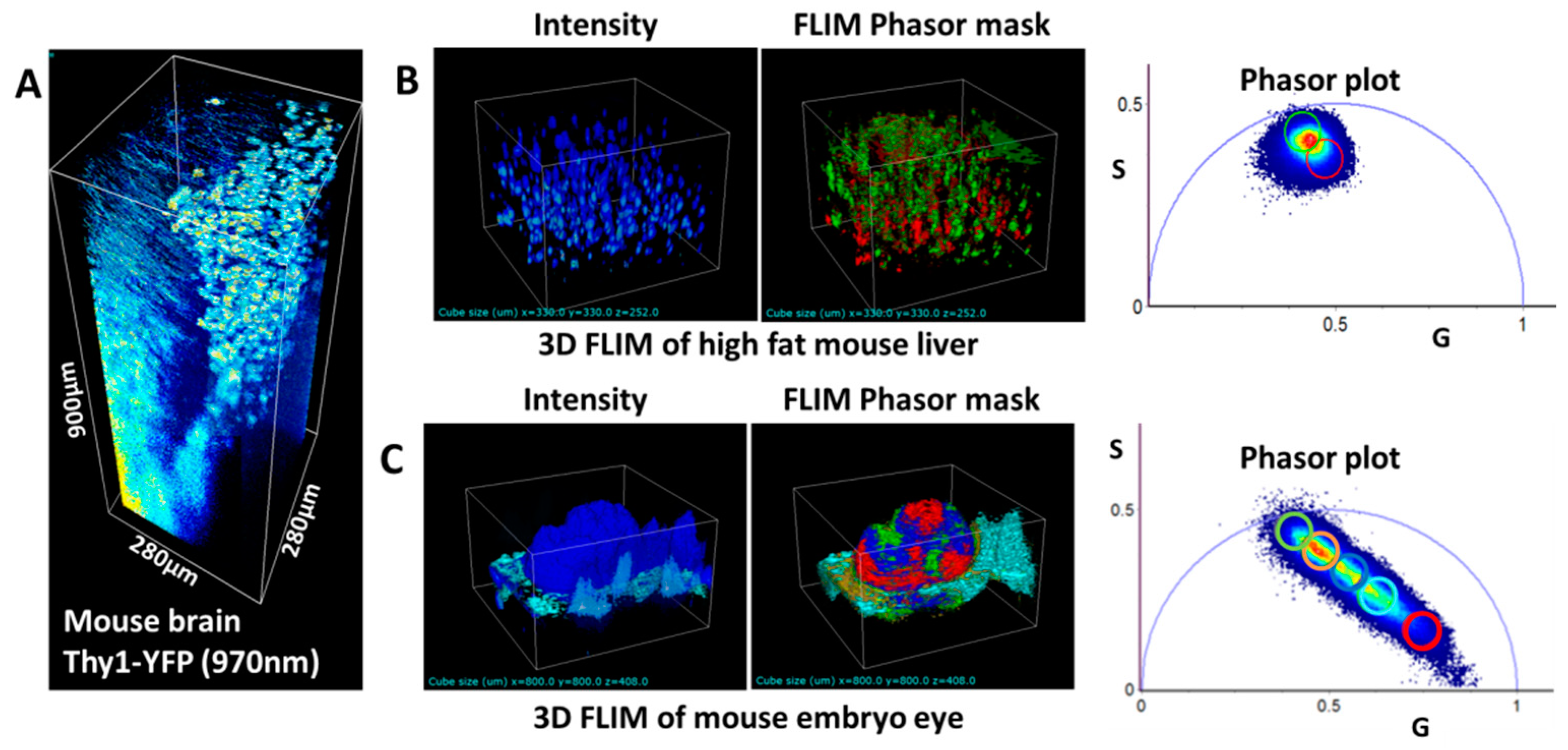
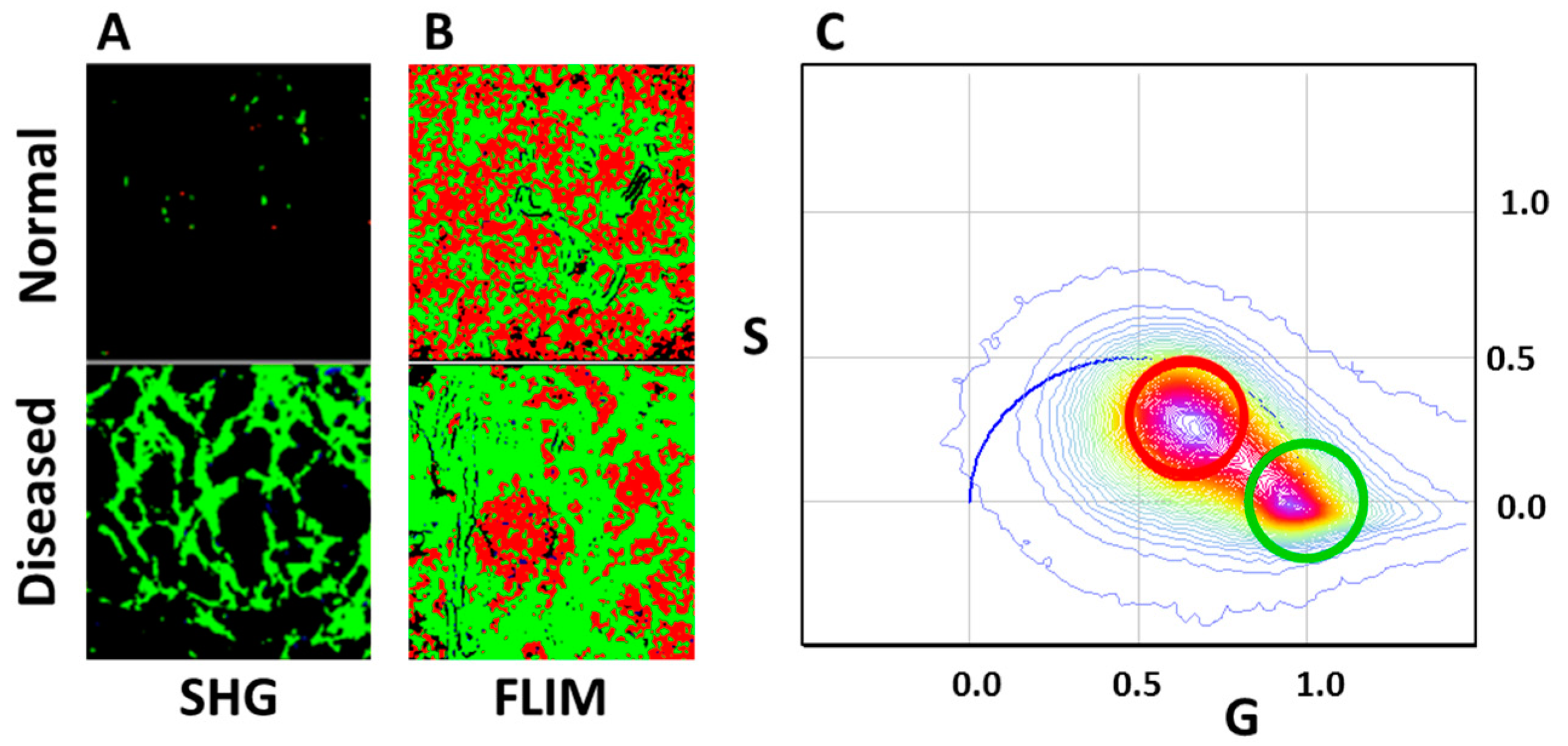
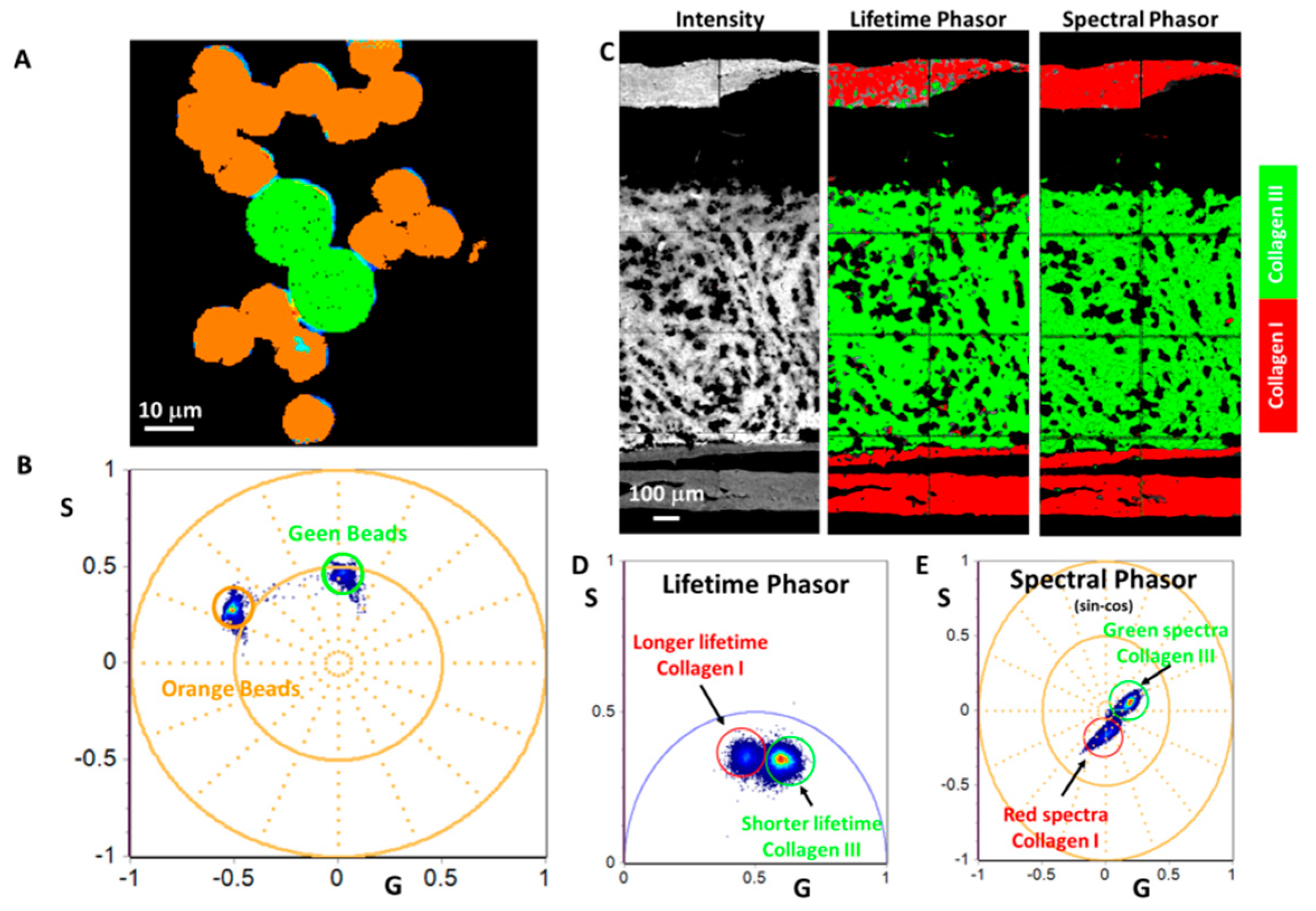
3.2. SHG and THG Imaging
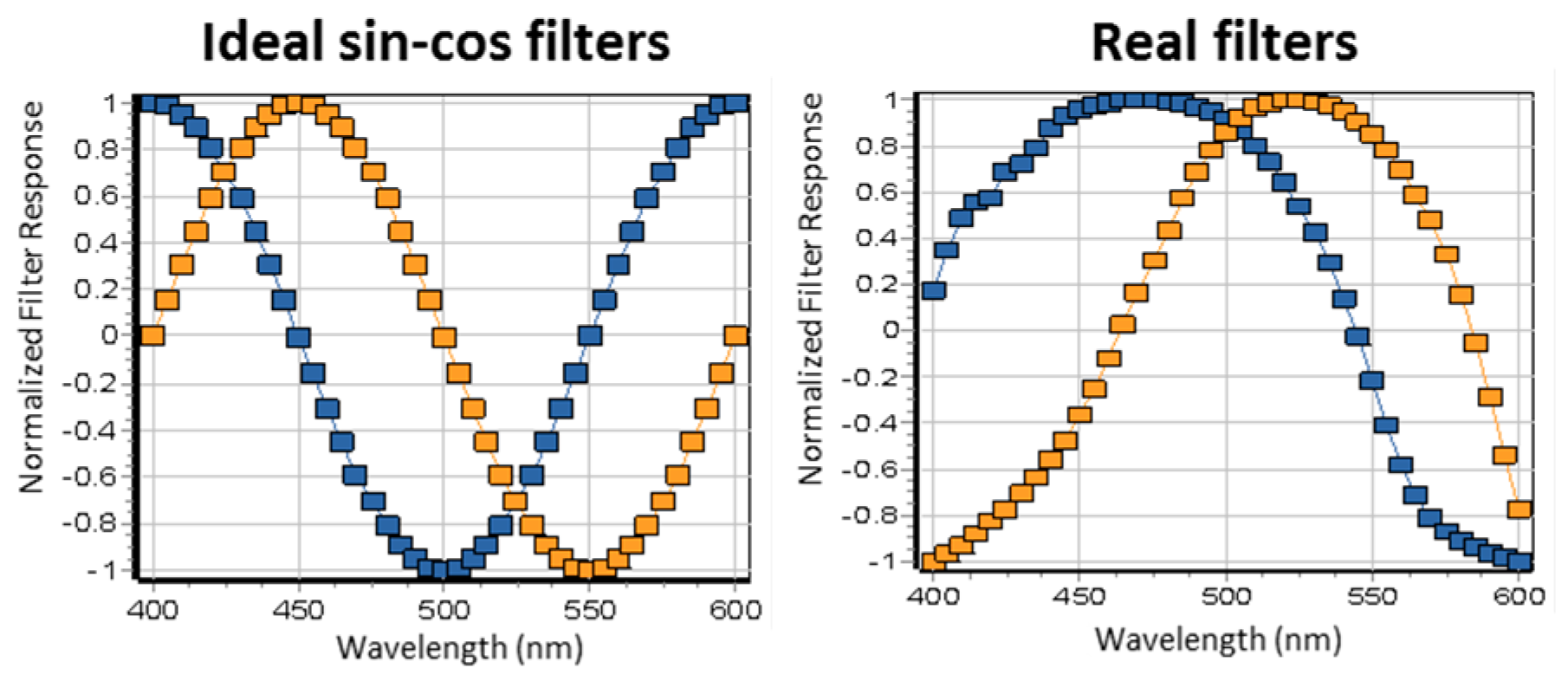
3.3. Hyperspectral Imaging Using Two Filters
4. Discussion
5. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Malley, D. Imaging in Depth: Controversies and Opportunities. In Methods in Cell Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2008; Volume 89, pp. 95–128. [Google Scholar]
- Theer, P.; Hasan, M.T.; Denk, W. Two-photon imaging to a depth of 1000 µm in living brains by use of a Ti:Al2O3 regenerative amplifier. Opt. Lett. 2007, 28, 1022–1024. [Google Scholar] [CrossRef]
- Kobat, D.; Durst, M.E.; Nishimura, N.; Wong, A.W.; Schaffer, C.B.; Xu, C. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt. Express 2009, 17, 13354–13364. [Google Scholar] [CrossRef] [PubMed]
- Cicchi, R.; Pavone, F.S.; Massi, D.; Sampson, D.D. Contrast and depth enhancement in two-photon microscopy of human skin ex vivo by use of optical clearing agents. Opt. Express 2005, 13, 2337–2344. [Google Scholar] [CrossRef]
- Riley, J.D.; Knutson, J.R.; Combs, C.A.; Smirnov, A.V.; Balaban, R.S.; Gandjbakhche, A.H. Optimization of multiphoton excitation microscopy by total emission detection using a parabolic light reflector. J. Microsc. 2007, 228, 330–337. [Google Scholar]
- Crosignani, V.; Dvornikov, A.S.; Gratton, E. Enhancement of imaging depth in turbid media using a wide area detector. J. Biophotonics 2011, 4, 592–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosignani, V.; Dvornikov, A.; Aguilar, J.S.; Stringari, C.; Edwards, R.; Mantulin, W.W.; Gratton, E. Deep tissue fluorescence imaging and in vivo biological applications. J. Biomed. Opt. 2012, 17, 116023. [Google Scholar] [CrossRef] [PubMed]
- Crosignani, V.; Jahid, S.; Dvornikov, A.S.; Gratton, E. A deep tissue fluorescence imaging system with enhanced SHG detection capabilities. Microsc. Res. Tech. 2014, 77, 368–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvornikov, A.; Gratton, E. Imaging in turbid media: A transmission detector gives 2-3 order of magnitude enhanced sensitivity compared to epi-detection schemes. Biomed. Opt. Express 2016, 7, 3747–3755. [Google Scholar] [CrossRef]
- Campagnola, P.J.; Dong, C.Y. Second harmonic generation microscopy: Principles and applications to disease diagnosis. Laser Photonics Rev. 2011, 5, 13–26. [Google Scholar] [CrossRef]
- Barad, Y.; Eisenberg, H.; Horowitz, M.; Silberberg, Y. Nonlinear scanning laser microscopy by third harmonic generation. Appl. Phys. Lett. 1997, 70, 922–924. [Google Scholar] [CrossRef] [Green Version]
- Zipfel, W.R.; Williams, R.M.; Webb, W.W. Nonlinear magic: Multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Tserevelakis, G.J.; Megalou, E.V.; Filippidis, G.; Petanidou, B.; Fotakis, C.; Tavernarakis, N. Label-free imaging of lipid depositions in C. elegans using third-harmonic generation microscopy. PLoS ONE 2014, 9, e84431. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Raggio, C.; Campagnola, P.J. Second-harmonic generation circular dichroism studies of osteogenesis imperfecta. Opt. Lett. 2012, 37, 3837–3839. [Google Scholar] [CrossRef] [PubMed]
- Nadiarnykh, O.; LaComb, R.B.; Brewer, M.A.; Campagnola, P.J. Alterations of the ECM in Ovarian Carcinogenesis Studied by Second Harmonic Generation Imaging Microscopy. BMC Cancer 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Ajeti, V.; Nadiarnykh, O.; Ponik, S.M.; Keely, P.J.; Eliceiri, K.W.; Campagnola, P.J. Structural changes in mixed Col I/Col V collagen gels probed by SHG microscopy: Implications for probing stromal alterations in human breast cancer. Biomed. Opt. Express 2011, 2, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, S.V.; Kenny, A.M.; Walsh, S.J.; Zubrowski, B.; Joseph, C.; Scranton, V.L.; Kuchel, G.A.; Dauser, D.; Xu, M.; Pilbeam, C.C.; et al. Measurement of muscle disease by quantitative second-harmonic generation imaging. J. Biomed. Opt. 2008, 13, 044018. [Google Scholar] [CrossRef]
- Ranjit, S.; Dvornikov, A.; Stakic, M.; Hong, S.H.; Levi, M.; Evans, R.M.; Gratton, E. Imaging Fibrosis and Separating Collagens using Second Harmonic Generation and Phasor Approach to Fluorescence Lifetime Imaging. Sci. Rep. 2015, 5, 13378. [Google Scholar] [CrossRef]
- Ranjit, S.; Dvornikov, A.; Levi, M.; Furgeson, S.; Gratton, E. Characterizing fibrosis in UUO mice model using multiparametric analysis of phasor distribution from FLIM images. Biomed. Opt. Express 2016, 7, 3519–3530. [Google Scholar] [CrossRef]
- Aguilar-Arnal, L.; Ranjit, S.; Stringari, C.; Orozco-Solis, R.; Gratton, E.; Sassone-Corsi, P. Spatial dynamics of SIRT1 and the subnuclear distribution of NADH species. Proc. Natl. Acad. Sci. USA 2016, 113, 12715–12720. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Digman, M.A.; Malacrida, L.; Gratton, E. Measurements of absolute concentrations of NADH in cells using the phasor FLIM method. Biomed. Opt. Express 2016, 7, 2441–2452. [Google Scholar] [CrossRef] [Green Version]
- Bastiaens, P.I.H.; Squire, A. Fluorescence lifetime imaging microscopy: Spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999, 9, 48–52. [Google Scholar] [CrossRef]
- Van Munster, E.B.; Gadella, T.W.J. Suppression of photobleaching-induced artifacts in frequency-domain FLIM by permutation of the recording order. Cytom. Part A 2004, 58, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.; Bergmann, A.; Hink, M.A.; König, K.; Benndorf, K.; Biskup, C. Fluorescence Lifetime Imaging by Time-Correlated Single-Photon Counting. Microsc. Res. Tech. 2004, 63, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Jameson, D.M.; Gratton, E.; Hall, R.D. The Measurement and Analysis of Heterogeneous Emissions by Multifrequency Phase and Modulation Fluorometry. Appl. Spectrosc. Rev. 1984, 20, 55–106. [Google Scholar] [CrossRef] [Green Version]
- Digman, M.A.; Caiolfa, V.R.; Zamai, M.; Gratton, E. The Phasor Approach to Fluorescence Lifetime Imaging Analysis. Biophys. J. 2008, 94, L14–L16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fereidouni, F.; Bader, A.N.; Gerritsen, H.C. Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 2012, 20, 12729–12741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malacrida, L.; Astrada, S.; Briva, A.; Bollati-Fogolín, M.; Gratton, E.; Bagatolli, L.A. Spectral phasor analysis of LAURDAN fluorescence in live A549 lung cells to study the hydration and time evolution of intracellular lamellar body-like structures. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 2625–2635. [Google Scholar] [CrossRef]
- Malacrida, L.; Gratton, E.; Jameson, D.M. Model-free methods to study membrane environmental probes: A comparison of the spectral phasor and generalized polarization approaches. Methods Appl. Fluoresc. 2015, 3, 047001. [Google Scholar] [CrossRef] [PubMed]
- Malacrida, L.; Gratton, E. LAURDAN fluorescence and phasor plots reveal the effects of a H2O2 bolus in NIH-3T3 fibroblast membranes dynamics and hydration. Free Radic. Biol. Med. 2018, 128, 144–156. [Google Scholar] [CrossRef]
- Sameni, S.; Malacrida, L.; Tan, Z.; Digman, M.A. Alteration in Fluidity of Cell Plasma Membrane in Huntington Disease Revealed by Spectral Phasor Analysis. Sci. Rep. 2018, 8, 734. [Google Scholar] [CrossRef] [Green Version]
- Dvornikov, A.; Gratton, E. Hyperspectral imaging in highly scattering media by the spectral phasor approach using two filters. Biomed. Opt. Express 2018, 9, 4833–4840. [Google Scholar] [CrossRef] [PubMed]
- Ayers, F.; Grant, A.; Kuo, D.; Cuccia, D.J.; Durkin, A.J. Fabrication and characterization of silicone-based tissue phantoms with tunable optical properties in the visible and near infrared domain. Proc. SPIE 2008, 6870. [Google Scholar] [CrossRef]
- Ranjit, S.R.; Dvornikov, A.; Dobrinskikh, E.; Wang, X.; Luo, Y.; Levi, M.; Gratton, E. Measuring the effect of a Western diet on liver tissue architecture by FLIM autofluorescence and harmonic generation microscopy. Biomed. Opt. Express 2017, 8, 371–378. [Google Scholar]
- Negrean, A.; Mansvelder, H.D. Optimal lens design and use in laser-scanning microscopy. Biomed. Opt. Express 2014, 5, 1588–1609. [Google Scholar] [CrossRef] [PubMed]
- Suhalim, J.L.; Chung, C.Y.; Lilledahl, M.B.; Lim, R.S.; Levi, M.; Tromberg, B.J.; Potma, E.O. Characterization of cholesterol crystals in atherosclerotic plaques using stimulated Raman scattering and second-harmonic generation microscopy. Biophys. J. 2012, 102, 1988–1995. [Google Scholar] [CrossRef]
- Ranjit, S.; Dobrinskikh, E.; Montford, J.; Dvornikov, A.; Lehman, A.; Orlicky, D.J.; Nemenoff, R.; Gratton, E.; Levi, M.; Furgeson, S. Label-free fluorescence lifetime and second harmonic generation imaging microscopy improves quantification of experimental renal fibrosis. Kidney Int. 2016, 90, 1123–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjin, G.; Xu, P.; Kable, S.H.; Kable, E.P.W.; Burgess, J.K. Quantification of collagen I in airway tissues using second harmonic generation. J. Biomed. Opt. 2014, 19, 36005. [Google Scholar] [CrossRef]
- Sun, C.-K.; Yu, C.-H.; Tai, S.-P.; Kung, C.-T.; Wang, I.-J.; Yu, H.-C.; Huang, H.-J.; Lee, W.-J.; Chan, Y.-F. In vivo and ex vivo imaging of intra-tissue elastic fibers using third-harmonic-generation microscopy. Opt. Express 2007, 15, 11167–11177. [Google Scholar]
- Débarre, D.; Supatto, W.; Pena, A.M.; Fabre, A.; Tordjmann, T.; Combettes, L.; Schanne-Klein, M.C.; Beaurepaire, E. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat. Methods 2006, 3, 47–53. [Google Scholar] [CrossRef]
- Theer, P.; Denk, W. On the fundamental imaging-depth limit in two-photon microscopy. J. Opt. Soc. Am. A 2006, 23, 3139–3149. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dvornikov, A.; Malacrida, L.; Gratton, E. The DIVER Microscope for Imaging in Scattering Media. Methods Protoc. 2019, 2, 53. https://doi.org/10.3390/mps2020053
Dvornikov A, Malacrida L, Gratton E. The DIVER Microscope for Imaging in Scattering Media. Methods and Protocols. 2019; 2(2):53. https://doi.org/10.3390/mps2020053
Chicago/Turabian StyleDvornikov, Alexander, Leonel Malacrida, and Enrico Gratton. 2019. "The DIVER Microscope for Imaging in Scattering Media" Methods and Protocols 2, no. 2: 53. https://doi.org/10.3390/mps2020053
APA StyleDvornikov, A., Malacrida, L., & Gratton, E. (2019). The DIVER Microscope for Imaging in Scattering Media. Methods and Protocols, 2(2), 53. https://doi.org/10.3390/mps2020053



