Impacts of Sourdough Technology on the Availability of Celiac Peptides from Wheat α- and γ-Gliadins: In Silico Approach
Abstract
1. Introduction
2. Materials and Methods
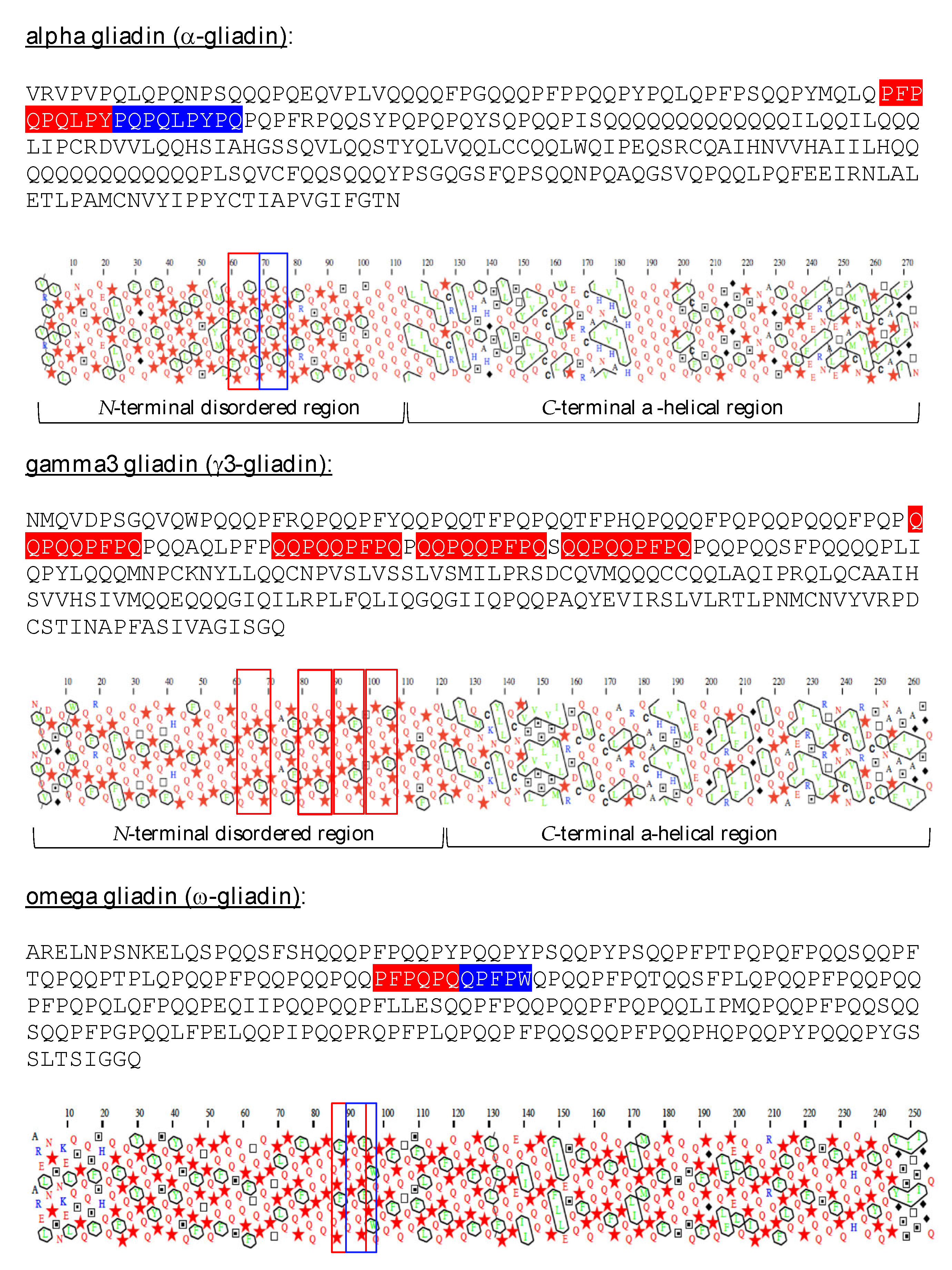
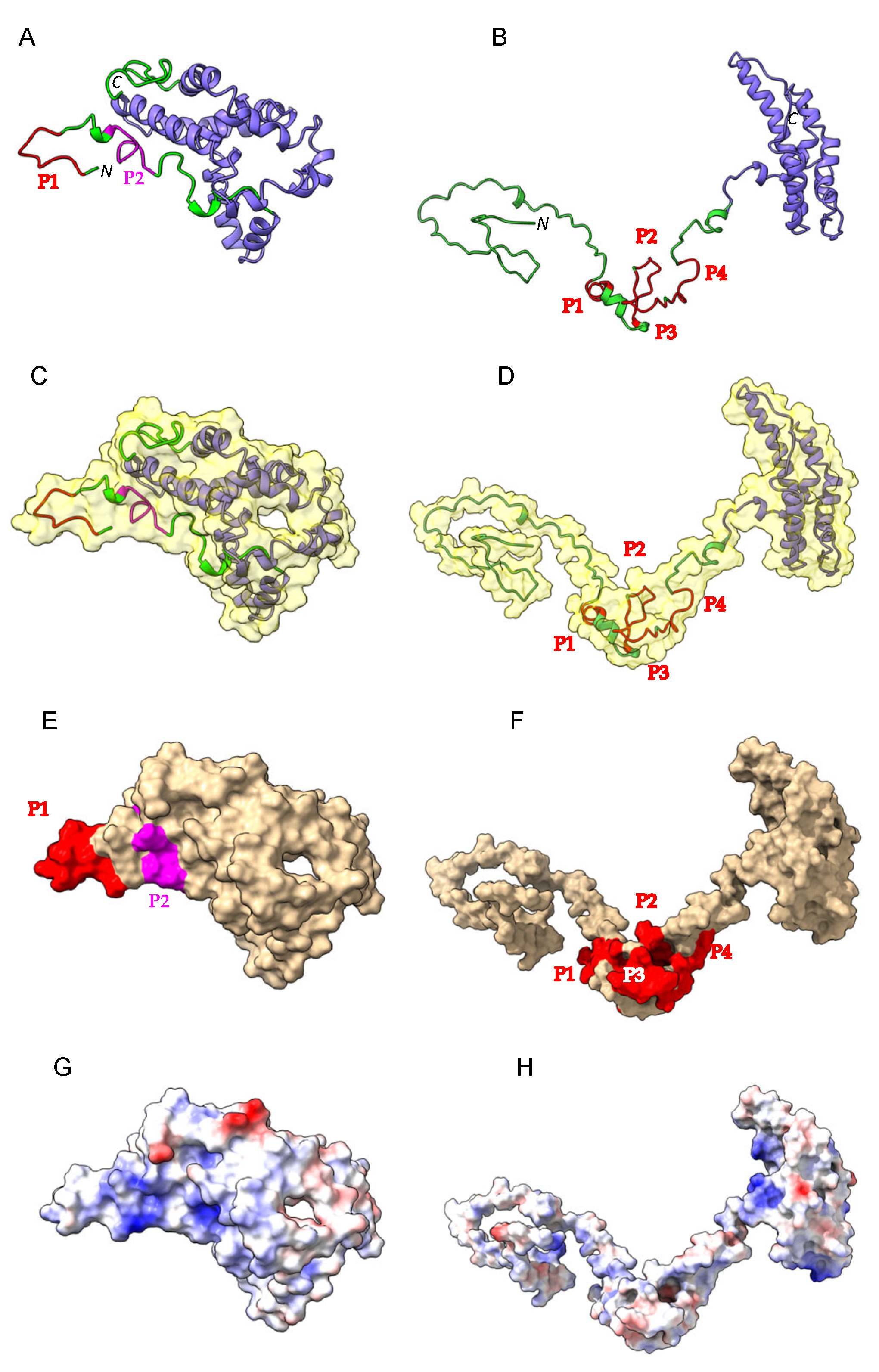
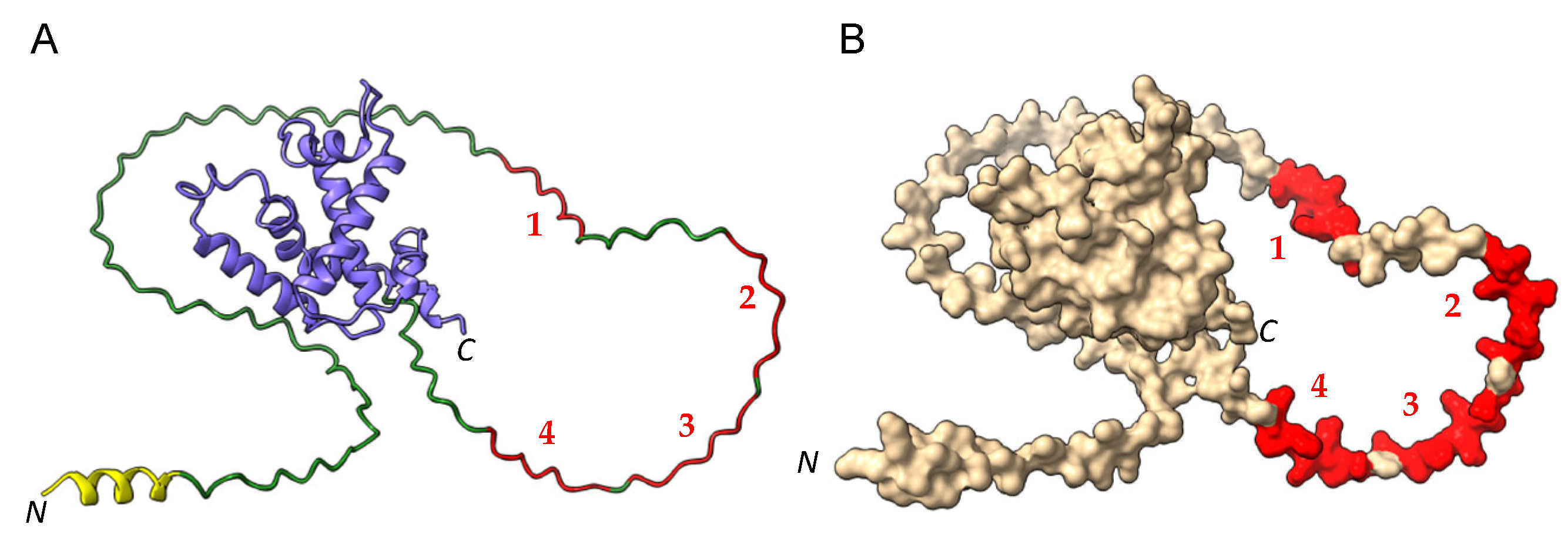
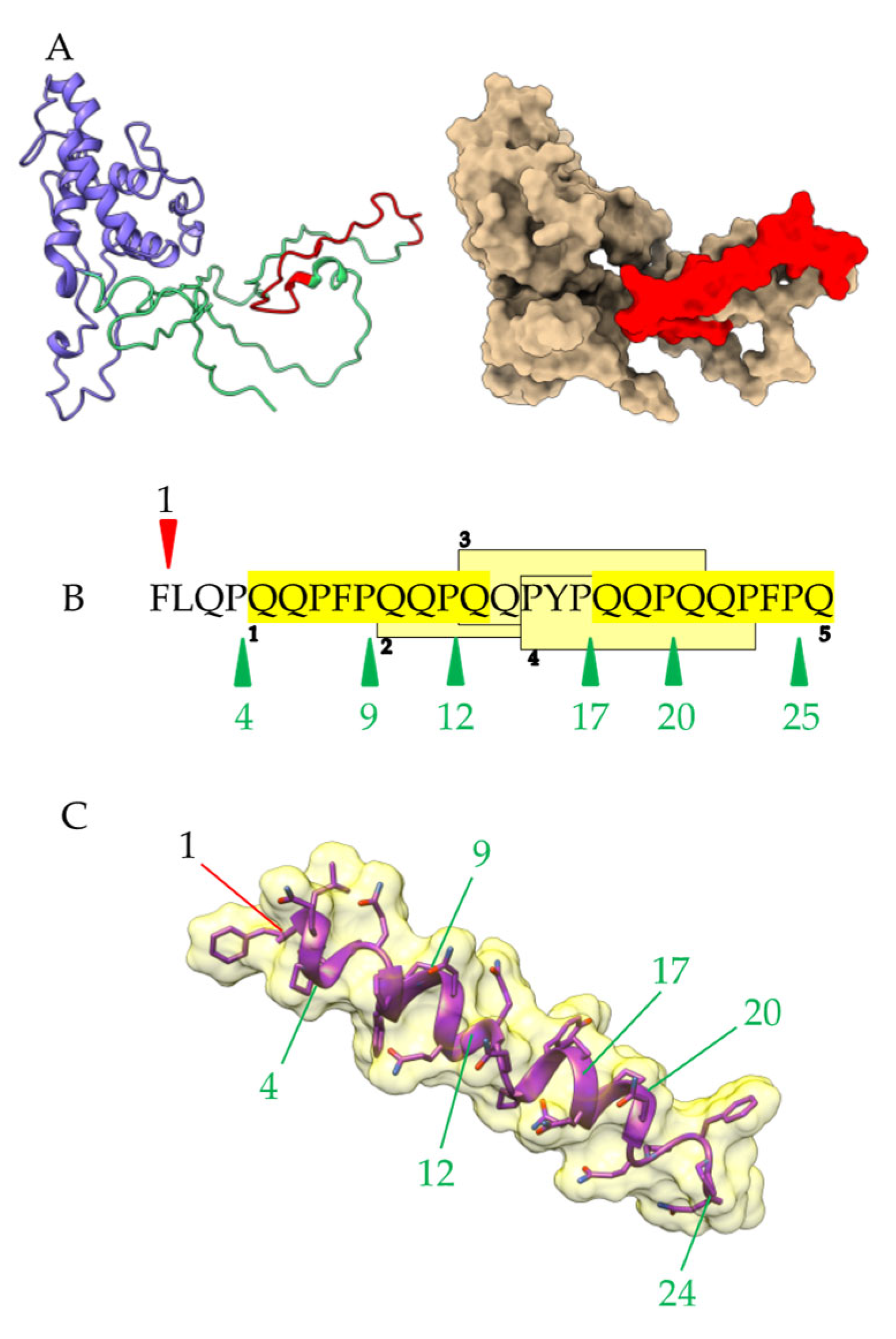
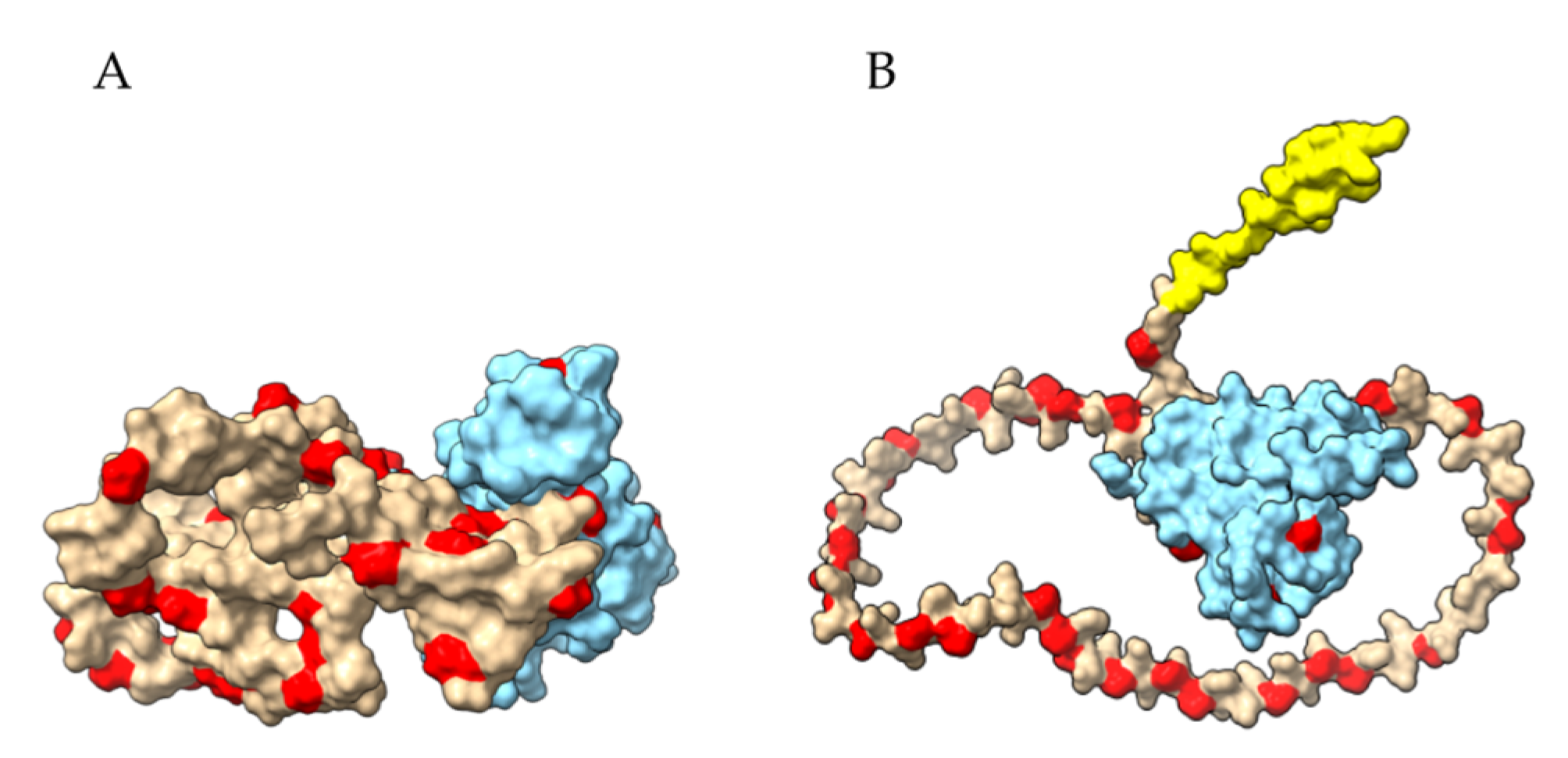
2.1. Three-Dimensional Structure of α- and γ-Gliadin
2.2. Proteolysis of α- and γ3-Gliadin by Digestive Proteases
2.3. Proteolysis of α- and γ3-Gliadin by Sourdough Proteases
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raiteri, A.; Granito, A.; Giamperoli, A.; Catenaro, T.; Negrini, G.; Tovoli, F. Current guidelines for the management of celiac disease: A systematic review with comparative analysis. World J. Gastroenterol. 2022, 28, 154–176. [Google Scholar] [CrossRef] [PubMed]
- Standard for Foods for Special Dietary Use for Persons Intolerant to Gluten. CXS 118-1979, Adopted in 1979. Available online: http://www.fao.org/fao-who-codexalimentarius (accessed on 20 July 2022).
- Barak, S.; Mudgil, D.; Khatkar, B.S. Biochemical and Functional Properties of Wheat Gliadins: A Review. Crit. Rev. Food Sci. Nutr. 2015, 55, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Sahli, L.; Boire, A.; Solé-Jamault, V.; Rogniaux, H.; Giuliani, A.; Roblin, P.; Renard, D. New exploration of the γ-gliadin structure through its partial hydrolysis. Int. J. Biol. Macromol. 2020, 165, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Matsuo, H.; Chinuki, Y.; Takahashi, H.; Dahlström, J.; Tanaka, A. Food-dependent exercise-induced anaphylaxis—Importance of omega-5 gliadin and HMW-glutenin as causative antigens for wheat-dependent exercise-induced anaphylaxis. Allergol. Int. 2009, 58, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Thiele, C.; Grassl, S.; Gänzle, M. Gluten Hydrolysis and Depolymerization during Sourdough Fermentation. J. Agric. Food Chem. 2004, 52, 1307–1314. [Google Scholar] [CrossRef]
- Vermeulen, N.; Pavlovic, M.; Ehrmann, M.A.; Gänzle, M.G.; Vogel, R.F. Functional Characterization of the Proteolytic System of Lactobacillus sanfranciscensis DSM 20451 T during Growth in Sourdough. Appl. Environ. Microbiol. 2005, 71, 6260–6266. [Google Scholar] [CrossRef]
- Mandile, R.; Picascia, S.; Parrella, C.; Camarca, A.; Gobbetti, M.; Greco, L.; Troncone, R.; Gianfrani, C.; Auricchio, R. Lack of immunogenicity of hydrolysed wheat flour in patients with coeliac disease after a short-term oral challenge. Aliment. Pharmacol. Ther. 2017, 46, 440–446. [Google Scholar] [CrossRef]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural Basis for Gluten Intolerance in Celiac Sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef]
- Shan, L.; Qiao, S.-W.; Arentz-Hansen, H.; Molberg, Ø.; Gray, G.M.; Sollid, L.M.; Khosla, C. Identification and Analysis of Multivalent Proteolytically Resistant Peptides from Gluten: Implications for Celiac Sprue. J. Proteome Res. 2005, 4, 1732–1741. [Google Scholar] [CrossRef]
- Coto, L.; Mendia, I.; Sousa, C.; Bai, J.C.; Cebolla, A. Determination of gluten immunogenic peptides for the management of the treatment adherence of celiac disease: A systematic review. World J. Gastroenterol. 2021, 27, 6306–6321. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and listing of celiac disease-related gluten epitopes recognized by CD4+ T cells. Immunogenetics 2020, 72, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Gaboriaud, C.; Bissery, V.; Benchetrit, T.; Mornon, J.P. Hydrophobic cluster analysis: An efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987, 224, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, I.; Labesse, G.; Durand, P.; Poupon, A.; Canard, L.; Chomilier, J.; Henrissat, B.; Mornon, J.P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): Current status and perspectives. Cell. Mol. Life Sci. 1997, 53, 621–645. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Gourinath, S.; Alam, N.; Srinivasan, A.; Betzel, C.; Singh, T.P. Structure of the bifunctional inhibitor of trypsin and α-amylase from ragi seeds at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2000, 56, 287–293. [Google Scholar] [CrossRef]
- Strobl, S.; Maskos, K.; Wiegand, G.; Huber, R.; Gomis-Rüth, F.X.; Glockshuber, R. A novel strategy for inhibition of α-amylases: Yellow meal worm α-amylase in complex with the Ragi bifunctional inhibitor at 2.5 Å resolution. Structure 1998, 6, 911–921. [Google Scholar] [CrossRef]
- Oda, Y.; Matsunaga, T.; Fukuyama, K.; Miyazaki, T.; Morimoto, T. Tertiary and quaternary structures of 0.19 α-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 Å resolution. Biochemistry 1997, 36, 13503–13511. [Google Scholar] [CrossRef]
- Møller, M.S.; Vester-Christensen, M.B.; Jensen, J.M.; Hachem, M.A.; Henriksen, A.; Svensson, B. Crystal Structure of Barley Limit Dextrinase-Limit Dextrinase Inhibitor (LD-LDI) Complex Reveals Insights into Mechanism and Diversity of Cereal Type Inhibitors. J. Biol. Chem. 2015, 290, 12614–12629. [Google Scholar] [CrossRef]
- Mueller, G.A.; Gosavi, R.A.; Pomés, A.; Wünschmann, S.; Moon, A.F.; London, R.E.; Pedersen, L.C. Ara h 2: Crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy 2011, 66, 878–885. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017, 24, 954–964. [Google Scholar] [CrossRef]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. Author Correction: 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2020, 27, 683. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Quarsten, H.; Bergseng, E.; Khosla, C.; Sollid, L.M. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. USA 2004, 101, 4175–4179. [Google Scholar] [CrossRef]
- Broughton, S.E.; Petersen, J.; Theodossis, A.; Scally, S.W.; Loh, K.L.; Thompson, A.; van Bergen, J.; Kooy-Winkelaar, Y.; Henderson, K.N.; Beddoe, T.; et al. Biased T Cell Receptor Usage Directed against Human Leukocyte Antigen DQ8-Restricted Gliadin Peptides Is Associated with Celiac Disease. Immunity 2012, 37, 611–621. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Gobbetti, M.; Rizzello, C.G.; Di Cagno, R.; De Angelis, M. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol. 2014, 37, 30–40. [Google Scholar] [CrossRef]
- Ogilvie, O.J.; Gerrard, J.A.; Roberts, S.; Sutton, K.H.; Larsen, N.; Domigan, L.J. A Case Study of the Response of Immunogenic Gluten Peptides to Sourdough Proteolysis. Nutrients 2021, 13, 1906. [Google Scholar] [CrossRef]
- Shan, L.; Marti, T.; Sollid, L.M.; Gray, G.M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for celiac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Di Cagno, R.; Camarca, A.; Silano, M.; Losito, I.; De Vincenzi, M.; De Bari, M.D.; Palmisano, F.; Maurano, F.; et al. Highly Efficient Gluten Degradation by Lactobacilli and Fungal Proteases during Food Processing: New Perspectives for Celiac Disease. Appl. Environ. Microbiol. 2007, 73, 4499–4507. [Google Scholar] [CrossRef]
- Stepniak, D.; Spaenij-Dekking, L.; Mitea, C.; Moester, M.; De Ru, A.; Baak-Pablo, R.; van Veelen, P.; Edens, L.; Koning, F. Highly efficient gluten degradation with a newly identified prolyl endoprotease: Implications for celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G621–G629. [Google Scholar] [CrossRef]
- De Angelis, M.; Cassone, A.; Rizzello, C.G.; Gagliardi, F.; Minervini, F.; Calasso, M.; Di Cagno, R.; Francavilla, R.; Gobbetti, M. Mecanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl. Environ. Microbiol. 2010, 76, 508–518. [Google Scholar] [CrossRef]
- Heredia-Sandoval, N.G.; de la Barca, A.M.C.; Islas-Rubio, A.R. Gluten degradation in wheat flour with Aspergillus niger prolyl-endopeptidase to prepare a gluten-reduced bread supplemented with an amaranth blend. J. Cereal Sci. 2016, 71, 73–77. [Google Scholar] [CrossRef]
- Shetty, R.; Vestergaard, M.; Jessen, F.; Hägglund, P.; Knorr, V.; Koehler, P.; Prakash, H.S.; Hobley, T.J. Discovery, cloning and characterization of proline specific prolyl endopeptidase, a gluten degrading thermos-stable enzyme from Sphaerobacter thermophiles. Enzym. Microb. Technol. 2017, 107, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Sakandar, H.A.; Usman, K.; Imran, M. Isolation and characterization of gluten-degrading Enterococcus mundtii and Wickerhamomyces anomalus, potential probiotic strains from indigenously fermented sourdough (Khamir). LWT 2018, 91, 271–277. [Google Scholar] [CrossRef]
- Yu, J.; Wu, J.; Xie, D.; Du, L.; Tang, Y.-J.; Xie, J.; Wei, D. Characterization and rational design for substrate specificity of a prolyl endopeptidase from Stenotrophomonas maltophilia. Enzym. Microb. Technol. 2020, 138, 109548. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.S.; Bersanetti, P.A.; de Araújo Morandim-Giannetti, A. Peptidases production by fungi obtained from Manihot esculenta Crantz waste and its application in gluten hydrolysis. Biocat. Agric. Biotechnol. 2021, 37, 102184. [Google Scholar] [CrossRef]
- Montserrat, V.; Bruins, M.J.; Edens, L.; Koning, F. Influence of dietary components on Aspergillus niger prolyl endoprotease mediated gluten degradation. Food Chem. 2015, 174, 440–445. [Google Scholar] [CrossRef]
- Di Cagno, R.; De Angelis, M.; Auricchio, S.; Greco, L.; Clarke, C.; De Vincenzi, M.; Giovannini, C.; D’Archivio, M.; Landolfo, F.; Parrilli, G.; et al. Sourdough Bread Made from Wheat and Nontoxic Flours and Started with Selected Lactobacilli Is Tolerated in Celiac Sprue Patients. Appl. Environ. Microbiol. 2004, 70, 1088–1096. [Google Scholar] [CrossRef]
- Di Cagno, R.; Barbato, M.; Di Camillo, C.; Rizzello, C.G.; De Angelis, M.; Giuliani, G.; De Vincenzi, M.; Gobbetti, M.; Cucchiara, S. Gluten-free Sourdough Wheat Baked Goods Appear Safe for Young Celiac Patients: A Pilot Study. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 777–783. [Google Scholar] [CrossRef]
- Calasso, M.; Vincentini, O.; Valitutti, F.; Felli, C.; Gobbetti, M.; Di Cagno, R. The sourdough fermentation may enhance the recovery from intestinal inflammation of coeliac patients at the early stage of the gluten-free diet. Eur. J. Nutr. 2012, 51, 507–512. [Google Scholar] [CrossRef]
- Jouanin, A.; Gilissen, L.J.W.J.; Boyd, L.A.; Cockram, J.; Leigh, F.J.; Wallington, E.J.; van den Broeck, H.C.; van der Meer, I.M.; Schaart, J.G.; Visser, R.G.F.; et al. Food processing and breeding strategies for coeliac-safe and healthy wheat products. Food Res. Int. 2018, 110, 11–21. [Google Scholar] [CrossRef]
- García-Molina, M.D.; Giménez, M.J.; Sánchez-León, S.; Barro, F. Gluten free wheat: Are we there? Nutrients 2019, 11, 487. [Google Scholar] [CrossRef] [PubMed]
- Jouanin, A.; Gilissen, L.J.; Schaart, J.G.; Leigh, F.J.; Cockram, J.; Wallington, E.J.; Boyd, L.A.; van den Broeck, H.C.; van der Meer, I.M.; America, A.H.P.; et al. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure—Reviewing methods to screen for coeliac safety. Front. Nutr. 2020, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Gil Humanes, J.; Piston, F.; Hernando, A.; Alvarez, J.B.; Shewry, P.R.; Barro, F. Silencing of γ-gliadins by RNA interference (RNAi) in bread wheat. J. Cereal Sci. 2008, 48, 565–568. [Google Scholar] [CrossRef]
- Becker, D.; Wieser, H.; Koehler, P.; Folck, A.; Mühling, K.H.; Zörb, C. Protein composition and techno-functional properties of transgenic wheat with reduced a-gliadin content obtained by RNA interference. J. Appl. Bot. Food Qual. 2012, 85, 23. [Google Scholar]
- Altenbach, S.B.; Chang, H.C.; Yu, X.B.; Seabourn, B.W.; Green, P.H.; Alaedini, A. Elimination of omega-1,2 gliadins from breade wheat (Triticum aestivum) flour: Effects of immunogenic potential and end-use quality. Front. Plant Sci. 2019, 10, 580. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Pistón, F.; Tollefsen, S.; Sollid, L.M.; Barro, F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc. Natl. Acad. Sci. USA 2010, 107, 17023–17028. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-León, S.; Gil Humanes, J.; Ozuna, C.V.; Gimenez, M.J.; Sousa, C.; Voytas, D.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Jouanin, A.; Schaart, J.G.; Boyd, L.A.; Cockram, J.; Leigh, F.J.; Bates, R.; Wallington, E.J.; Visser, R.G.F.; Smulders, M.J.M. Ourlook for coeliac disease patients: Towards bread wheat with hypoimmunogenic gluten by gene editing of α- and γ-gliadin gene families. BMC Plant Biol. 2019, 19, 333. [Google Scholar] [CrossRef]
- Jouanin, A.; Born, T.; Boyd, L.A.; Cockram, J.; Leigh, F.; Santos, B.A.C.M.; Visser, R.G.F.; Smulders, M.J.M. Development of the GlutEnSeq capture system for sequencing gluten gene families in hexaploidy bread wheat with deletions and mutations induced by γ-irradiation of CRISPR/Cas9. J. Cereal Sci. 2019, 88, 157–166. [Google Scholar] [CrossRef]
- Jouanin, A.; Tenorio-Berrio, R.; Schaart, J.G.; Leigh, F.; Visser, R.G.; Smulders, M.J. Optimization of droplet digital PCR for determining copy number variation of α-gliadin genes in mutant and gene-dited polyploid bread wheat. J. Cereal Sci. 2020, 92, 102903. [Google Scholar] [CrossRef]



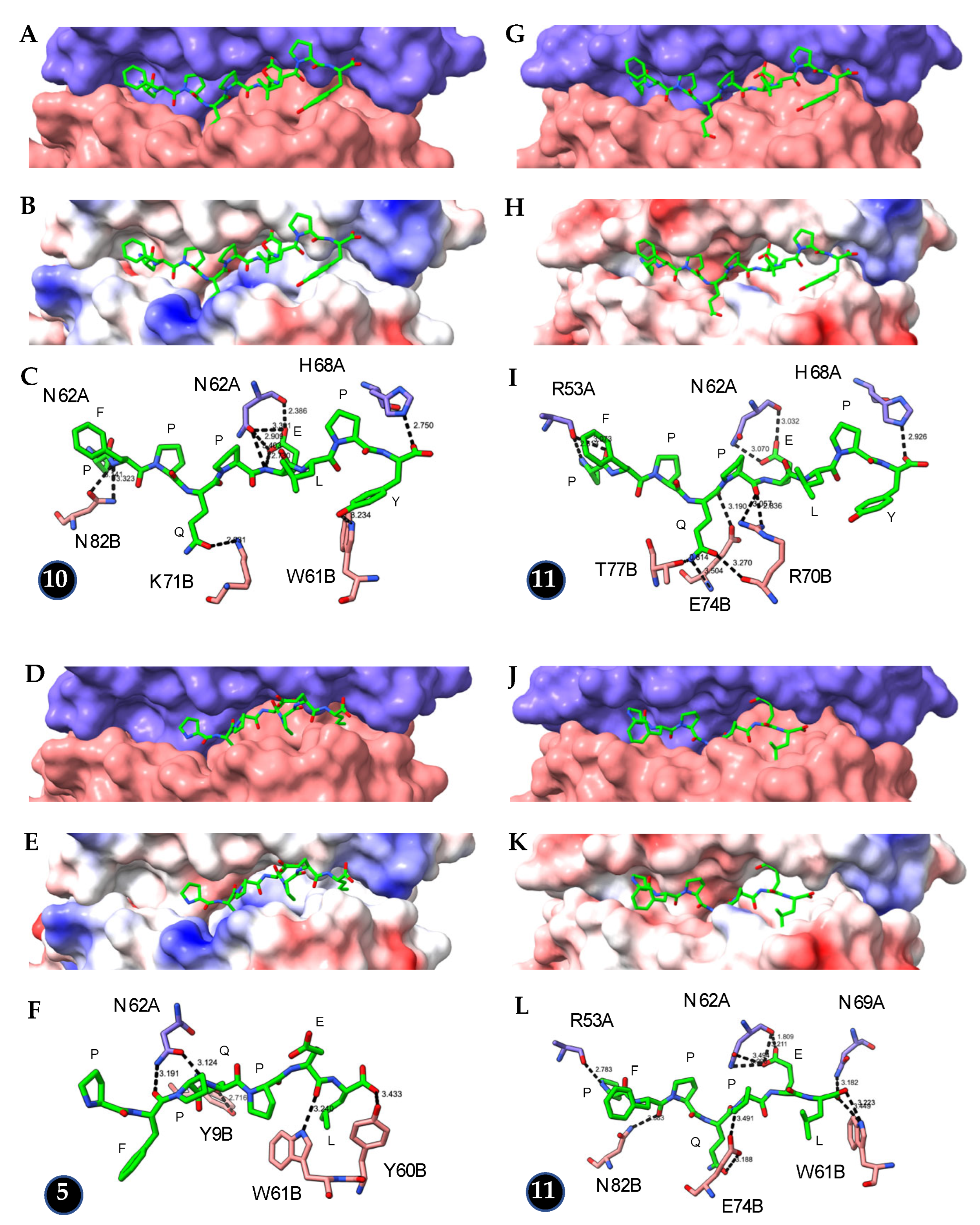



| Peptide | HLA-DQ2/DQ8 | Hydrogen * Bonds | Docking Score |
|---|---|---|---|
| PFPQPELPY | HLA-DQ2 | 10 | 230 |
| PFPQPEL | HLA-DQ2 | 5 | 207 |
| PFPQPELPY | HLA-DQ8 | 11 | 245 |
| PYPQPEL | HLA-DQ8 | 11 | 252 |
| QGSVQPQQL | HLA-DQ2 | 11 | 221 |
| QGSVQPQQ | HLA-DQ2 | 11 | 217 |
| QGSVQPQQL | HLA-DQ8 | 16 | 243 |
| QGSVQPQQ | HLA-DQ8 | 17 | 241 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Benoist, H.; Rougé, P. Impacts of Sourdough Technology on the Availability of Celiac Peptides from Wheat α- and γ-Gliadins: In Silico Approach. Allergies 2023, 3, 39-57. https://doi.org/10.3390/allergies3010004
Barre A, Benoist H, Rougé P. Impacts of Sourdough Technology on the Availability of Celiac Peptides from Wheat α- and γ-Gliadins: In Silico Approach. Allergies. 2023; 3(1):39-57. https://doi.org/10.3390/allergies3010004
Chicago/Turabian StyleBarre, Annick, Hervé Benoist, and Pierre Rougé. 2023. "Impacts of Sourdough Technology on the Availability of Celiac Peptides from Wheat α- and γ-Gliadins: In Silico Approach" Allergies 3, no. 1: 39-57. https://doi.org/10.3390/allergies3010004
APA StyleBarre, A., Benoist, H., & Rougé, P. (2023). Impacts of Sourdough Technology on the Availability of Celiac Peptides from Wheat α- and γ-Gliadins: In Silico Approach. Allergies, 3(1), 39-57. https://doi.org/10.3390/allergies3010004








