Comparative Transcriptome Analyses Reveal Different Regulatory Mechanisms in Ecological Adaptation between Chrysanthemum vestitum and Chrysanthemum mongolicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Morphological Analysis
2.3. RNA Extraction, Library Construction, and Illumina Sequencing
2.4. Sequence Assemble and DEG Analysis
2.5. Validation of DEG Expression
3. Results and Discussion
3.1. SEM Analysis of Leaf Surface Micro-Morphology of C. mongolicum and C. vestitum
3.2. High-Throughput Transcriptome Sequencing and Read Assembly
3.3. Differential Gene Expression Analysis
3.4. Enrichment Analysis of DEG
3.5. Gene Validation and Expression Analysis
3.6. DEGs Involved in Flavonoid Biosynthesis
3.7. Candidate Genes Involved in Terpenoid Biosynthesis
3.8. Candidate Genes Involved in Starch and Sucrose Metabolism Pathway
3.9. Biosynthesis of Unsaturated Fatty Acids
3.10. Candidate Genes Involved in ABC Transporters
3.11. DEGs Involved in Plant-Pathogen Interaction
3.12. DEGs Involved in Phenylpropanoid Biosynthesis
3.13. Transcription Factors Analysis of C. vestitum and C. mongolicum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shih, Z.; Christopher, J.H.; Michael, G.G. Chrysanthemum. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2011; Volume 20, pp. 669–676. [Google Scholar]
- Li, J.; Wan, Q.; Guo, Y.P.; Abbott, R.J.; Rao, G.Y. Should I stay or should I go: Biogeographic and evolutionary history of a polyploid complex (Chrysanthemum indicum complex) in response to Pleistocene climate change in China. New Phytol. 2014, 201, 1031–1044. [Google Scholar] [CrossRef] [PubMed]
- Su, J.S.; Jiang, J.F.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.M.; Chen, F.D. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.P.; Chen, M.M.; Wei, J.X.; Zhao, L.; Liu, P.L.; Dai, S.L.; Wen, J. Origin of Chrysanthemum cultivars—Evidence from nuclear low-copy LFY gene sequences. Biochem. Syst. Ecol. 2016, 65, 129–136. [Google Scholar] [CrossRef]
- Ma, Y.P.; Zhao, L.; Zhang, W.J.; Zhang, Y.H.; Xing, X.; Duan, X.X.; Hu, J.; Harris, A.J.; Liu, P.L.; Dai, S.L.; et al. Origins of cultivars of Chrysanthemum—Evidence from the chloroplast genome and nuclear LFY gene. J. Syst. Evol. 2020, 58, 925–944. [Google Scholar] [CrossRef]
- Song, C.; Liu, Y.F.; Song, A.P.; Dong, G.Q.; Zhao, H.B.; Sun, W.; Ramakrishnan, S.; Wang, Y.; Wang, S.B.; Li, T.Z.; et al. The Chrysanthemum nankingense genome providesinsights into the evolution and diversification of Chrysanthemum flowers and medicinal traits. Mol. Plant 2018, 11, 1482–1491. [Google Scholar] [CrossRef]
- Zhang, W.J.; Xu, H.Y.; Duan, X.X.; Hu, J.; Li, J.J.; Zhao, L.; Ma, Y.P. Characterizing the leaf transcriptome of Chrysanthemum rhombifolium (Ling et C. Shih), a drought resistant, endemic plant from China. Front. Genet. 2021, 12, 625985. [Google Scholar] [CrossRef]
- Wen, X.H.; Li, J.Z.; Wang, L.L.; Lu, C.F.; Gao, Q.; Xu, P.; Pu, Y.; Zhang, Q.L.; Hong, Y.; Hong, L.; et al. The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic. Res. 2022, 9, uhab022. [Google Scholar] [CrossRef]
- Zhao, H.E.; Liu, Z.H.; Hu, X.; Yin, J.L.; Li, W.; Rao, G.Y.; Zhang, X.H.; Huang, C.L.; Anderson, N.; Zhang, Q.X.; et al. Chrysanthemum genetic resources and related genera of Chrysanthemum collected in China. Genet. Resour. Crop Evol. 2009, 56, 937–946. [Google Scholar] [CrossRef]
- Zhao, H.B.; Chen, F.D.; Chen, S.M.; Wu, G.S.; Guo, W.M. Molecular phylogeny of Chrysanthemum, Ajania and its allies (Anthemideae, Asteraceae) as inferred from nuclear ribosomal ITS and chloroplast trnL-F IGS sequences. Plant Syst. Evol. 2010, 284, 153–169. [Google Scholar] [CrossRef]
- Li, J.; Wan, Q.; Abbott, R.J.; Rao, G.Y. Geographical distribution of cytotypes in the Chrysanthemum indicum complex as evidenced by ploidy level and genome-size variation. J. Syst. Evol. 2013, 51, 196–204. [Google Scholar] [CrossRef]
- Parkinson, J.; Blaxter, M. Expressed sequence tags: An overview. Methods Mol. Biol. 2009, 533, 1–12. [Google Scholar]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Wu, Q.J.; Chen, Z.D.; Sun, W.J.; Deng, T.T.; Chen, M.J. De novo sequencing of the leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. baijiguan. Front. Plant Sci. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Kashif, M.; Sang, Y.M.; Mo, S.M.; Rehman, S.U.; Khan, S.; Khan, M.R.; He, S.; Jiang, C.J. Deciphering the biodesulfurization pathway employing marine mangrove Bacillus aryabhattai strain NM1-A2 according to whole genome sequencing and transcriptome analyses. Genomics 2023, 115, 110635. [Google Scholar] [CrossRef] [PubMed]
- Liaquat, F.; Munis, M.F.H.; Arif, S.; Haroon, U.; Shi, J.; Saqib, S.; Zaman, W.; Che, S.; Liu, Q. PacBio single-molecule long-read sequencing reveals genes tolerating manganese stress in Schima superba saplings. Front. Genet. 2021, 12, 635043. [Google Scholar] [CrossRef]
- Usma, A.; Ahmad, M.; Zafar, M.; Sultana, S.; Ullah, F.; Saqib, S.; Ayaz, A.; Zaman, W. Palynological study of weed flora from Potohar plateau. Agronomy 2022, 12, 2500. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 261, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Xu, H.Y.; Li, X.Y.; Wang, L.J.; Wang, X.; Liu, Y.Q.; Ma, Y.P. Molecular cloning and functional analysis of a Chrysanthemum vestitum GME homolog that enhances drought tolerance in transgenic tobacco. Sci. Rep. 2022, 12, 13551. [Google Scholar] [CrossRef]
- Xu, H.Y.; Li, J.J.; Wang, L.J.; Li, X.Y.; Liu, Y.Q.; Wang, X.; Gao, T.T.; Ma, Y.P. Integrated transcriptomic and metabolomics analysis reveals abscisic acid signal transduction and sugar metabolism pathways as defense responses to cold stress in Argyranthemum frutescens. Environ. Exp. Bot. 2023, 205, 105115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 254, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Světlíková, P.; Hájek, T.; Těšitel, J. Hydathode trichomes actively secreting water from leaves play a key role in the physiology and evolution of root-parasitic rhinanthoid Orobanchaceae. Ann. Bot. 2015, 116, 61–68. [Google Scholar] [CrossRef]
- Fich, E.A.; Fisher, J.; Zamir, D.; Rose, J.K.C. Transpiration from tomato fruit occurs primarily via trichome-associated transcuticular polar pores. Plant Physiol. 2020, 184, 1840–1852. [Google Scholar] [CrossRef] [PubMed]
- Fernández, V.; Sancho-Knapik, D.; Guzmán, P.; Peguero-Pina, J.J.; Gil, L.; Karabourniotis, G.; Khayet, M.; Fasseas, C.; Heredia-Guerrero, J.A.; Heredia, A.; et al. Wettability, polarity, and water absorption of Holm Oak leaves: Effect of leaf side and age. Plant Physiol. 2014, 166, 168–180. [Google Scholar] [CrossRef]
- Dhawan, S.S.; Shukla, P.; Gupta, P.; Lal, R.K. A cold-tolerant evergreen interspecific hybrid of Ocimum kilimandscharicum and Ocimum basilicum: Analyzing trichomes and molecular variations. Protoplasma 2016, 253, 845–855. [Google Scholar] [CrossRef]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Wang, X.; Mahmood Ur, R.; Fiaz, S.; Azeem, F.; Shaheen, T. Morphological and physiological response of Helianthus annuus L. to drought stress and correlation of wax contents for drought tolerance traits. Arab. J. Sci. Eng. 2022, 47, 6747–6761. [Google Scholar] [CrossRef]
- Nichols, S.N.; Hofmann, R.W.; Williams, W.M. Physiological drought resistance and accumulation of leaf phenolics in white clover interspecific hybrids. Environ. Exp. Bot. 2015, 119, 40–47. [Google Scholar] [CrossRef]
- Murphy, K.M.; Zerbe, P. Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 2020, 172, 112289. [Google Scholar] [CrossRef] [PubMed]
- Savoi, S.; Wong, D.C.J.; Arapitsas, P.; Miculan, M.; Bucchetti, B.; Peterlunger, E.; Fait, A.; Mattivi, F.; Castellarin, S.D. Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.). BMC Plant Biol. 2016, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Bai, M.D.; Hao, G.W.; Guo, H.P.; Fu, B.C. Transcriptomics analysis of field-droughted pear (Pyrus spp.) reveals potential drought stress genes and metabolic pathways. PeerJ 2022, 10, e12921. [Google Scholar] [CrossRef]
- Duan, X.X.; Zhang, W.J.; Li, J.J.; Xu, H.Y.; Hu, J.; Zhao, L.; Ma, Y.P. Comparative metabolomics analysis revealed biomarkers and distinct flavonoid biosynthesis regulation in Chrysanthemum mongolicum and C. rhombifolium. Phytochem. Anal. 2022, 33, 373–385. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.J.; Liu, Z.G.; Wang, S.B.; Huang, B.S.; Liu, Y.F. Comparative transcriptome analysis of three chrysanthemums provides insights into flavonoid and terpenoid biosynthesis. J. Plant Biol. 2021, 64, 389–401. [Google Scholar] [CrossRef]
- Thirumurugan, D.; Cholarajan, A.; Raja, S.S.; Vijayakumar, R. An introductory chapter: Secondary metabolites. In Secondary Metabolites: Sources and Applications; IntechOpen: London, UK, 2018; pp. 1–21. [Google Scholar]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.Y.; Zhang, Y.N.; Zhu, L.; Liu, N.N.; Bai, H.T.; Sun, G.F.; Zhang, J.Z.; Shi, L. Chromosome-level assembly and analysis of the Thymus genome provide insights into glandular secretory trichome formation and monoterpenoid biosynthesis in thyme. Plant Commun. 2022, 3, 100413. [Google Scholar] [CrossRef] [PubMed]
- Luu, V.T.; Weinhold, A.; Ullah, C.; Dressel, S.; Schoettner, M.; Gase, K.; Gaquerel, E.; Xu, S.; Baldwin, I.T. O-Acyl Sugars protect a wild tobacco from both native fungal pathogens and a specialist herbivore. Plant Physiol. 2017, 174, 370–386. [Google Scholar] [CrossRef] [PubMed]
- Champagne, A.; Boutry, M. Proteomics of terpenoid biosynthesis and secretion in trichomes of higher plant species. Biochim. Biophys. Acta BBA Proteins Proteom. 2016, 1864, 1039–1049. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Vriet, C.; Edwards, A.; Smith, A.M.; Trevor, L.; Wang, T.L. Sucrose and Starch Metabolism. In The Lotus Japonicus Genome, Compendium of Plant Genomes; Satoshi, T., Jens, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 97–115. [Google Scholar]
- Zheng, Z.F.; Xu, X.P.; Crosley, R.A.; Greenwalt, S.A.; Sun, Y.J.; Blakeslee, B.; Wang, L.Z.; Ni, W.T.; Sopko, M.S.; Yao, C.L.; et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010, 153, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Lu, J.; Liu, Y.J.; Hu, D.G.; Hao, Y.J. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase Genes. Plant Physiol. 2017, 174, 2348–2362. [Google Scholar] [CrossRef]
- Kazaz, S.; Miray, R.; Lepiniec, L.; Baud, S. Plant monounsaturated fatty acids: Diversity, biosynthesis, functions and uses. Prog. Lipid Res. 2022, 85, 101138. [Google Scholar] [CrossRef]
- Guo, Q.; Li, T.; Qu, Y.; Liang, M.Z.; Ha, Y.M.; Zhang, Y.; Wang, Q. New research development on trans fatty acids in food: Biological effects, analytical methods, formation mechanism, and mitigating measures. Prog. Lipid Res. 2023, 89, 101199. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty acid- and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- He, M.; Qin, C.X.; Wang, X.; Ding, N.Z. Plant unsaturated fatty acids: Biosynthesis and regulation. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Sachdev, A.; Praveen, S. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant 2021, 171, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Charkowski, A. The role of ATP-binding Cassette (ABC) transporters in Bacterial phytopathogenesis. Phytopathology 2020, 111, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Malik, W.A.; Wang, X.G.; Wang, X.L.; Shu, N.; Cui, R.F.; Chen, X.G.; Wang, D.L.; Lu, X.K.; Yin, Z.J.; Wang, J.J.; et al. Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromol. 2020, 153, 470–491. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Lee, J.; Noh, E.W.; Lee, Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005, 138, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.H.; Zhao, S.Q.; Hao, Y.Q.; Gu, S.; Zhang, Y.; Qi, B.X.; Xing, Y.; Qin, L. Transcriptome analysis reveals key genes involved in the resistance to Cryphonectria parasitica during early disease development in Chinese chestnut. BMC Plant Biol. 2023, 23, 79. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef]
- Shah, M.; Alharby, H.F.; Hakeem, K.R.; Ali, N.; Rahman, I.U.; Munawar, M.; Anwar, Y. De novo transcriptome analysis of Lantana camara L. revealed candidate genes involved in phenylpropanoid biosynthesis pathway. Sci. Rep. 2020, 10, 13726. [Google Scholar] [CrossRef]
- Chen, Y.F.; Yi, N.; Yao, S.B.; Zhuang, J.H.; Fu, Z.P.; Ma, J.; Yin, S.X.; Jiang, X.L.; Liu, Y.J.; Gao, L.P.; et al. CsHCT-mediated lignin synthesis pathway involved in the response of tea plants to biotic and abiotic stresses. J. Agric. Food Chem. 2021, 69, 10069–10081. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.; Qin, Y. Plant low temperature stress: Signaling and response. Agronomy 2022, 12, 702. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Tweneboah, S.; Oh, S.K. Biological roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in solanaceous crops. J. Plant Biotechnol. 2017, 44, 1–11. [Google Scholar] [CrossRef]
- Yao, P.F.; Sun, Z.X.; Li, C.L.; Zhao, X.R.; Li, M.F.; Deng, R.Y.; Huang, Y.J.; Zhao, H.X.; Chen, H.; Wu, Q. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Niu, Y.L.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
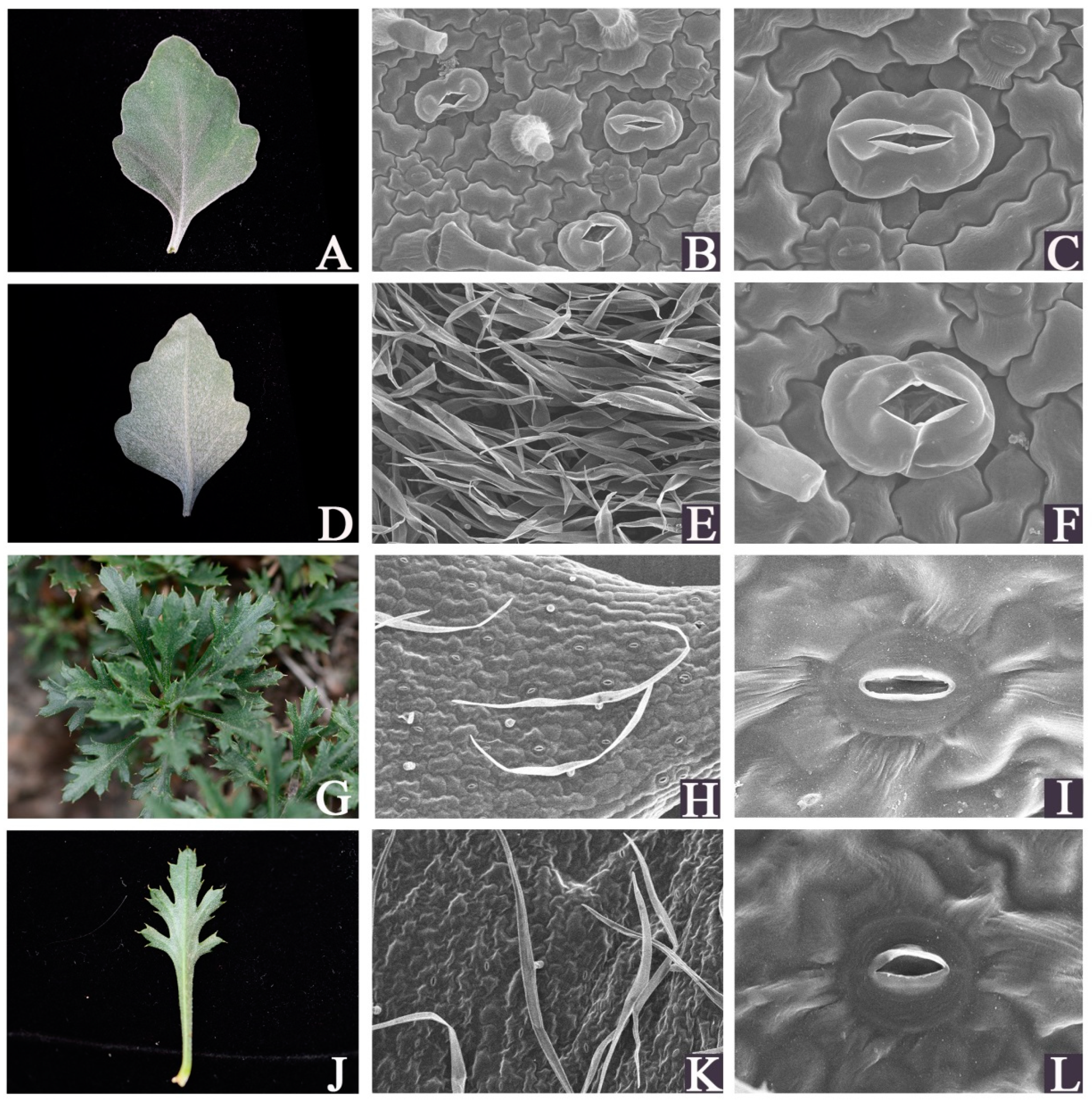

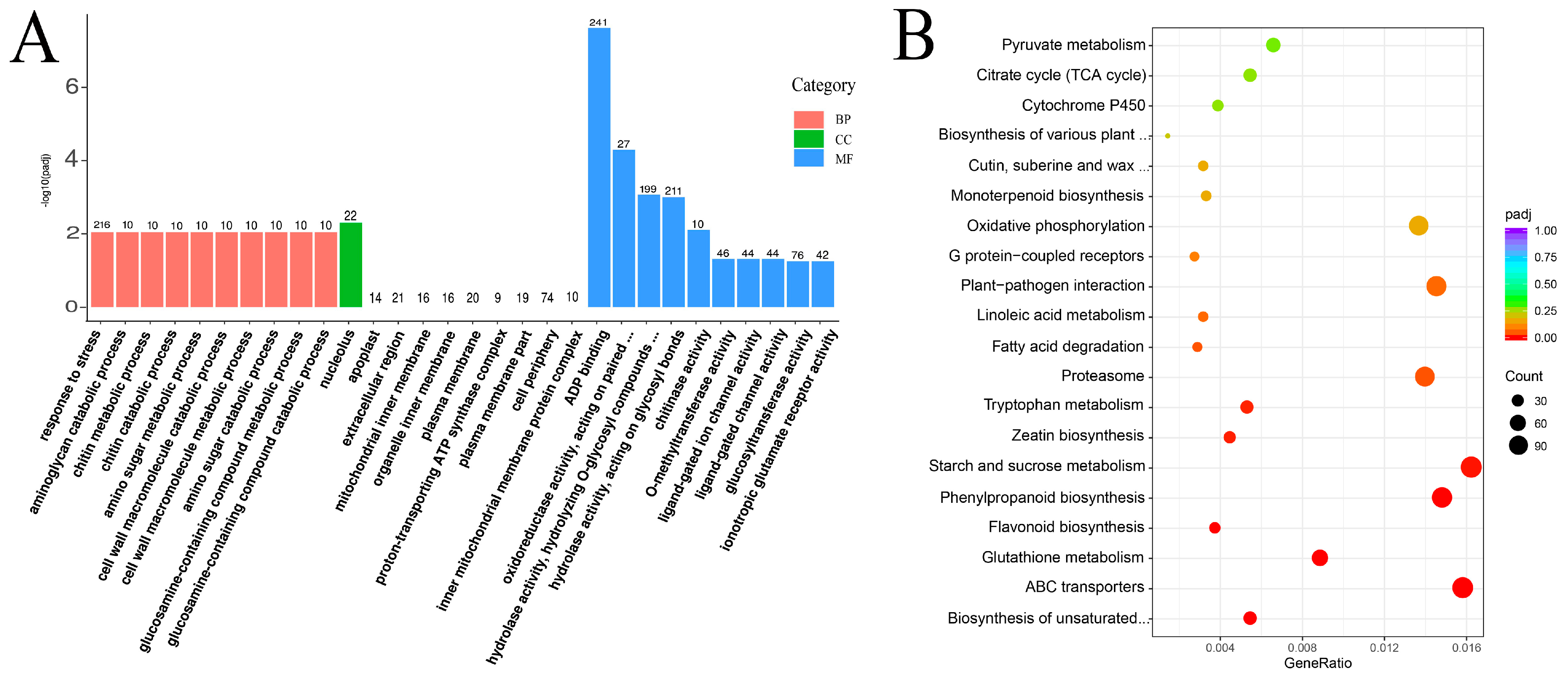

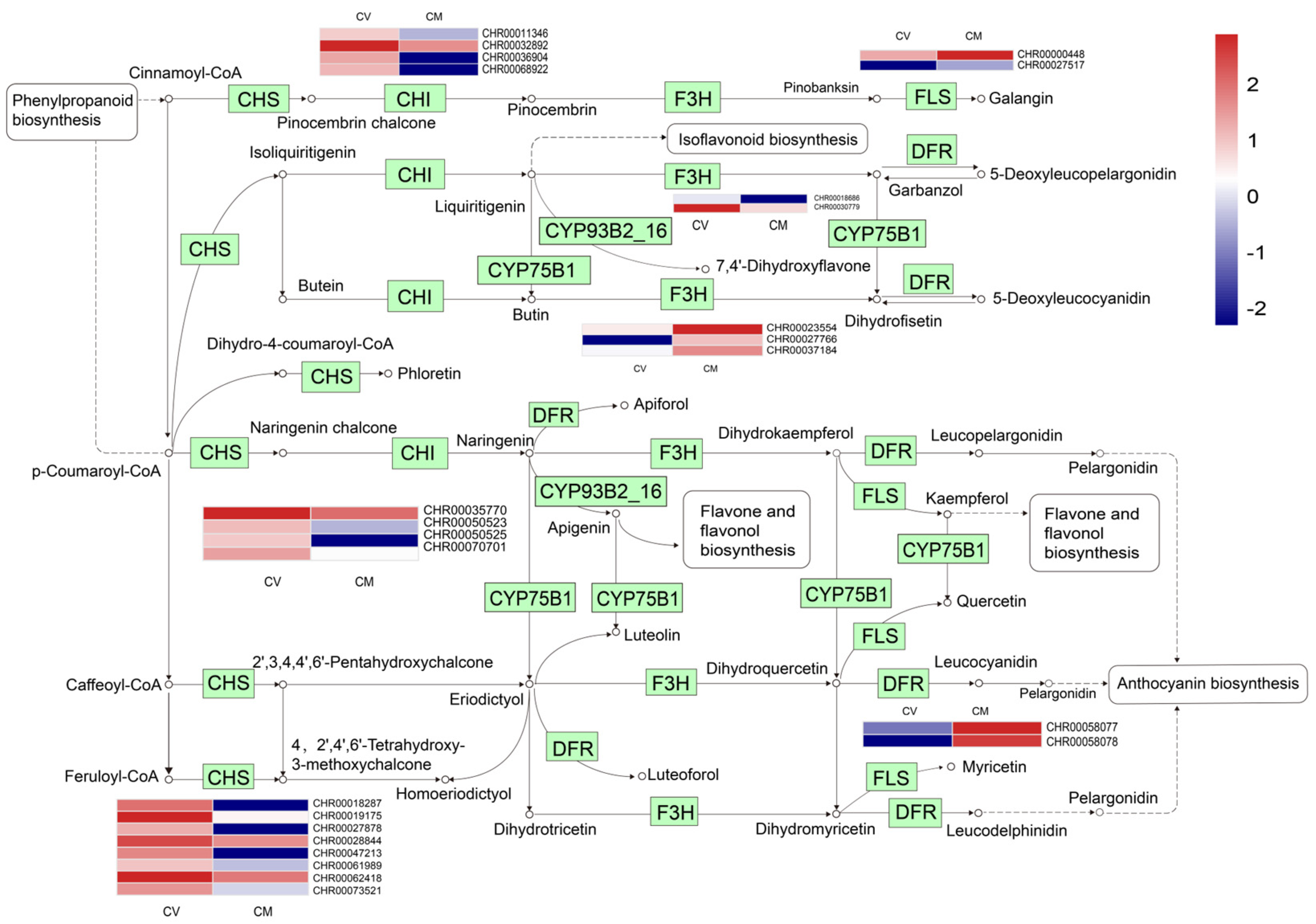
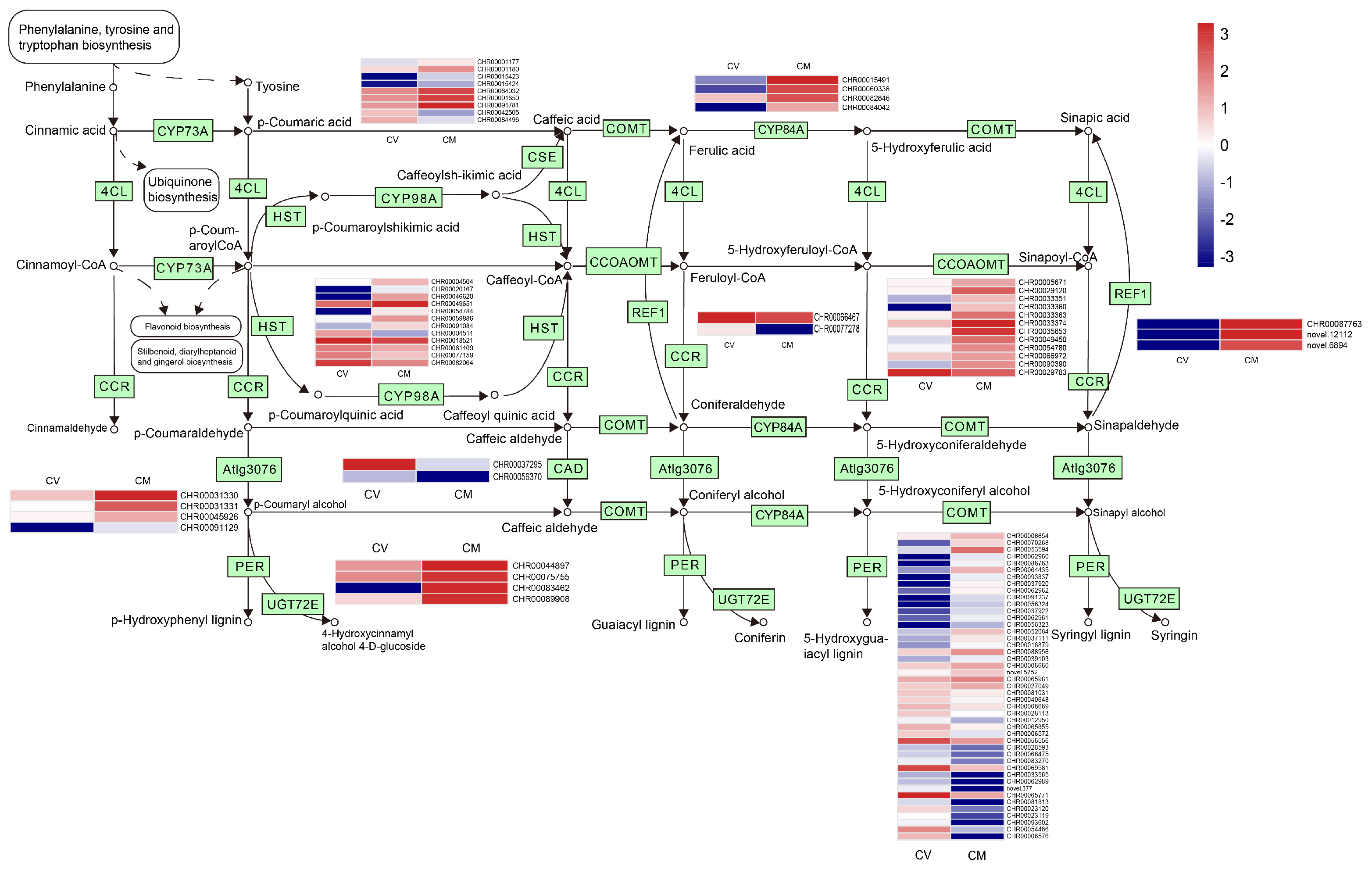
| Sample | Raw-Reads | Clean-Reads | GC Content (%) | Q20 (%) | Mapping Rate % |
|---|---|---|---|---|---|
| V1 | 47,996,046 | 46,458,626 | 42.86 | 97.87 | 68.96 |
| V2 | 50,286,250 | 48,704,408 | 43.10 | 97.76 | 64.26 |
| V3 | 48,940,764 | 47,877,184 | 42.82 | 97.9 | 67.03 |
| M1 | 50,463,176 | 49,115,504 | 42.71 | 97.93 | 65.36 |
| M2 | 48,187,394 | 46,899,782 | 42.74 | 97.86 | 62.89 |
| M3 | 50,427,536 | 49,386,860 | 42.69 | 97.81 | 63.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Meng, Y.; Lin, J. Comparative Transcriptome Analyses Reveal Different Regulatory Mechanisms in Ecological Adaptation between Chrysanthemum vestitum and Chrysanthemum mongolicum. Horticulturae 2023, 9, 868. https://doi.org/10.3390/horticulturae9080868
Ma Y, Meng Y, Lin J. Comparative Transcriptome Analyses Reveal Different Regulatory Mechanisms in Ecological Adaptation between Chrysanthemum vestitum and Chrysanthemum mongolicum. Horticulturae. 2023; 9(8):868. https://doi.org/10.3390/horticulturae9080868
Chicago/Turabian StyleMa, Yueping, Yuan Meng, and Junjie Lin. 2023. "Comparative Transcriptome Analyses Reveal Different Regulatory Mechanisms in Ecological Adaptation between Chrysanthemum vestitum and Chrysanthemum mongolicum" Horticulturae 9, no. 8: 868. https://doi.org/10.3390/horticulturae9080868
APA StyleMa, Y., Meng, Y., & Lin, J. (2023). Comparative Transcriptome Analyses Reveal Different Regulatory Mechanisms in Ecological Adaptation between Chrysanthemum vestitum and Chrysanthemum mongolicum. Horticulturae, 9(8), 868. https://doi.org/10.3390/horticulturae9080868





