Abstract
Panax ginseng is an ancient and very potent herb, which has a long history of medicinal use, and recent studies have shown that ginsenosides are the main active substances in its pharmacological effects. However, the saponin content of wild ginseng and cultivated ginseng can hardly meet the market supply, and the ginseng adventitious root suspension culture technology can produce ginsenosides in a targeted manner. The length of culture time is an important factor affecting the growth and development of plants and the accumulation of secondary metabolites. After transcriptome sequencing of ginseng adventitious root material at different culture times, the results showed that a total of 5784 differentially expressed genes were screened, which contained 239 transcription factors. KEGG analysis showed that these differentially expressed genes were mainly enriched in metabolic pathways and biosynthesis of secondary metabolites. A proposed temporal analysis of differentially expressed genes among the two groups distributed the differentially expressed genes under nine clusters, and the differentially expressed genes under different clusters had the same expression trends, indicating that these genes can be jointly involved in specific biological processes. Extraction of ginsenosides from ginseng adventitious roots using water-saturated n-butanol and detection of ginsenoside content by high-performance liquid chromatography revealed a significant increase in total saponins and protopanaxadiol ginsenosides (particularly significant for ginsenosides Rd and Rb1), an increase in bioaccumulation of some protopanaxatriol ginsenosides, and a decrease in some protopanaxatriol ginsenosides (S-Rh1, R-Rg3, and Rf) saponin content decreased. We also found seven genes involved in ginsenoside biosynthesis and that the changes in these genes’ expression may be related to the accumulation of ginsenosides.
1. Introduction
Panax ginseng has been regarded as an extraordinary herb since ancient times, and the ancient Chinese book Sheng Nong’s Herbal Classic has recorded its effects dating back more than 5000 years. It strengthens the spirit, fixes the soul, stops panic and palpitations, removes evil spirits, brightens the eyes, makes the mind happy, educates the mind, lightens the body and prolongs life when taken for a long time [1]. Recent studies have shown that ginsenosides are triterpene saponins, which are the main active substances for biological and pharmacological effects [2]. Ginsenosides can be divided into two categories according to their glycosidic skeletons, dammarane and oleanolic ginsenosides. The protopanaxadiol ginsenosides (PPD) and the protopanaxatriol ginsenosides (PPT) belonged to dammarane ginsenosides, and Ro belonged to oleanolic ginsenosides [3]. To date, nearly 200 ginsenosides have been reported; some of them are considered as major ginsenosides, such as Rb1, Rb2, Rc, Rd, Re, and Rg1 [4]. Studies have shown that ginsenosides are effective in regulating blood pressure homeostasis [5], improving myocardial function [6], have anti-fatigue [7], anti-diabetic [8], vasodilatory [9] and antidepressant effects [10], and effectively stimulate memory as well as prevent cancer and aging processes [11].
The time required to grow ginseng in the field is long and affected by various environmental factors such as soil, shade, climate, pathogens and pests [12], and the low content of ginsenosides in ginseng makes it difficult to meet the demands of the market environment. The use of cell suspension culture technology, using the adventitious roots of ginseng after healing-tissue induction as tissue material, allows for the targeted, stable, and rapid production of the desired ginsenosides [13]. The suspension culture cycle of ginseng adventitious roots is an important indicator of ginsenoside factory production.
The synthesis of ginsenosides undergoes the mevalonate (MVA) pathway and methylerythritol phosphate (MEP) pathway to generate isopentenyl pyrophosphate (IPP), which undergoes allyltransferase and terpene cyclase to generate 2,3-oxidosqualene, which undergoes cyclization, hydroxylation, and glycosylation modifications, which in turn generate specific ginsenosides. This metabolic process is regulated by multiple genes, among which P450 and UGT are key players in the structural diversity of triterpenoid saponins [14], and the secondary metabolism of the plant is regulated at the transcriptional level. Available studies have shown that several transcription factors have been identified from ginseng that may be involved in the biosynthesis of ginsenosides, which include MYB [15], WRKY [16], and MADS [17]. However, due to the complexity of the ginsenoside synthesis pathway, more genetic data support and functional validation of candidate genes are needed to reveal this synthesis pathway. Therefore, this study aimed to screen key genes involved in ginsenoside biosynthesis in ginseng adventitious roots based on differential gene expression data and ginsenoside phenotype data under different culture times.
The importance of screening functional genes associated with ginsenoside biosynthesis is due to the broad prospects of ginsenosides in medicine, health products and cosmetics. Therefore, in this study, we used RNA sequencing technology to detect the saponin content and screen the differentially expressed genes in ginseng adventitious roots at different culture times, and used GO functional annotation, KEGG analysis, and proposed temporal analysis to provide a reference for exploring the gene expression patterns and functions of related genes in ginseng adventitious roots at different culture times, and to establish a database of transcripts in ginseng adventitious roots The data were used to support the establishment of a transcript database in ginseng adventitious roots.
2. Materials and Methods
2.1. Collection of Samples from Three Different Periods
In this study, the ginseng adventitious roots were selected and identified as stable cultures in our laboratory. The ginseng adventitious roots were cultured in 15 L air-lift bioreactor with B5 liquid me-dium under the following conditions: ventilation rate of 0.05 vvm, culture temperature of 22 °C and darkness. Samples were taken at 15 days, 30 days and 40 days with three biological replicates in each group, and the samples were stored in a deep-freezer at −80 °C after freezing in liquid nitrogen.
2.2. RNA Extraction, Library Construction and Sequencing
The Trizol kit (Invitrogen, Carlsbad, CA, USA) was selected to extract the ginseng adventitious roots RNA. After extraction of total RNA, mRNA is purified and genomic DNA is removed. The purified mRNA was then cut into short fragments using fragmentation buffer and reverse-transcribed into cDNAs using random primers. Second-strand cDNA was synthesized using DNA polymerase I, ribonuclease H, dNTP and buffer. Then, QiaQuick PCR extraction kit (Qiagen, Venlo, The Netherlands) was used for purification, end repair, addition of A bases, and ligation to the Illumina sequencing junction. The size of the ligated products was selected by agarose gel electrophoresis, and polymerase chain reaction amplification and sequencing were performed with Illumina Nove-seq 6000 from Gene Denovo Biotechnology (Gene Denovo, CA, Guangzhou, China).
2.3. Analysis of Differentially Expressed Genes (DEGs)
Based on the expression data of Unigenes, differential expression analysis of unigene in ginseng adventitious roots at different time points was performed using the DESeq2 package [18] in R language, and genes that met both false discovery rate (FDR) < 0.05 and fold change ≥ 2 were considered as differentially expressed genes. Gene expression is spatio–temporal specific, and to understand the expression patterns of differentially expressed genes at different treatment times, a heat map of expression patterns of differentially expressed genes was drawn using R language.
2.4. Functional Enrichment Analysis of Differentially Expressed Genes
To have a better understanding of the functions of differentially expressed genes, GO functional annotation of differentially expressed genes was performed using Blast2GO version 6.0.3 [19], which annotates differentially expressed genes to three ontologies describing the molecular function (MF), the cellular component (CC), and the biological process (BP) in which the gene is involved.
2.5. KEGG Enrichment Analysis of Differentially Expressed Genes
In living organisms, genes usually coordinate and interact with each other in the exercise of their biological functions. To further understand the biological functions of differentially expressed genes in ginseng, differentially expressed genes were subjected to KEGG analysis and hypergeometric tests and applied to identify differentially expressed genes that were significantly enriched to specific pathways. The differentially expressed genes were identified in the Kyoto Encyclopedia of Genes and Genomes (https://www.kegg.jp/, accessed on 1 December 2022) to identify the biochemical metabolic pathways and signal transduction pathways in which these differentially expressed genes are involved.
2.6. Proposed Chronological Analysis of Differentially Expressed Genes
Based on the expression data of differentially expressed genes, the TC-seq packages in R software version 4.2.2 were used for the proposed temporal analysis of differentially expressed genes, and set statistical analysis of differentially expressed genes under different clusters performed using GraphPad prism version 8.3.0.
2.7. Extraction and Content Determination of Ginsenosides
Samples were taken at 15, 30, and 40 days, baked to a constant weight at 40 °C, ground and prepared for ginsenoside extraction; 60 mL of distilled water were added to a conical flask and the sample was dissolved. The dissolved sample was transferred to a separatory funnel A; 60 mL of ether was added to the separatory funnel A and mixed well with the dissolved sample, then the gas was evacuated. The further procedure: shake the funnel well (about 15 min) and let it stand until the ether and aqueous layers are completely separated; transfer the aqueous layer to a new partition funnel B, add 60 mL of aqueous saturated n-butanol, mix well and leave for several minutes until the upper clear layer (aqueous saturated n-butanol layer) is completely separated from the lower liquid layer (aqueous layer); collect the upper clear layer in a beaker, continue adding the aqueous saturated n-butanol solution to the lower clear layer, mix well, leave it, partition it and separate the lower liquid layer (emulsion layer) slowly, finally leaving only the upper layer of liquid. Transfer the upper layer of liquid collected in the beaker to the separating funnel B, add 50 mL of distilled water, mix well and let stand until the upper layer (n-butanol layer) and the lower layer of liquid (aqueous layer) are completely separated. The upper clear layer was transferred to a weighed round bottom flask and concentrated under vacuum at 60 °C until the liquid disappeared, followed by drying of the round bottom flask in a drying oven at 105 °C for 1 h and leaving it to stand at room temperature for 1 h before weighing. The content of water-saturated n-butanol was then calculated. The dried n-butanol extract was completely dissolved in 10 mL of methanol and then filtered through 0.45-μm membrane filter paper. Ginsenosides were detected using high-performance liquid chromatography (HPLC) [20,21].
2.8. Real-Time Quantitative RT-PCR Validation of Key Enzyme Genes at Different Culture Times
Based on the results of GO functional annotation, we performed the fluorescent quantitative RT-PCR on transcripts annotated to key enzyme genes involved in ginsenoside biosynthesis-related genes. The Actin 1 gene was selected as the internal reference gene, and the assay was performed. The cDNA from ginseng adventitious roots were used as templates, and the fluorescent quantitative RT-PCR system using SYBR premix EX Taq II (Takara, Dalian, China) included 5.0 μL SYBR premix Ex Taq II, 0.2 μL ROX II, 0.4 μL design primer, 0.5 μL cDNA, and 3.5 μL ribonuclease-free water. Reactions were as follows: predenaturation at 95 °C for 30 s; PCR at 95 °C for 5 s and 60 °C for 34 s for 40 cycles; melting at 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. The 2−ΔΔCt method was used to analysis the real-time quantitative RT-PCR results in this study [22,23].
3. Result
3.1. DEGS Screening of Ginseng Adventitious Roots under Different Culture Times
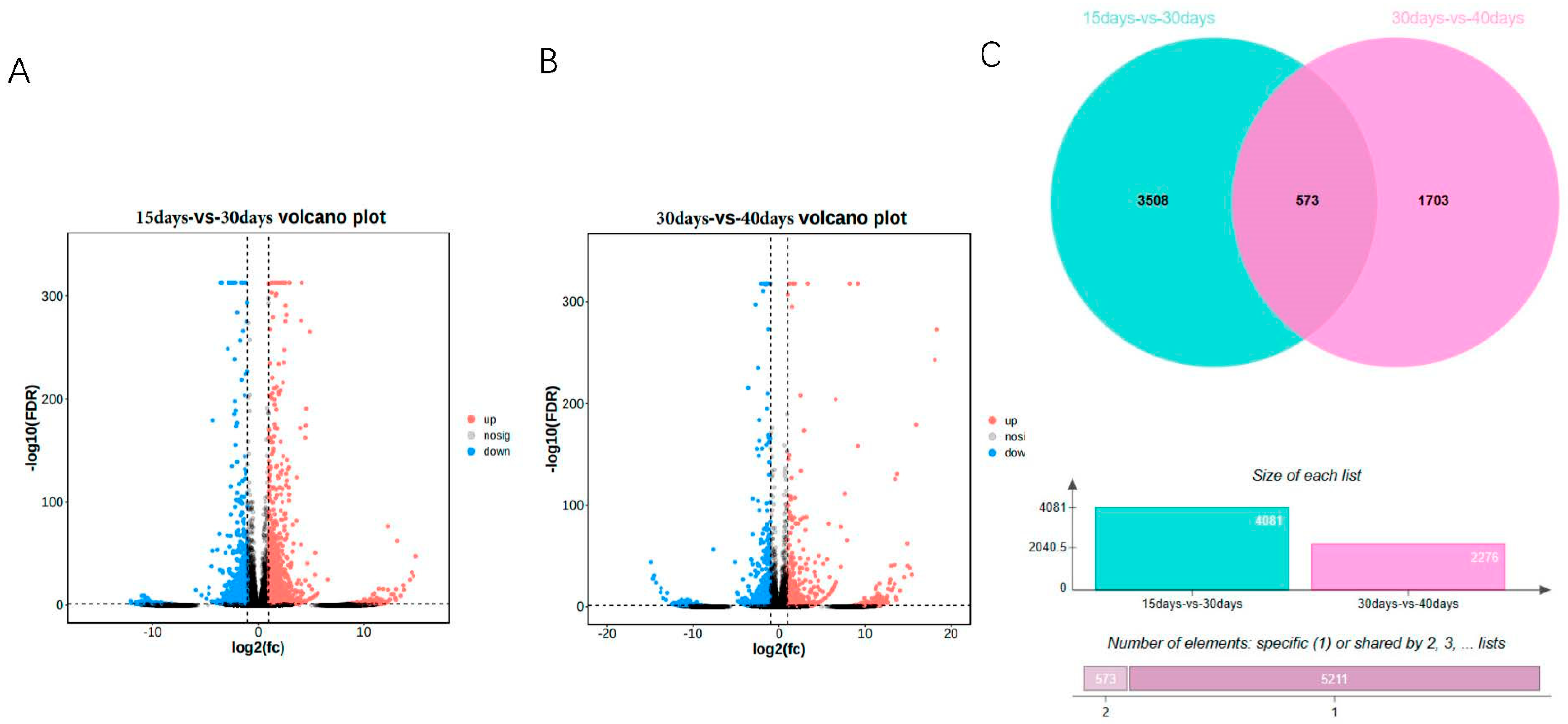
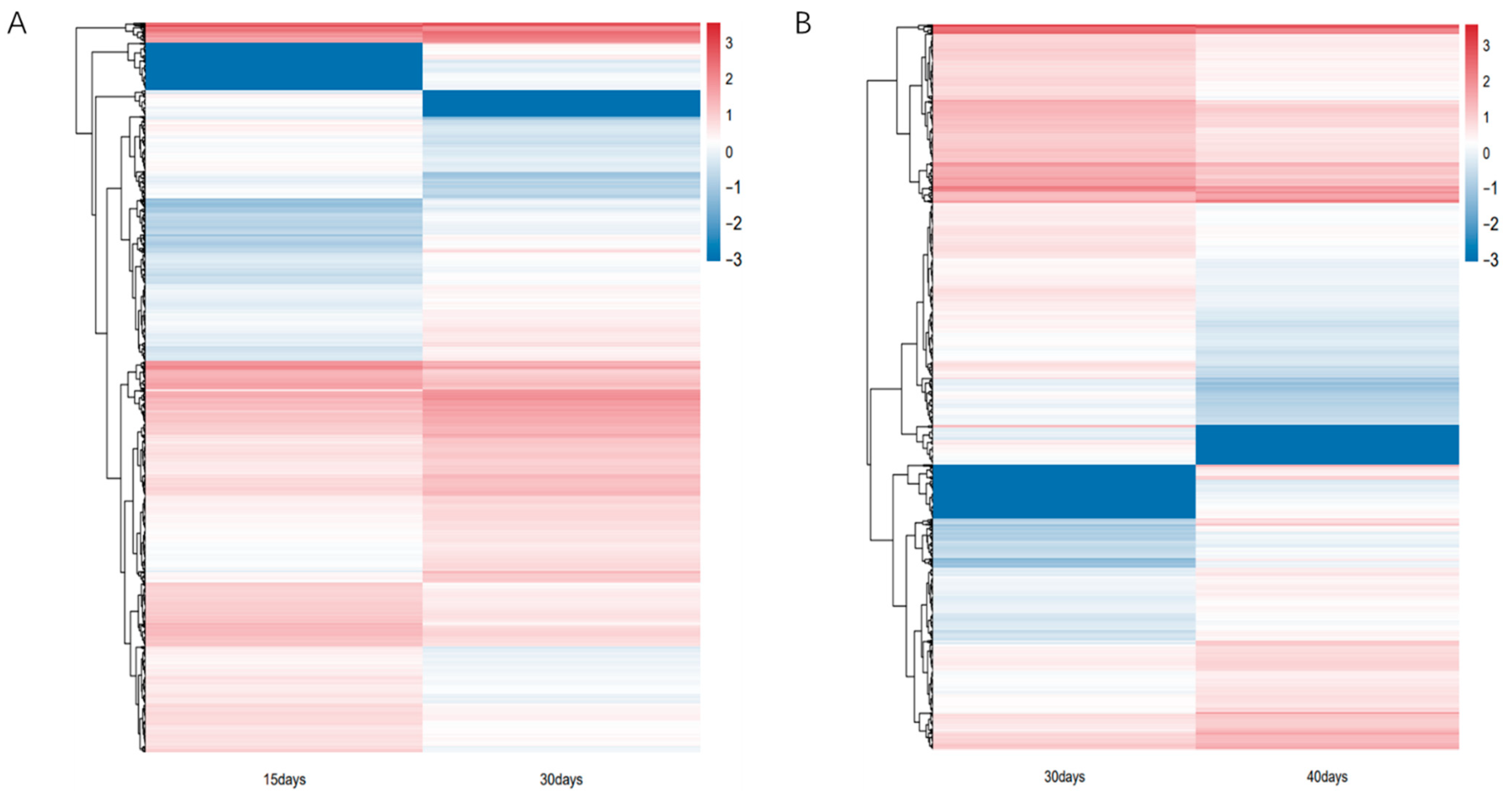
In this study, we analyzed the transcripts of ginseng adventitious roots at different culture times, grouped according to three different time points, and found that differential expression analysis was performed by DEseq2. The differentially expressed genes were 4081, 2334 up-regulated genes and 1747 down-regulated genes at 15 days and 30 days (Figure 1A, Table S1), and 2276, 932 up-regulated genes and 1344 down-regulated genes at 30 days and 40 days (Figure 1B). A total of 5784 differentially expressed genes were obtained by combining the data of differentially expressed genes between different groups. Using the differentially expressed genes between different groups, the Venn diagram was plotted and it was found that 573 genes had differential expression effects between two groups; 5211 genes were expressed between one group only, among which 3508 genes were expressed between 15 days vs. 30 days and 1703 genes were expressed between 30 days vs. 40 days. Finally, the heat map of differentially expressed genes between different groups was plotted by R language (Figure 2). The heat map results showed that differential genes were differentially expressed at different culture times in the ginseng adventitious roots.
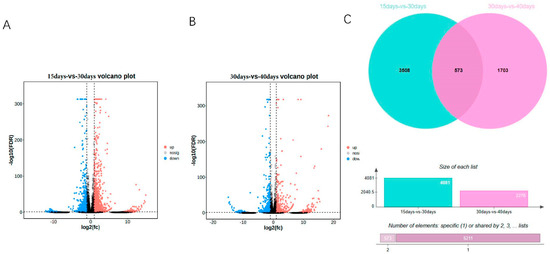
Figure 1.
Volcano map of differentially expressed genes in adventitious roots of ginseng at different times. (A) 15 days vs. 30 days volcano plot. (B) 30 days vs. 40 days volcano plot. (C) Venn diagram of differentially expressed genes between groups. FDR < 0.05 and fold change ≥ 2 were considered as DEGs.
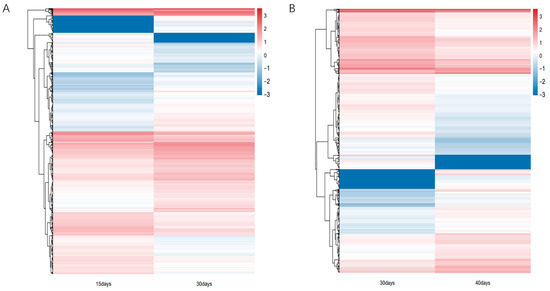
Figure 2.
Heatmap of differential gene expression patterns. (A) Heatmap of differential gene expression patterns of ginseng adventitious roots between 15 and 30 days of culture. (B) Heatmap of differential gene expression patterns of ginseng adventitious roots between 30 and 40 days of culture.
3.2. GO Functional Annotation of Differentially Expressed Genes
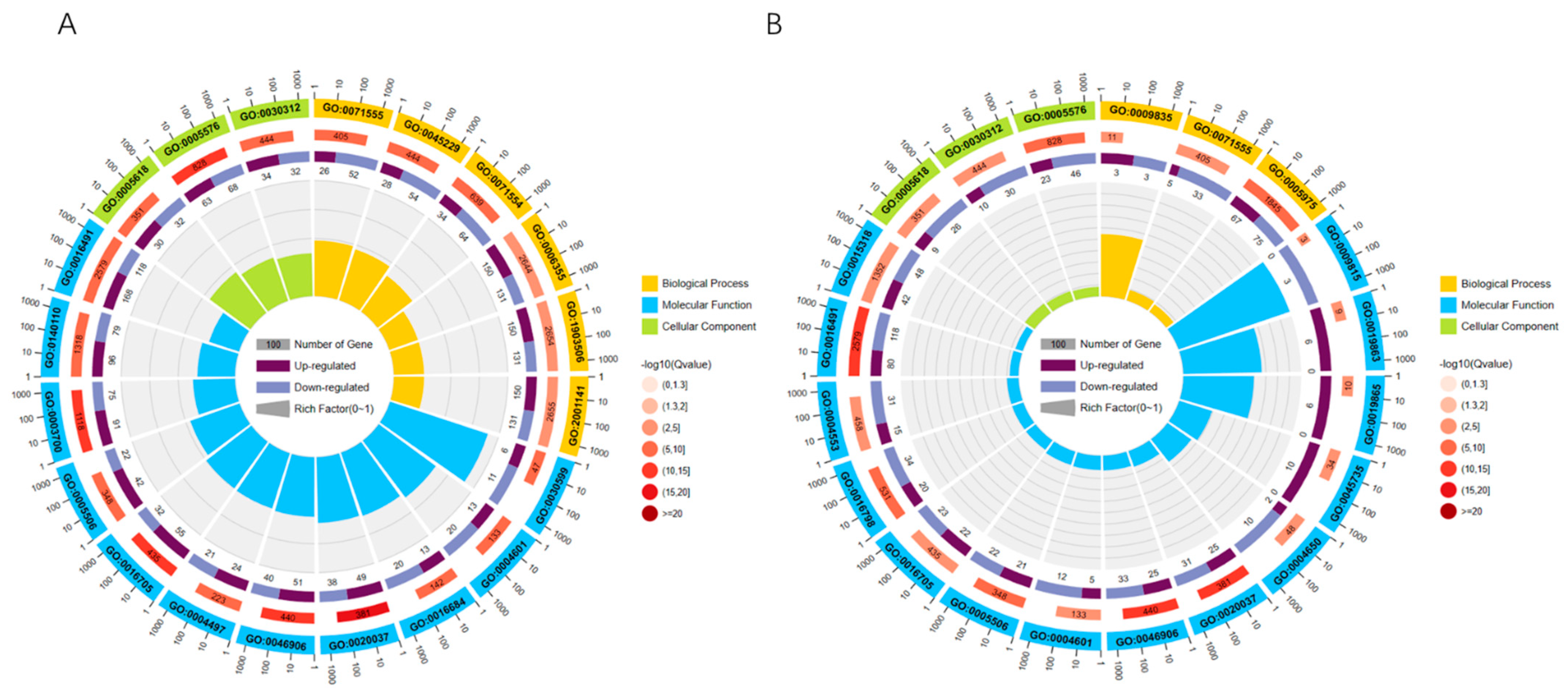
To better clarify the functions of differentially expressed genes, GO functional annotation of differentially expressed genes revealed that 5784 differentially expressed genes were annotated under a total of 711 GO entries, of which 169 GO entries were significantly enriched in 15 days vs. 30 days, including 51 biological process, 8 cellular component, and 110 molecular functions. The top 20 enriched GO terms were selected to plot the GO enrichment circle using R language (Figure 3A). In 30 days vs. 40 days, there are 48 GO entries significantly enriched, which include 12 BP, 6 CC, and 30 MF. The top 20 GO terms were selected and the GO enrichment circles were plotted using R language (Figure 3B). Under level 2, differentially expressed genes are mainly annotated under 48 GO entries, which include metabolic process (GO:0008152), growth (GO:0040007), developmental process (GO:0032502), signaling (GO: 0023052), transcription regulator activity (GO:0140110), binding (GO:0005488), protein-containing complex (Cellular Component), and other GO terms.
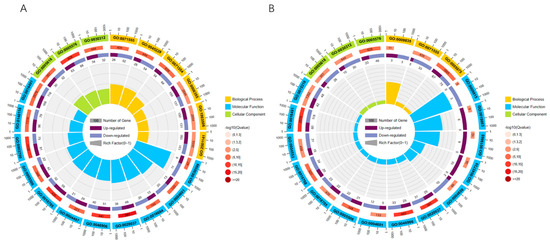
Figure 3.
GO enrichment circle graph: (first circle: top 20 GO terms enriched; second circle: number of this GO term in the differential gene background and Q value; third circle: bar graph of the proportion of up- and down-regulated differential genes; fourth circle: Rich Factor value of each GO term.) (A) 15 days vs. 30 days GO enrichment circle graph. (B) 30 days vs. 40 days GO enrichment circle graph.
3.3. KEGG Analysis of Differentially Expressed Genes
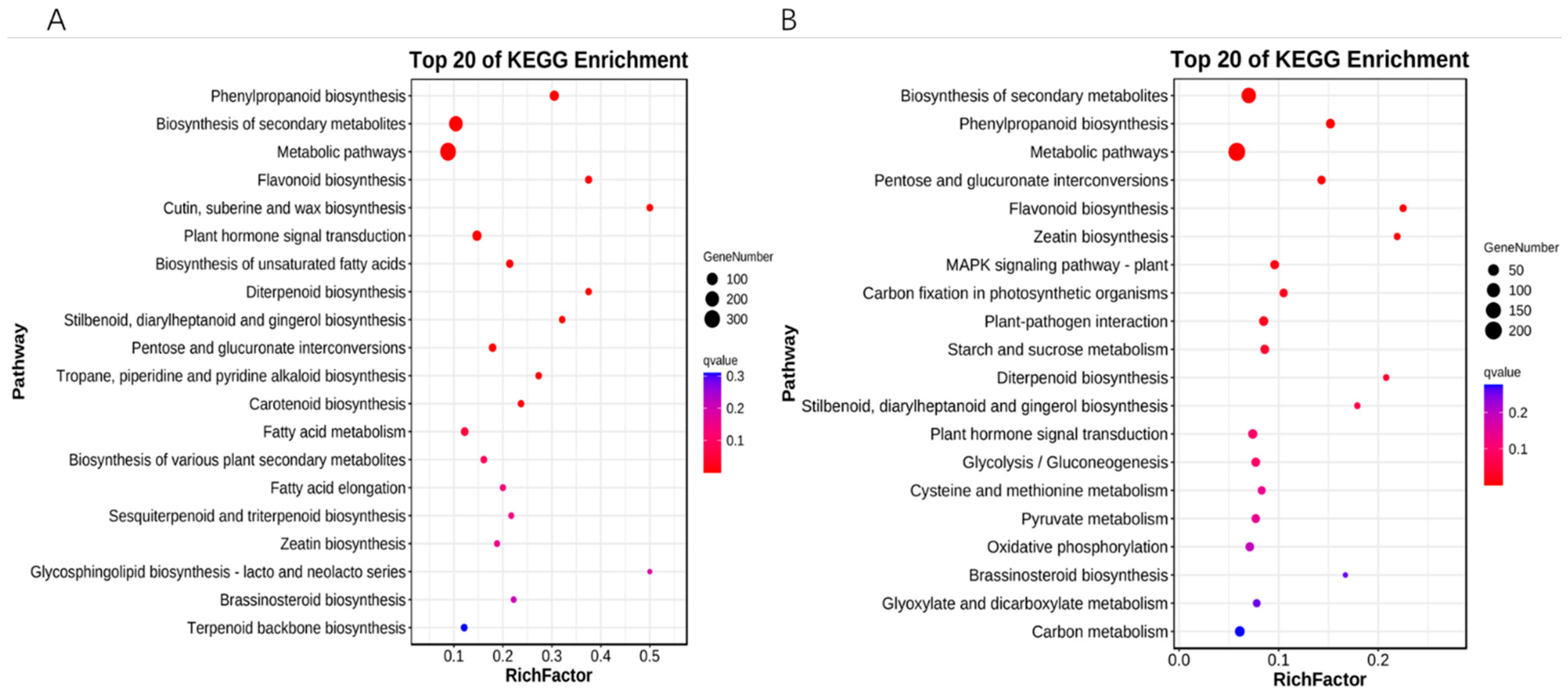
Annotation of the signaling pathways of differentially expressed genes in the KEGG database revealed that 15 days vs. 30 days differentially expressed genes were significantly enriched in 12 metabolic pathways, which included phenylpropanoid biosynthesis, biosynthesis of secondary metabolites, metabolic pathways, flavonoid biosynthesis, cutin, suberine and wax biosynthesis, plant hormone signal transduction, biosynthesis of unsaturated fatty acids, diterpenoid biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis, pentose and glucuronate interconversions, tropane, piperidine and pyridine alkaloid biosynthesis, carotenoid biosynthesis (Figure 4A); 30 days and 40 days differentially expressed genes were significantly enriched in 11 metabolic pathways: biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, pentose and glucuronate interconversions, flavonoid biosynthesis, zeatin biosynthesis, MAPK signaling pathway—plant, carbon fixation in photosynthetic organisms, plant-pathogen interaction, starch and sucrose metabolism, and diterpenoid biosynthesis (Figure 4B).
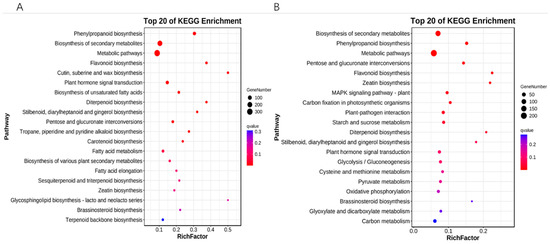
Figure 4.
KEGG enrichment analysis of differentially expressed genes. (A) 15 days vs. 30 days KEGG enrichment analysis bubble plot. (B) 30 days vs. 40 days KEGG enrichment analysis bubble plot. The vertical coordinate is the pathway, the horizontal coordinate is the enrichment factor, the size indicates quantity, and the redder the color the smaller the q-value.
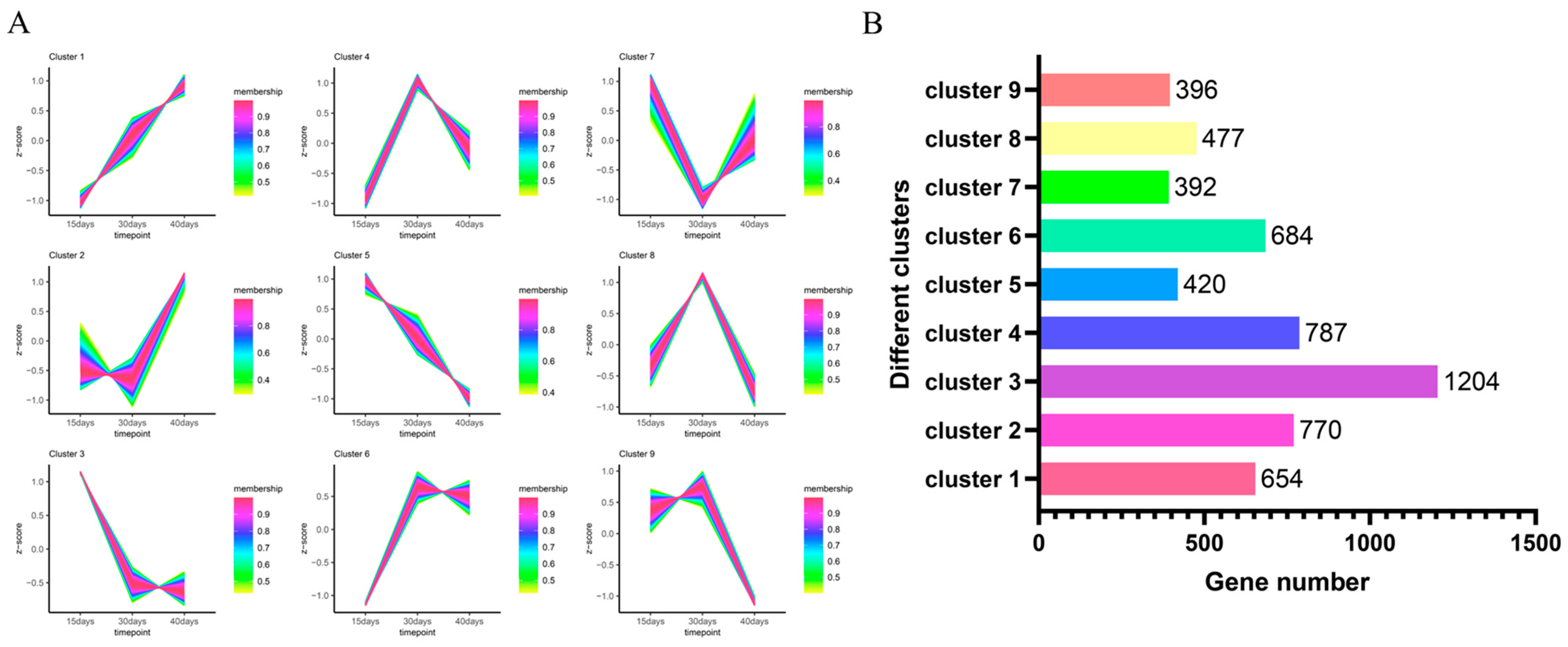
3.4. Proposed Chronological Analysis of Differentially Expressed Genes
The proposed temporal analysis of differentially expressed genes revealed that the expression of 5784 genes formed different expression trends under nine clusters (Figure 5A). After counting the number of genes under different clusters (Figure 5B), it was found that the largest number of differentially expressed genes was found under cluster 3, with 1204 differentially expressed genes having the same expression pattern, and the smallest number of differentially expressed genes was found under cluster 7, with 392 differentially expressed genes having similar expression trends.
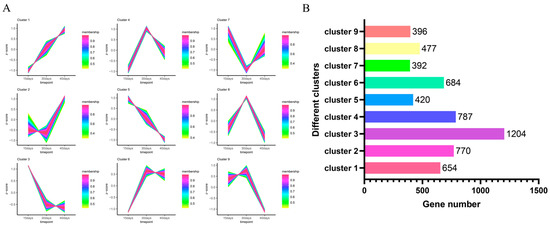
Figure 5.
Proposed temporal analysis of 5784 differentially expressed genes. (A) Proposed temporal permit analysis of differentially expressed genes under nine clusters. (B) The number of differentially expressed genes under different clusters.
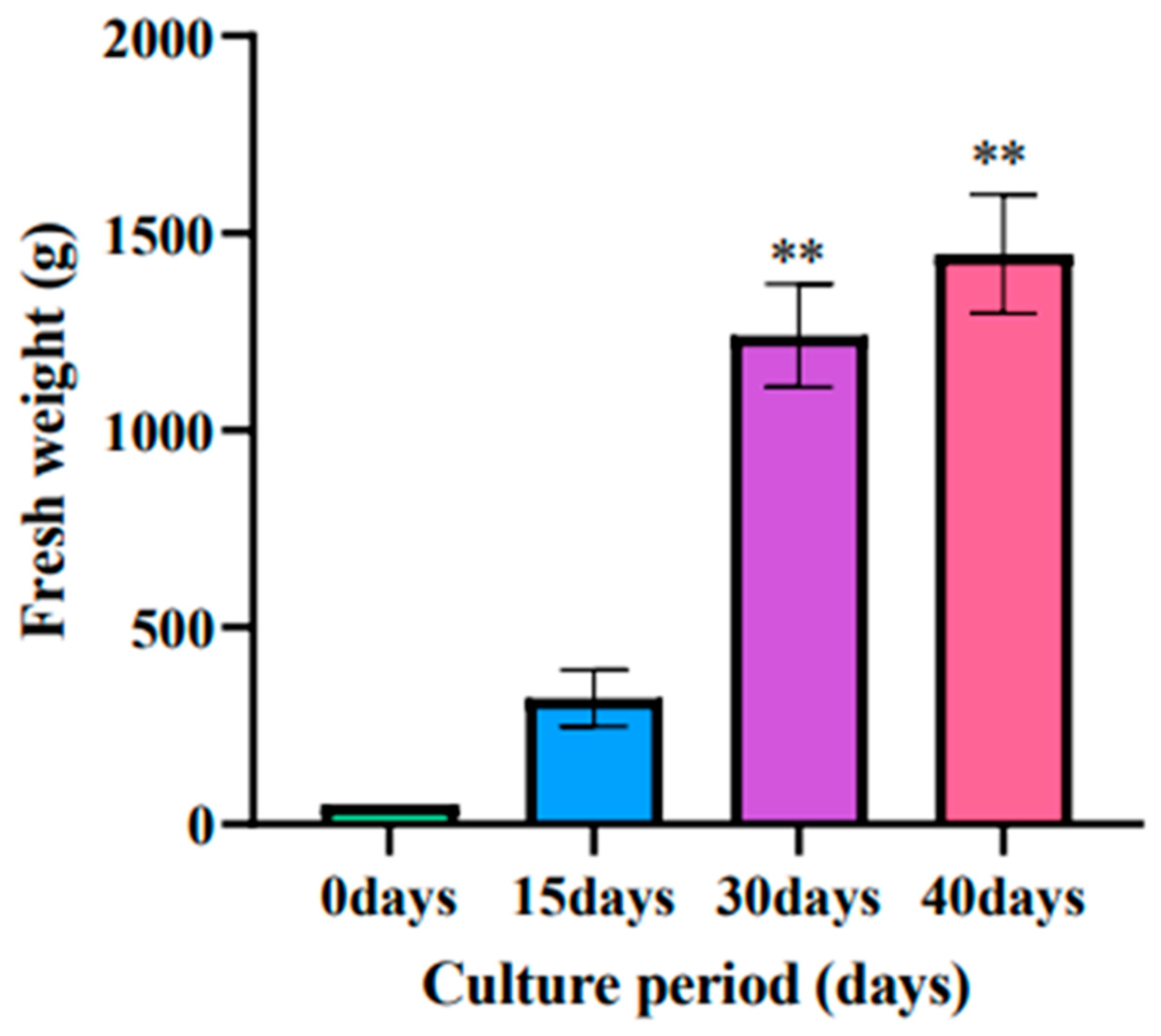
3.5. Effect of Culture Time on the Biomass of Adventitious Roots of Ginseng
With the increase of culture time, the biomass accumulation of ginseng adventitious roots also increased, and the fresh weight of ginseng adventitious roots reached 320 g after 15 days of culture, 1240 g after 30 days of culture, and 1446 g after 40 days of culture (Figure 6).
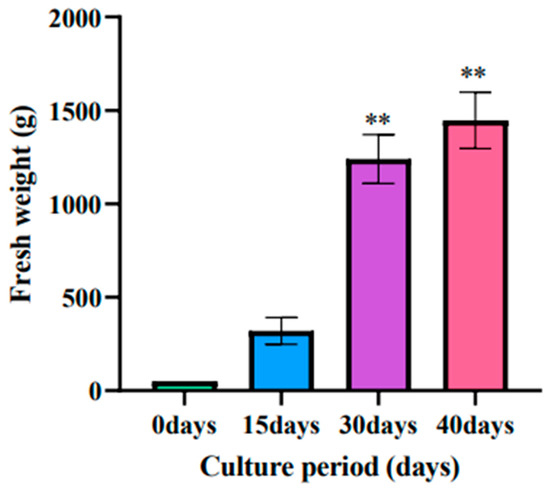
Figure 6.
Changes in the fresh weight of ginseng adventitious roots in ginseng shake bottles at different times. ** as significant at p ≤ 0.01.
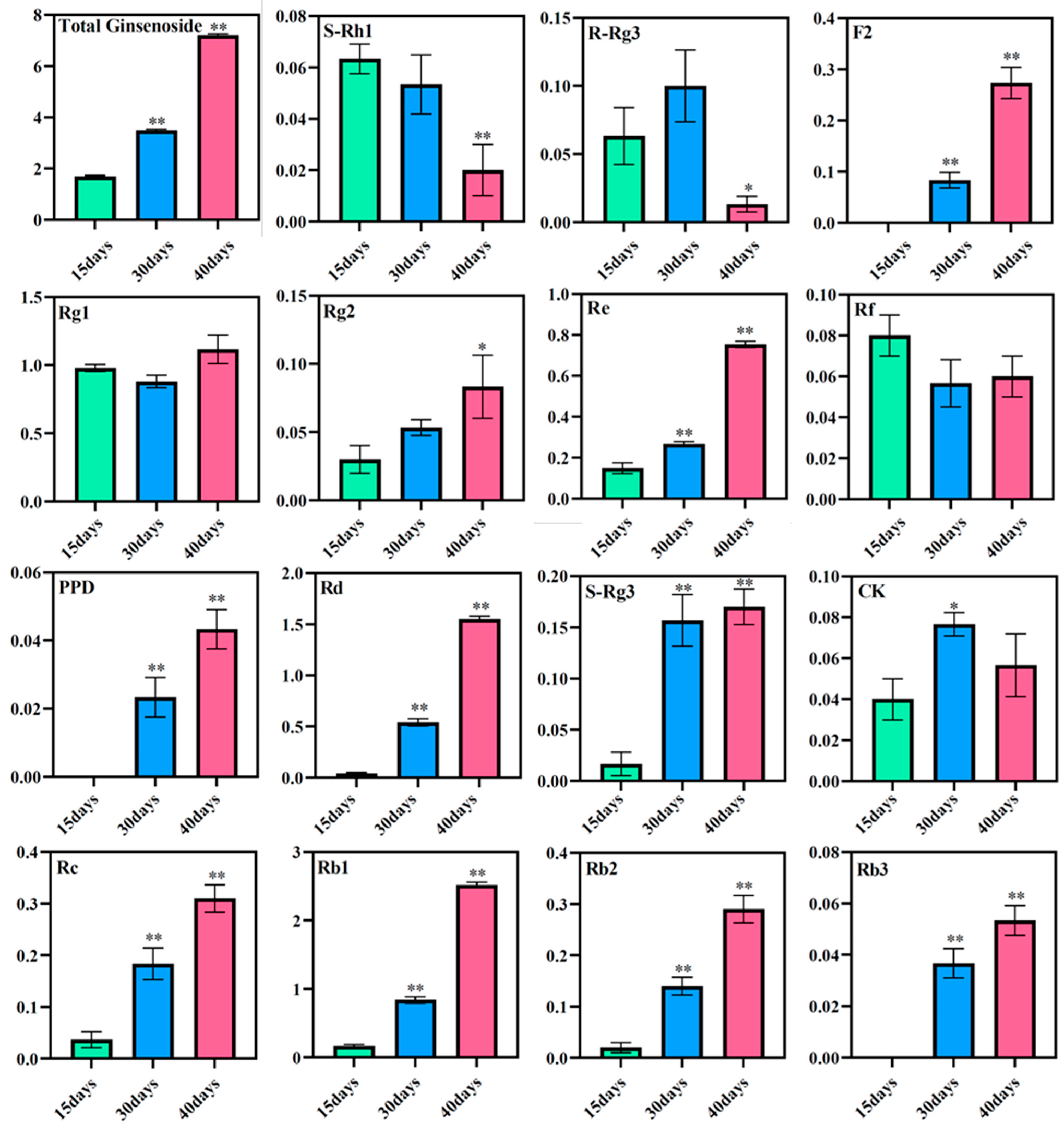
3.6. Expression of Ginsenosides at Different Culture Times
The total saponin content in the ginseng adventitious roots was found to increase with increasing culture time using high-performance liquid chromatography. The total saponin content in ginseng adventitious roots was 1.69 mg/g at 15 days of culture, 3.49 mg/g at 30 days of culture, and 7.22 mg/g after 40 days of culture. The contents of diol-type ginsenosides PPD, Rd, S-Rg3, Rc, Rb1, Rb2, and Rb3 increased with time, where the content of ginsenoside Rd and ginsenoside Rb1 changed most significantly. The content of ginsenoside Rd was 0.04 mg/g at 15 days of ginseng adventitious root culture and increased to 1.55 mg/g after 40 days of ginseng adventitious root culture, which was 38.75 times the original content. The content of ginsenoside Rb1 was 0.17 mg/g at 15 days of culture and 2.52 mg/g at 40 days of culture, which was 14.82 times higher than the original content. The content of triol-type ginsenosides Rg1, Rg2, and Re also increased with the increase of culture time. However, the content of ginsenosides S-Rh1, R-Rg3, and Rf decreased after 40 days of culture compared to 15 days of culture, and R-Rg3 reached its maximum after 30 days of culture (Figure 7).
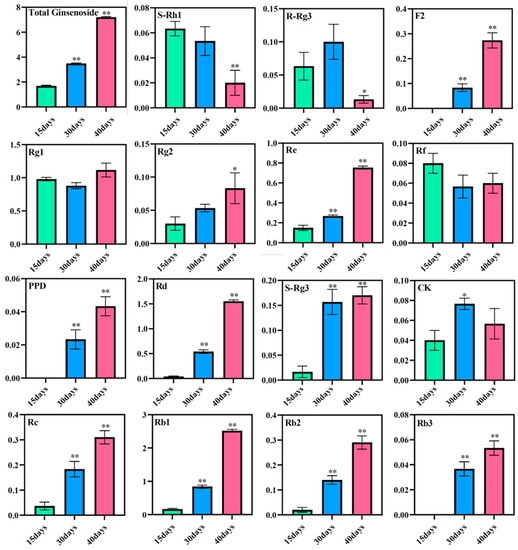
Figure 7.
Changes in ginsenoside content in adventitious roots of ginseng at different culture times. * as significant at p ≤ 0.05, ** as significant at p ≤ 0.01.
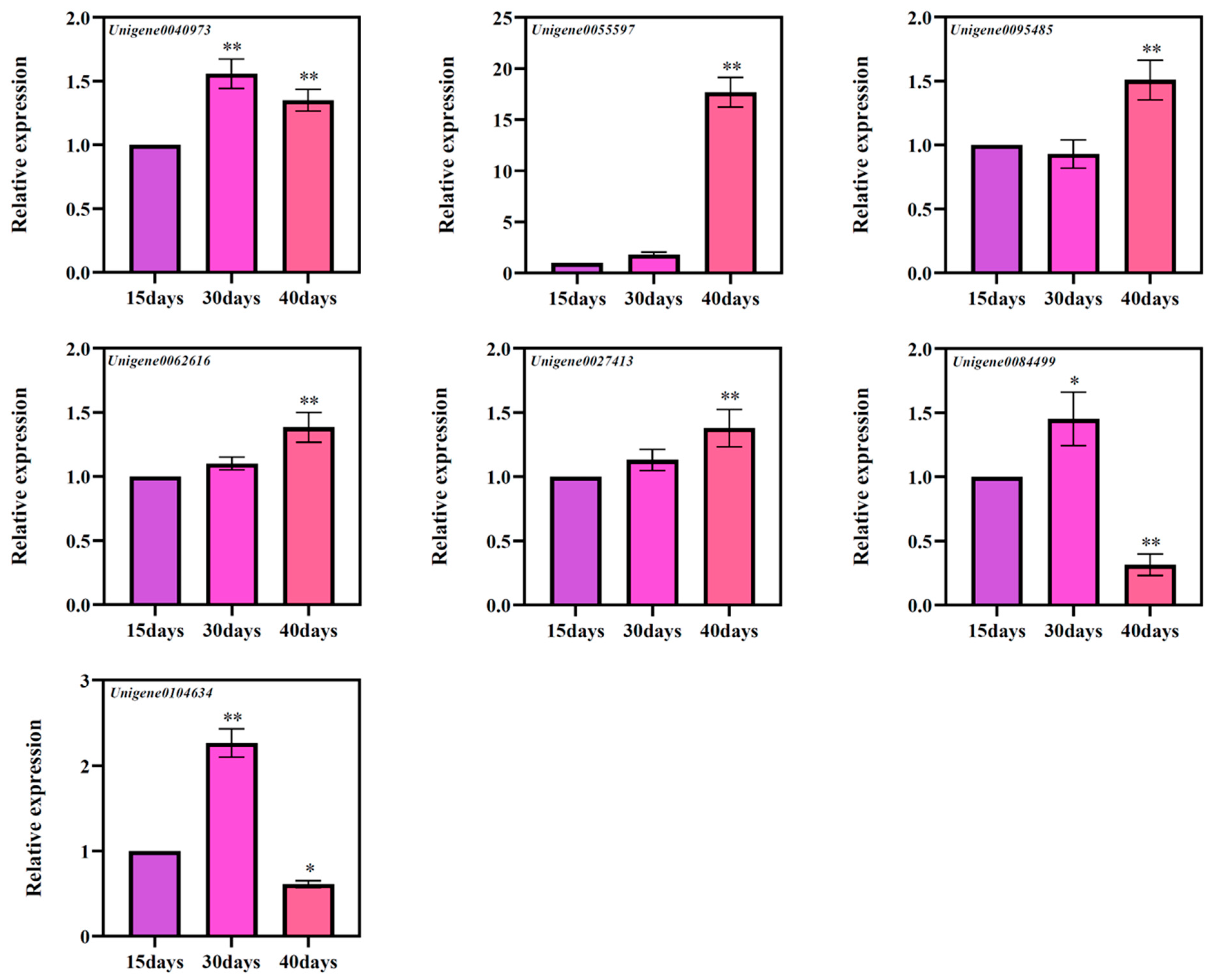
3.7. Gene Expression of Candidate Genes Involved in Ginsenoside Biosynthesis
Based on the results of GO annotation, a total of seven genes were screened, namely Unigene0040973 (CYP716A47), Unigene0055597 (CYP716A52v2), Unigene0095485 (CYP716A52v2), Unigene0062616 (CYP716A53v2), Unigene0027413 (UGT74AE2), Unigene0084499 (UGT74AE2), and Unigene0104634 (UGT74AE2) [20,21,22]. According to the results of fluorescence quantitative PCR, it was found that these seven genes have different expression patterns at different times (Figure 8). Among them, Unigene0055597, Unigene0062616, and Unigene0027413 genes showed a significant increase in the levels of these three genes with the increase of culture times, and at three different culture times, Unigene0040973, Unigene0084499, and Unigene0104634 genes expression levels showed a trend of increasing and then decreasing expression. Expression of Unigene0095485 showed a transient decrease at 30 days and a significant change after 40 days of culture compared to 15 days of culture. Therefore, we speculate that the changes in these genes may be related to the accumulation of ginsenosides.
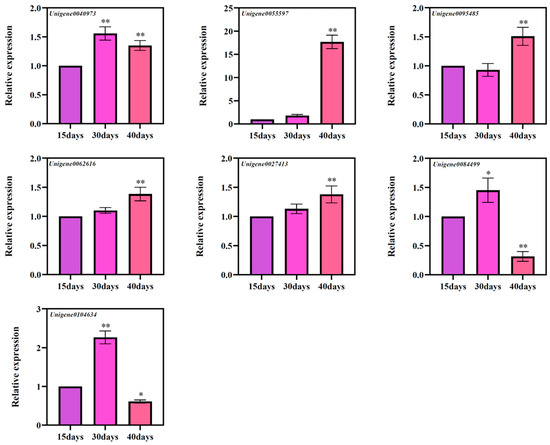
Figure 8.
The expressions of seven candidate genes’ ginsenoside biosynthesis at different culture times of the ginseng adventitious roots. * as significant at p ≤ 0.05, ** as significant at p ≤ 0.01.
4. Discussion
In the suspension culture of ginseng adventitious roots, the length of culture time is an important indicator of the change of ginsenoside content in ginseng adventitious roots. In this study, we analyzed the differentially expressed genes of adventitious roots at different culture times (15 days, 30 days, and 40 days) and examined the ginsenoside content at different times to provide data for the subsequent verification of the functions of related and ginsenoside biosynthesis genes.
As a preferred herb with high nutritional and pharmacological values, ginseng is favored in today’s society for its tonic effects and for the treatment of many diseases [24]. Ginsenoside content is an important index for evaluating the pharmacological value of ginseng. There were significant differences in the expression of relevant genes during suspension culture of ginseng adventitious roots under different time conditions. The number of differentially expressed genes after 15 days of culture versus 30 days of culture (4081 unigenes) was significantly more than the number of differentially expressed genes from 30 to 40 days of culture (2276 unigenes). This indicates that at the beginning of ginseng adventitious root culture, after a short delay period, it then enters the exponential growth period, during which the growth and development process in the plant is more vigorous and the metabolic level is more active. However, at the later stages of culture, cellular reproduction gradually slows down and metabolism is slower [25].
These differentially expressed genes were found to be mainly involved in plant growth and development processes and signaling, metabolism and other processes by GO annotation, and inducers as a specific biochemical signal that enables plant cells and tissue cultures to rapidly and efficiently trigger transcriptional reprogramming in plants, which in turn activates specific secondary metabolic pathways, thereby increasing plant secondary metabolite production [26]. Related studies have shown that treatment of ginseng adventitious root cultures with jasmonic acid can promote the accumulation of ginsenoside content in ginseng adventitious root cultures [27]. In this study, we identified among differentially expressed genes the related TIFY proteins Unigene0052304, Unigene0066465, Unigene0081716 that respond to jasmonic acid signaling. Unigene0077012 had been shown to be able to respond to jasmonic acid signaling and then regulate the synthesis of related secondary metabolites by regulating downstream transcription factors [28].
KEGG annotation revealed that differentially expressed genes were significantly enriched to signaling pathways such as biosynthesis of secondary metabolites, metabolic pathways, flavonoid biosynthesis, and interconversion of pentose and glucuronide. The results suggest that differentially expressed genes annotated to relevant metabolic pathways may be involved in the accumulation of ginsenosides, ginseng polysaccharides, flavonoids and other secondary metabolites in ginseng.
Related studies suggest that the synthesis of ginsenosides is regulated by numerous genes [29]. Among them, Cytochrome P450 monooxygenase (P450) is able to further modify the triterpene backbone in ginsenosides, which in turn generates structurally diverse triterpenoid compounds, and UDP-glycosyltransferase (UGT) is a key enzyme for glycosylation modification of natural ginsenoside biosynthesis [14]. The presence of a transcript (Unigene0104634) with high homology to the already identified UGT74AE2 gene between 15 and 30 days of culture suggests that Unigene0104634 may have the same function as UGT74AE2 and that Unigene0104634 may catalyze the C-3 hydroxylation of protopanaxadiol and compound K glycosylation to produce ginsenoside Rh2 and ginsenoside F2, respectively [30]. Between 30 and 40 days of culture, we found a transcript, Unigene0055597, which is highly homologous to the gene for β-amyrin 28-oxidase (CYP716A52v2) that has been verified [31], which may be involved in the synthesis of oleanolic acid-type triterpenoid ginsenosides, but unfortunately, we did not detect oleanolic acid Ro in the adventitious roots, which may be due to the low content of oleanolic acid-type ginsenosides. The accumulation of secondary metabolites in the plant is regulated at the transcriptional level [32]. Previous studies have shown that overexpression of the PgWRKY4X gene can significantly increase the expression of the PgSE gene, which in turn enhances the accumulation of ginsenoside production [16], and under jasmonic acid signaling, PgMYB2 can bind to the promoter of dammarendiol synthase (DS), leading to the speculation that PgMYB2 may be involved in the regulation of ginsenoside synthesis [15]. PgMADS41 and PgMADS44 are involved in the binding of SE-4, CYP716A52v2-4 and β-AS-13 promoters of ginsenoside Ro biosynthesis, which in turn regulates ginsenoside biosynthesis [17]. In our study, some transcription factors—bHLH (Unigene0017986 and Unigene0089919), MYB (Unigene0076906, Unigene0021549 and Unigene0024095), WRKY (Unigene0022251 and Unigene0090069)—can play a role in the biosynthesis of ginsenosides.
We also found seven ginsenoside biosynthesis pathway P450 and UGT genes (Unigene0040973, Unigene0055597, Unigene0095485, Unigene0062616, Unigene0027413, Unigene0084499, and Unigene0104634) [20,21,22], these genes had been reported to be involved in ginsenoside biosynthesis function for expression studies that these genes expression changes can be related to the accumulation of ginsenosides in ginseng adventitious roots.
Co-expression networks are a powerful tool for understanding gene regulation. They have been used to identify novel regulation and functions of genes involved in plant development and its response to the environment [33]. In this study, the differentially expressed genes were subjected to a proposed temporal analysis, and differentially expressed genes formed different expression patterns under nine clusters; differentially expressed genes under the same cluster had similar expression patterns, and these genes with the same expression pattern may regulate the expression of receptor genes at a specific time, in a specific space, and thus affect the organism during reproductive growth, nutritional growth, and secondary metabolism levels.
5. Conclusions
Traditional ginseng production methods have led to an unstable and uncontrollable quality of ginseng, which can no longer meet the increasing demand for ginsenosides in the market. With the rapid development of plant cell engineering technology, ginseng adventitious root isolation culture technology has gradually matured, establishing a new way to solve the problem of ginsenoside production through a ginseng cell factory. In this study, the transcriptome data of ginseng adventitious roots at three culture times were analyzed using RNA-seq technology to obtain differentially expressed genes and establish the gene expression patterns of ginseng adventitious roots at different culture times. This study provides a dynamic analysis of gene expression at different culture times in the suspension culture of ginseng adventitious roots and can provide a theoretical basis and genetic resources for further research on important functional genes, as well as a possibility for ginsenoside synthesis in plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9070762/s1, Table S1: The differentially expressed genes at different culture times in adventitious roots of Panax ginseng.
Author Contributions
Formal analysis, X.L. and Y.Z.; Funding acquisition, H.C.; Methodology, X.L., Y.Z., K.W. and M.L.; Software, K.W. and M.L.; Writing—original draft, H.C. and K.W.; Writing—review & editing, H.C. and K.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an award from Jilin Provincial Department of Industry and Information Technology (2017), Department of Science and Technology of Jilin Province (20200801063GH).
Data Availability Statement
The transcriptome data of ginseng adventitious roots (15 days, 30 days, and 40 days) in this study been deposited in NCBI under accession number BioProject PRJNA980200, BioSample SAMN35622170—SAMN35622178, and SRA SRR24827690—SRR24827698.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kuang, H. Chemistry of Chinese Materiel Medica; China Traditional Chinese Medicine Publishing House: Beijing, China, 2003. (In Chinese) [Google Scholar]
- Kim, S.K.; Park, J.H. Trends in ginseng research in 2010. J. Ginseng. Res. 2011, 35, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, P.; Yang, T.-J.; Song, Y.H. Genes and Regulatory Mechanisms for Ginsenoside Biosynthesis. J. Plant Biol. 2023, 66, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ratan, Z.A.; Haidere, M.F.; Hong, Y.H.; Park, S.H.; Lee, J.-O.; Lee, J.; Cho, J.Y. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng. Res. 2021, 45, 199–210. [Google Scholar] [CrossRef]
- Qin, N.; Gong, Q.H.; Wei, L.W.; Wu, Q.; Huang, X.N. Total ginsenosides inhibit the right ventricular hypertrophy induced by monocrotaline in rats. Biol. Pharm. Bull. 2008, 31, 1530–1535. [Google Scholar] [CrossRef]
- Zhou, H.; Hou, S.Z.; Luo, P.; Zeng, B.; Wang, J.R.; Wong, Y.F.; Jiang, Z.H.; Liu, L. Ginseng protects rodent hearts from acute myocardial ischemia-reperfusion injury through GR/ER-activated RISK pathway in an endothelial NOS-dependent mechanism. J. Ethnopharmacol. 2011, 135, 287–298. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Fan, Y.; Yan, C.; Liu, D.; Cheng, H.; Gao, X.; Zhou, Y. Anti-fatigue activity of the water-soluble polysaccharides isolated from Panax ginseng C. A. Meyer. J. Ethnopharmacol. 2010, 130, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.S.; Xie, J.T.; Mehendale, S. Ginseng and Diabetes. Am. J. Chin. Med. 2005, 33, 397–404. [Google Scholar]
- Rimar, S.; Lee-Mengel, M.; Gillis, C.N. Pulmonary protective and vasodilator effects of a standardized Panax ginseng preparation following artificial gastric di-gestion. Pulm. Pharmacol. 1996, 9, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Chen, Y.; Liu, X.; Wang, Q.; Wang, L.; Jia, W.; Wang, Y. Antidepressant effects of ginseng total saponins in the forced swimming test and chronic mild stress models of depression—ScienceDirect. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2009, 33, 1417–1424. [Google Scholar] [CrossRef]
- Xiao, D.; Yue, H.; Xiu, Y.; Sun, X.; Wang, Y.; Liu, S. Accumulation characteristics and correlation analysis of five ginsenosides with different cultivation ages from different regions. J. Ginseng. Res. 2015, 39, 338–344. [Google Scholar] [CrossRef]
- Paek, K.Y.; Murthy, H.N.; Hahn, E.J.; Zhong, J.J. Large scale culture of ginseng adventitious roots for production of ginsenosides. Adv. Biochem. Eng. Biotechnol. 2009, 113, 151–176. [Google Scholar]
- Le, K.-C.; Jeong, C.-S.; Lee, H.; Paek, K.-Y.; Park, S.-Y. Ginsenoside accumulation profiles in long- and short-term cell suspension and adventitious root cultures in Panax ginseng. Hortic. Environ. Biotechnol. 2018, 60, 125–134. [Google Scholar] [CrossRef]
- Seki, H.; Tamura, K.; Muranaka, T. P450s and UGTs: Key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 2015, 56, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Luo, T.; Guo, X.; Zou, X.; Zhou, D.; Afrin, S.; Li, G.; Zhang, Y.; Zhang, R.; Luo, Z. PgMYB2, a MeJA-responsive transcription factor, positively regulates the dammarenediol synthase gene expression in Panax ginseng. Int. J. Mol. Sci. 2019, 20, 2219. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Wang, J.; Sun, J.; He, J.; Paek, K.-Y.; Park, S.-Y.; Huang, L.; Gao, W. A WRKY transcription factor, PgWRKY4X, positively regulates ginsenoside biosynthesis by activating squalene epoxidase transcription in Panax ginseng. Ind. Crop. Prod. 2020, 154, 112671. [Google Scholar] [CrossRef]
- Jiao, H.; Hua, Z.; Zhou, J.; Hu, J.; Zhao, Y.; Wang, Y.; Yuan, Y.; Huang, L. Genome-wide analysis of Panax MADS-box genes reveals role of PgMADS41 and PgMADS44 in modulation of root development and ginsenoside synthesis. Int. J. Biol. Macromol. 2023, 233, 123648. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Zhao, M.; Lin, Y.; Wang, Y.; Li, X.; Han, Y.; Wang, K.; Sun, C.; Wang, Y.; Zhang, M. Transcriptome analysis identifies strong candidate genes for ginsenoside biosynthesis and reveals its underlying molecular mechanism in Panax ginseng C.A. Meyer. Sci. Rep. 2019, 9, 615. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Lin, Y.; Wang, Y.; Wang, K.; Sun, C.; Lu, T.; Zhang, M. Structural variation, functional differentiation, and activity correlation of the Cytochrome P450 gene superfamily revealed in ginseng. Plant Genome 2018, 11, 170106. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Zhao, M.; Wang, K.; Sun, C.; Zhu, L.; Han, Y.; Chen, P.; Lei, J.; Wang, Y.; et al. Integrative transcriptome analysis identifies new oxidosqualene cyclase genes involved in ginsenoside biosynthesis in Jilin ginseng. Genomics 2021, 113, 2304–2316. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Pan, Z.; Yu, J.; Zhu, L.; Zhao, M.; Wang, Y.; Chen, P.; Liu, C.; Hu, J.; Liu, T.; et al. Transcriptome-wide characterization, evolutionary analysis, and expression pattern analysis of the NF-Y transcription factor gene family and salt stress response in Panax ginseng. BMC Plant Biol. 2022, 22, 320. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H. A review on the medicinal potentials of ginseng and ginsenosides on cardiovascular diseases. J. Ginseng Res. 2014, 38, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, A.C. Plant Cell Suspension Cultures. Tissue Cult. Methods Appl. 1973, 215–219. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- Yu, K.W.; Gao, W.; Hahn, E.J.; Paek, K.Y. Jasmonic acid improves ginsenoside accumulation in adventitious root culture of Panax ginseng C.A. Meyer. Biochem. Eng. J. 2002, 11, 211–215. [Google Scholar] [CrossRef]
- Ma, P.; Pei, T.; Lv, B.; Wang, M.; Dong, J.; Liang, Z. Functional pleiotropism, diversity, and redundancy of Salvia miltiorrhiza Bunge JAZ family proteins in jasmonate-induced tanshinone and phenolic acid biosynthesis. Hortic. Res. 2022, 9, uhac166. [Google Scholar] [CrossRef]
- Kim, N.H.; Jayakodi, M.; Lee, S.C.; Choi, B.S.; Jang, W.; Lee, J.; Kim, H.H.; Waminal, N.E.; Lakshmanan, M.; van Nguyen, B.; et al. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 2018, 16, 1904–1917. [Google Scholar] [CrossRef]
- Jung, S.C.; Kim, W.; Park, S.C.; Jeong, J.; Park, M.K.; Lim, S.; Lee, Y.; Im, W.T.; Lee, J.H.; Choi, G. Two ginseng UDP-Glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014, 55, 2177–2188. [Google Scholar] [CrossRef]
- Jung-Yeon, H.; Min-Jun, K.; Yong-Wook, B.; Hwan-Su, H.; Yong-Eui, C. The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2013, 53, 2034–2046. [Google Scholar]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Riken, J.; Nakamichi, N.; Hvidsten, T.; Ahnert, S.E. Co-expression networks from gene expression variability between genetically identical seedlings can reveal novel regula-tory relationships. Front. Plant Sci. 2021, 11, 599464. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).