Abstract
Peonies are significant ornamental plants that are primarily propagated through distant cross-breeding to create new varieties. However, hybrid failure is a critical issue that impedes the advancement of breeding. Numerous studies have demonstrated that endogenous hormones in the seed embryo constitute a significant factor in embryo failure. Nevertheless, it is still unknown how plant hormones control the development of peony embryos at the molecular level. In this study, we characterized the endogenous hormone levels in peony seeds of hybrid-aborted, hybrid-normal, and self-crossing normal after 26 days of pollination (DAP). Our findings show that the hybrid-aborted embryos had significantly higher amounts of ABA, IAA, and GA. In addition, the ratio of GA+IAA/ABA was lower than that of the hybrid-normal seeds and higher than that of the self-crossing normal seeds. To further investigate the mechanism of hormone control on peony embryo development, we conducted a transcriptome sequencing analysis of the three seed types. Results revealed that differentially expressed genes involved in phytohormone metabolism and signal transduction significantly enriched the aborted embryos. Furthermore, we examined the expression levels of six hormones in different seeds and used the Gene Common Expression Trend analysis to analyze genes highly correlated with phytohormone in the KEGG pathway. We used protein interaction networks to explore the interactions between proteins in the hormone pathway in aborted embryos. Then we identified key genes and transcription factors (TFs) such as Abscisic acid-insensitive 5 (ABI5), Auxin Response Factor 5 (ARF5), Gibberellin Insensitive Dwarf 1 (GID1), Arabidopsis Response Regulator4 (ARR4), Jasmonate-zim-domain protein 1 (JAZ1), Brassinazole-resistant 1 (BZR1), etc., whose functions require further investigation. Our findings establish a foundation for the metabolic regulation of peony hybrid embryo abortion via networks regulating phytohormone signaling. However, further research is needed to determine the exact mechanisms by which hormones regulate peony embryo development and to explore new methods for improving the success rate of hybridization.
1. Introduction
The embryo, which originates from a zygote containing fundamental plant tissue and is surrounded by nutritive endosperm and maternal organization, plays a crucial role in plant reproduction and serves as the beginning of plant life [1]. However, the development of angiosperm embryos from fertilization onward involves a complex biological process that undergoes a series of changes [2]. Any developmental errors at any point can lead to the abortion of an embryo. Embryo abortion results in seeds that are unable to develop into complete plants, reduces seed production, and negatively impacts plant breeding [3]. Additionally, there are various types of embryonic abortion including abnormalities in the embryo’s body structure, death at a critical stage of development, or the failure of the seed to grow into a typical plant after fertilization [4]. Researching embryo abortion can help promote the normal development and efficient growth of embryos, improving the number and quality of seeds. Particularly, research on embryo abortion in distant hybridization can enhance the germplasm resources of species and breed superior plants [5].
There are several factors that contribute to embryo abortion including physiological and biochemical, embryological, external environment, and molecular regulation [6,7,8,9,10]. Endogenous hormones, which are part of the physiological and biochemical indicators, have been extensively studied as they are involved in various processes of plant growth and development, and their effects on embryonic development may lead to embryo failure [11]. Indole-3-acetic acid (IAA) and Gibberellin acid (GA) promote embryonic cell growth and development, advance protein synthesis, and provide more energy to the embryo [12,13]. Cytokinines (CTK) and Brassinosteroid (BR) regulate the size and number of embryonic cells and have an important role [14,15]. High levels of Abscisic acid (ABA) and Jasmonic acid (JA) inhibit amylase synthesis and activity and may induce cell death [16,17]. Studies have shown that the level and balance of endogenous hormones significantly differ between aborted and regular fruit, suggesting that imbalances in hormone levels may be a critical factor in embryonic abortion [18]. In particular, embryonic development is hypothesized to be influenced by the ratio of growth-promoting substances to growth-inhibiting substances. A lower ratio, especially in the early stage of embryonic development, means that the balance of hormones is disrupted, leading to embryo abortion [19]. Furthermore, research has identified genes related to plant hormones that may also be associated with embryonic abortion [20,21].
Despite the identification of certain genes, the relationship between the transcriptional regulatory network of peony in plant hormone signal transduction and embryo abortion remains poorly understood. Recently, transcriptomes analyzed via high-throughput sequencing have emerged as a method to explore embryo abortion and have revealed insights into the metabolic pathways, hormone balance, signal transduction cascade, and other plant protection mechanisms involved in this process [22]. Among the genes related to phytohormone pathways, those associated with ABA, including 9-cis-epoxycarotenoid dioxygenase (NCED), ABRE binding factors 2 (ABF2), Abscisic acid-insensitive 4 (ABI4), and ABI5, are substantially expressed in embryos and endosperm [23,24,25]. ABA first binds to PYR1/PYL/RCAR (PYLs) and then forms a complex with type 2C protein phosphates (PP2Cs) protein which inhibits PP2Cs protein activity and further phosphorylates SNF l-related protein kinase 2 (SnRK2s) [26,27,28]. SnRK2 then undergoes autophosphorylation, controlling various physiological reactions, including ion channels, transcription factors such as ABI5 [29] and transporters [30,31]. ABI5 is a basic leucine zipper (bZIP) transcription factor and a crucial component of the ABA signal transduction system [32,33]. The bZIP transcription factor family is the most widespread and conserved class of eukaryotic transcription factors and primarily controls the buildup of stored proteins during seed maturation [34]. ABI5, which binds to the ABA-responsive cis-acting element (ABRE) of ABA-inducible gene promoters, regulates early seeds development and dilation and is essential for determining embryogenesis and regulating the transition from germination to vegetative growth [35].
Paeonia suffruticosa and P. lactiflora are two beloved and established floral species in China, known for their extensive history and geographical range [36,37]. Additionally, these hybrids have upright stalks, vibrant colors, and increased resilience to adversity stresses [38]. As prolonged intraspecific hybridization has limited P. suffruticosa’s potential for superior breeding, interspecific hybridization has become a strategy for creating new varieties. Moreover, the new flower varieties have not yet reached their full potential, and flower germplasm resources are underutilized. It is possible to significantly increase the preservation and use of germplasm resources by breeding efficient hybrids. However, in the case of distant hybridization, immature embryos often wither and perish soon after fertilization. Thus, understanding the molecular mechanisms behind embryo abortion in peonies is critical to the dominant breeding of hybrid varieties. To be clear, the technique for resolving embryo abortion in distant hybridization of peony is embryo rescue, which must be closely associated with the hormone content [39]. Additionally, it is still unknown how exactly endogenous hormones cause embryo abortion. Here, P. suffruticosa ‘Fengdanbai’ was chosen as the female and P. lactiflora ‘Fenyunu’ was the male due to the high oil value of the former. Additionally, we analyzed transcriptome sequences and determined hormone contents to compare the differences between normal and aborted embryos. Our study aims to explore the possibility and potential mechanisms of hormones’ role in clarifying the molecular mechanisms of embryo abortion in the distant hybridization of peonies. Through the study of key genes of related hormone regulation, embryo rescue, and breeding efficiency can be improved. In addition, this study offers a theoretical basis for cultivating efficient varieties and increasing the preservation and use of germplasm resources.
2. Materials and Methods
2.1. Plant Materials and Pretreatment
All the materials utilized in the experiments were grown under identical conditions at the Research Center of the High-quality flower and vegetable project, at Henan Agricultural University, located in Zhengzhou, Henan, China (34.78° N, 113.66° E). The male parent was P. lactiflora ‘Fenyunu,’ a typical early flowering pink single-petal variety from the herbaceous family. The female parent was a white-flowered deciduous shrub called P. suffruticosa ‘Fengdanbai.’ Pollen was collected from the male parent between 9 a.m. and 11 a.m. in May 2021, and its vitality was determined using in vitro culture before being stored at 4 °C. The following year, artificial pollination, repeated pollination, and bagging were all carried out. At the same time, the pollen of ‘Fengdanbai’ is collected and self-pollinated on the stigma. Prior studies conducted by the research group have shown significant differentiation among peony embryos at 26 days of pollination (DAP). Selected the ‘Fengdanbai’ seeds after self-crossing normal (Ps self normal), the hybrid-aborted seeds (Ps hybrid aborted), and hybrid-normal seeds (Ps hybrid normal) at the same stage. Embryos were collected at various stages of development, immediately frozen in liquid nitrogen, and then stored at −80 °C. Three biological replicates of three different samples were utilized for the RNA-seq analysis.
2.2. Determination of the Hormone Content
Using an enzyme-linked immunoassay (ELISA) and the methodology outlined by Dietz [40], the same weight of seeds is selected to determine the hormone concentration in various seed embryos. Stir the sample by adding a certain amount of PBS (pH 7.4) and using a homogenizer. After centrifugation for 20 min, collect the supernatant to obtain the solution. Dilute the sample 5-fold, then add the microplate reagent with the standard sample and incubate for one hour. After washing with washing solution, add both chromogen A and B to the wells. After 15 min of color development at 37 °C while in the dark, a stop solution is then added. Based on the regression equations between the optical density and concentration of the standard hormone, the concentrations of ABA, IAA, and GA were measured at 450 nm using an Infinite 200 PRO (Tecan, Austria). Shanghai Yuanju Biotechnology Center provided the enzyme-linked kit. The hormone content of the samples was determined according to the standard curve. The experiment was conducted with three biological replicates to ensure the accuracy and reproducibility of the results.
2.3. RNA Extraction and Transcriptome Sequencing
The RNA extraction method utilized was CTAB, and the concentration and purity of the extracted RNA were determined using a NanoDrop UV-Vis Spectrophotometer (Thermo Fisher Scientific, Madison, USA) with one μL of the sample. Following the directions provided by the manufacturer, cDNA was synthesized using the Vazyme Lamp® Master Mix (Dye Plus, Vazyme, Nanjing, China). Agarose gel electrophoresis was used to determine the integrity and concentration of the RNA and the cDNA libraries were generated by PCR amplification. The cDNA libraries were sequenced using the high-throughput sequencing platform Illumina NovaSeq6000 by Beijing BMK Biotechnology Inc. in Beijing, China (https://www.biocloud.net, accessed on 1 July 2022).
2.4. Transcriptome Assembly and Functional Annotation
Following RNA-Seq, the raw reads underwent further filtration to obtain clean reads by removing those containing more than 10% of unknown nucleotides and low-quality reads. The resultant high-quality Clean Data was obtained after eliminating low-quality reads, ploy-N, and adapters. The Q30 and GC content of clean reads were then computed. The Clean Data was then denovo assembled into a transcript using Trinity 2.5.1, which was then sequence-completed to obtain a Unigene library that served as the reference sequence for subsequent species analysis. To obtain functional annotation data, the unigene sequences were compared with the KOG, COG, NR, GO, Pfam, Swissprot, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using DIAMOND 2.0.4 software. KOBAS 3.0 was used to obtain KEGG Orthology results for Unigene. The HMMER 3.1b2 software was then employed to compare the amino acid sequence of Unigene with the Pfam database to obtain Unigene annotation data. The BLAST parameter E-value was not greater than 1 × 10−5, while the HMMER parameter E-value was not greater than 1 × 10−10. DNAMAN 9.0 was utilized to predict CDS by comparing the transcript and database using BLAST. The expression abundance of the corresponding Unigene is expressed as Fragments Per Kilobase of Transcript per Million Mapping Reads (FPKM) value.
2.5. Differentially Expressed Genes Analysis of the Data
We employed the DEGseq method to identify differentially expressed genes (DEGs) among the three groups (Ps hybrid aborted vs. Ps hybrid normal, Ps hybrid aborted vs. Ps self normal, Ps hybrid normal vs. Ps self normal), which involved contrasting gene expression patterns across different hybridization modes. This approach utilizes a statistical framework based on the negative binomial distribution to determine differential expression levels. To correct for multiple testing, we utilized the widely accepted Benjamini-Hochberg method in the differential expression analysis process. The corrected p-value, or False Discovery Rate (FDR), was then utilized as the primary index for identifying differentially expressed genes, minimizing the potential for false positives. We applied screening criteria for DEG identification including a difference multiple Fold Change (FC) of 1.5 or greater, which measures the expression ratio between two samples, and an FDR less than 0.05. We identified genes encoding TFs among the module genes using the PlantTFDB database (http://planttfdb.gao-lab.org, accessed on 10 November 2022). We visualized the gene co-expression network using Cytoscape 3.9.1 and calculated gene connectivity using the same software. Node size was positively associated with the degree of gene connectivity.
2.6. Quantitative Real-Time PCR Verification and Expression Analysis
The RNA extracted from hybrid embryos underwent reverse transcription using the HiScript® II 1st Strand cDNA Synthesis Kit (+gDNA wiper) from Vazyme (Nanjing, China). The group’s preliminary research found that the β-Tubulin gene was the most stable and the most suitable gene for peony internal reference [41]. The Prime Primer 5.0 program was used to create a specific primer using the β-Tubulin genes as an internal reference (Table S1). The real-time quantification of target genes was conducted using the Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Marsiling, Singapore), along with the SYBR® Green Pro Taq HS qPCR Kit II (Rox Plus) (AG11702, Changsha, China). The comparative threshold cycle (CT) method (2−△△CT formula) [42] was followed to calculate the relative expression levels of the target genes. The analysis was performed using three technical replicates and each biological sample was analyzed thrice.
3. Results
3.1. Peony Seeds Development and Quantitative Analyses of Hormone Levels
The seed embryos in self-crossing peony pods were evenly sized and arranged (Figure 1A). However, in the hybridized pods, the seeds were of varying sizes (Figure 1B). Both the hybrid-normal embryos and self-crossing normal embryos were well-developed, with the former being slightly larger. The hybrid-aborted embryos were smaller in size, both laterally and longitudinally than the normal embryos. It appears that the development of aborted embryos had ceased before 26 DAP, and the distinction between normal embryos and hybrid aborted peony embryos is apparent. The measurement of plant hormone content (Figure 1C) revealed that the ABA content in the aborted seed embryos was unusually high compared to that in the normal embryos, with the least amount of ABA found in the standard hybrid embryos. The levels of IAA and GA followed a similar trend as ABA. The ratio of GA+IAA/ABA in the aborted embryos was lower than that in hybrid-normal embryos but higher than that in self-crossing normal embryos. It can be inferred that there is a close relationship between embryo abortion and hormone content.
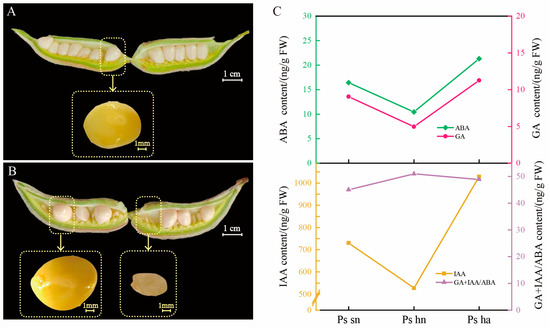
Figure 1.
The phenotype of seeds and seed pods, as well as the hormone content of seeds, were studied at 26 DAP in three groups. (A) Diagram of the fruiting pods and self-crossing normal seeds of P. suffruticosa ‘Fengdanbai’. (B) Diagram of the fruiting pods and seeds of the combinations P. suffruticosa ‘Fengdanbai’ × P. lactiflora ‘Fenyunu’, hybrid-normal seeds on the left and hybrid-aborted seeds on the right. (C) Folding line chart of plant hormones content for the three groups. Ps ha: the hybrid-aborted embryos, Ps hn: the hybrid-normal embryos, Ps sn: the self-crossing normal embryos.
3.2. Summary and Transcriptome Data Analysis of DEGs in RNA Sequencing
The transcriptomes of nine samples were sequenced, and a total of 93.85 Gb Clean Data was obtained, with each sample’s Clean Data reaching 10.01 Gb and the Q30 base ratio exceeding 92.79% (Table S2). A total of 69,816 unigenes with annotation information were retrieved with respect to seven databases (Table S3). To identify DEGs involved in peony embryo development, we analyzed differential expression in embryos with different developmental conditions and hybrid combinations including the (Ps hybrid aborted vs. Ps hybrid normal), (Ps hybrid aborted vs. Ps self normal), and (Ps hybrid normal vs. Ps self normal) groups. The (Ps hybrid aborted vs. Ps hybrid normal) group had the highest number of DEGs at 7777, with 3576 up-regulated and 4201 down-regulated. In contrast, the (Ps hybrid aborted vs. Ps self normal) group had 6163 DEGs, with 2849 up-regulated and 3314 down-regulated (Figure 2A). The (Ps hybrid normal vs. Ps self normal) group had 1820 DEGs, with 1025 up-regulated and 795 down-regulated. Additionally, it can also be observed that, in (Ps hybrid normal vs. Ps self normal), more genes were up-regulated than down-regulated, whereas in (Ps hybrid aborted vs. Ps hybrid normal) and (Ps hybrid aborted vs. Ps self normal), more genes were down-regulated than up-regulated. The (Ps hybrid aborted vs. Ps hybrid normal) and (Ps hybrid aborted vs. Ps self normal) groups showed the DEGs that compare aborted and normal embryos in different hybridization. Of these, there were 4482 DEGs in common between the (Ps hybrid aborted vs. Ps hybrid normal) and (Ps hybrid aborted vs. Ps self normal) groups (Figure 2B), which accounted for the largest proportion and may be the key to the difference between aborted embryos and normal embryos.
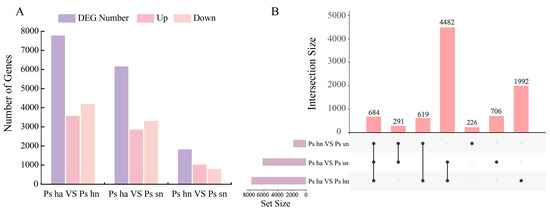
Figure 2.
DEGs expression patterns during different conditions of development. (A) The number of DEGs with up-regulated and down-regulated genes during embryonic developmental stages; (B) The UpSet graphic summarizes the intersection of DEGs for the three groups. The bars in the bottom left corner represent the number of differential genes in each group, and each line connected by circles in the graph on the right represents the number of differential genes that overlap in this combination. Ps ha: the hybrid-aborted embryos, Ps hn: the hybrid-normal embryos, Ps sn: the self-crossing normal embryos.
3.3. Functional Classification and Enrichment of DEGs
Upon comparison of the DEGs with the GO database for the three differential groupings, it was discovered that a total of 55,317 DEGs were annotated into 57 GO functional categories (Figure S1). The highest number of DEGs were annotated to cellular components, with a total of 2404 DEGs, among which the most common was “cells” and “cell part”. A total of 2605 DEGs were annotated to biological processes, with “cellular process” being the most frequently occurring term. Additionally, 3105 DEGs were annotated to molecular functions, with the highest number being “binding”. Through comparison with the KEGG database, it was found that 21,390 DEGs in the three differential groupings are involved in 133 metabolic pathways, which include genetic information processing, cellular processes, environmental information processing, metabolism, and organismal systems (Figure S2). The top 20 enriched KEGG metabolic pathways of DEGs were chosen in each of the three differential groups for mapping. Specifically, “Plant hormone signal transduction” (ko04075) (163 DEGs, Ps hybrid aborted vs. Ps hybrid normal; 131 DEGs, Ps hybrid aborted vs. Ps self normal) exhibited an extremely significant enrichment level relative to the other 19 terms, respectively (Figure 3A,B). This also indicates that the most prominent differential enrichment for the contrast between normal and abortion embryos was “Plant hormone signal transduction”. However, “Plant hormone signal transduction” pathways were not revealed by the DEGs enrichment between normal embryos, and “Phenylpropanoid biosynthesis” had an extremely significant enrichment level relative to the other 19 terms (Figure S4). It is highly likely that “Plant hormone signal transduction” is the crucial regulatory mechanism associated with embryo abortion.
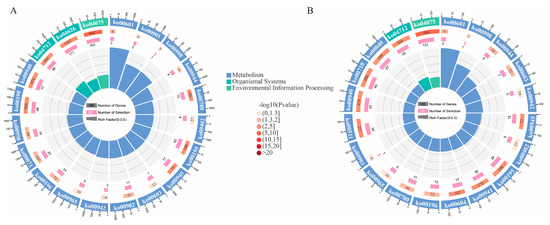
Figure 3.
Network diagram of differentially expressed genes and KEGG pathways. (A) DEGs in (Ps hybrid aborted vs. Ps hybrid normal). (B) DEGs in (Ps hybrid aborted vs. Ps self normal). From outside to inside, the first circle represents the top 20 enriched KEGG terms. Different colors represent different ontologies. Outside the first circle is the scale of the number of genes. The second circle is the number of DEGs and the p-value of the KEGG term. The third circle represents the number of genes in the term. The fourth circle is the Rich Factor value of each KEGG term and the background grid line, with each grid representing 0.1.
3.4. Analysis of DEGs in the Common Expression Patterns
The common expression patterns of DEGs in the three differential groups were analyzed, and mRNAs with the same expression trend were divided into one data set and plotted by analyzing the different patterns of variation in mRNA expression abundance between samples, in total dividing the data into seven clusters (Figure 4). Among the seven clusters, three clusters were found that were relatively consistent with the phytohormone phenotype. Aborted embryos were found to be significantly up-regulated in three clusters with 1012, and 3134 common expressed genes, respectively (Figure 4A,G). Separate KEGG enrichment maps of their common expressed genes are represented (Figure 4H,I). Still, their KEGG enrichment is mostly associated with “Plant hormone signal transduction”. In addition, 281 DEGs were also up-regulated in aborted seeds (Figure 4D), with the number of DEGs and KEGG enrichment mostly associated with “photosynthesis” (Figure S4). In the common expressions pattern, the vast majority of genes were still found to be significantly enriched in the “Plant hormone signal transduction”, “Plant-pathogen interaction”, and “MAPK signaling pathway-plant” pathway.
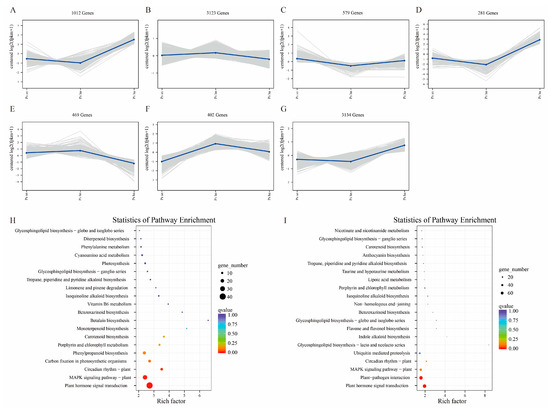
Figure 4.
The graph depicts the trend analysis of common gene expression and the KEGG enrichment analysis of three of seven clusters. (A–G) Graphs of gene common expression trends for the seven clusters, with the number of genes on the graph being the number of genes in the same expression trend. (H) KEGG enrichment analysis of 1012 DEGs in plot A. (I) KEGG enrichment analysis of 3134 DEGs in plot G. Ps ha: the hybrid-aborted embryos, Ps hn: the hybrid-normal embryos, and Ps sn: the self-crossing normal embryos.
3.5. Metabolic Pathway about Plant Hormone Signaling Pathways
Upon analysis of DEGs between aborted and normal embryos, it was observed that a significant number of DEGs were enriched in the “Plant hormone signal transduction” pathway. Through enrichment analysis of the KEGG pathway, a range of hormone signaling-related genes, including those encoding receptors and response factors, were identified (Figure 5). It was observed that the FPKM values exhibited significant differences between aborted and normal embryos, with the trend remaining consistent in normal embryos.
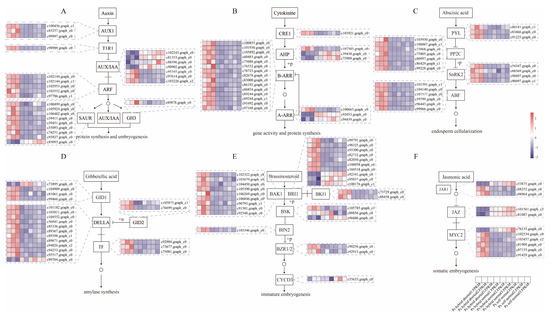
Figure 5.
The metabolic pathway for six phytohormones contains DEGs from three different types of embryos. Each hormone pathway (A–F) has a number of family genes that play a regulatory role, and the boxed sections are the genes corresponding to that family. Colors indicate normalized values of FPKM; red indicates up-regulated expression, and blue indicates down-regulated expression. The samples are arranged in the order of the boxes in the diagram, and the analysis has three biological replicates.
In the IAA pathway, almost all transcription factors belonging to the ARF family are up-regulated in hybrid-aborted embryos, while most genes in the IAA pathway are down-regulated. GH3, acting as a negative regulator of growth hormone, is up-regulated in hybrid-aborted embryos (Figure 5A). In the CTK pathway, all genes, except for the AHP family and A-ARR family genes, are up-regulated in hybrid-aborted embryos (Figure 5B). ABA promotes endosperm cellularization during embryonic development, and all DEGs are up-regulated in hybrid-aborted embryos but down-regulated in normal embryos (Figure 5C). In the GA signal pathway, which promotes amylase synthesis, DELLA proteins act as inhibitors of GA signaling. However, almost all DELLA family member genes are up-regulated in hybrid-aborted embryos (Figure 5D). BR promotes immature embryogenesis and the expression of the BZR1/2 gene as a major regulator of BR signaling is up-regulated in hybrid-aborted embryos (Figure 5E). JA plays an important role in plant development and response to abiotic stresses. Two JAZ family genes are down-regulated and five MYC2 genes are up-regulated in hybrid-aborted embryos (Figure 5F). It suggests that these genes may be associated with the occurrence of abortion in the embryo.
3.6. Analysis of Transcription Factors and Core Genes in Hormones Pathway
Transcription factors are crucial in regulating gene expression and play a pivotal role in embryonic development. Among the genes in the common expression trend (Figure 4A,G), 119 genes were found to be enriched in “Plant hormone signal transduction” pathways. These genes include 48 TFs and 9 transcriptional regulators (TRs) (Table S4). The majority of these transcription factors belong to other families such as GARP-G2-like (Golden2, ARR-B, Psr1-Golden2-Like), GRAS (GAI, RGA, SCR), bHLH (basic Helix-Loop-Hleix), bZIP (basic leucine zipper), and other families (Figure 6A). Using Cytoscape software, the 119 genes were visualized to form a protein interaction network consisting of 195 pairs of 87 genes, with each gene assigned a score based on the degree algorithm (Figure 6B). Notably, ARF5, GAI, and ABI5, which have the highest ranking in the protein interaction network, interact with all other proteins significantly. The genes closer to the center of the diagram have more interactions with other genes. Additionally, IAA, GA, BR, ABA, JA, and CTK signaling pathway genes were found to be involved in the network. Both ARF5, GAI, and ABI5 were highly expressed in aborted embryos, indicating that these phytohormone-related transcription factors regulate embryonic abortion and play a vital role in the transcriptional regulatory network.
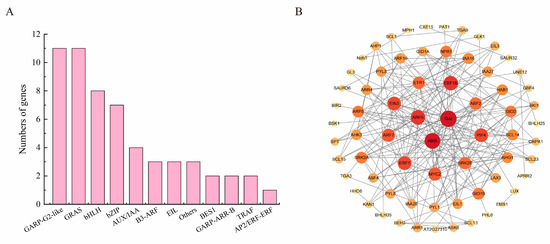
Figure 6.
A diagram of the key transcription factors involved in the plant hormone pathway, as well as the protein interaction network. (A) The families to which the 57 TFs and TRs belong and the number of genes in each family. (B) Diagram of the protein interaction network of DEGs involved in plant hormone signaling pathways. Each circle represents a gene’s protein, with darker hues signifying a larger proportion of the protein network. The stronger the protein interaction and the larger the protein proportion, the larger the circle and the redder the color.
3.7. Validation of DEGs Expression by qRT-PCR
Based on the above analysis, nine DEGs were selected for qRT-PCR analysis to verify the reliability of the RNA-seq data. They mainly involve the signaling pathways of ABA and GA-related plant hormones including some related transcription factors. qRT-PCR results were highly consistent with the values obtained by RNA-seq (Figure 7). This demonstrates the reliability and reproducibility of the RNA-seq analysis.
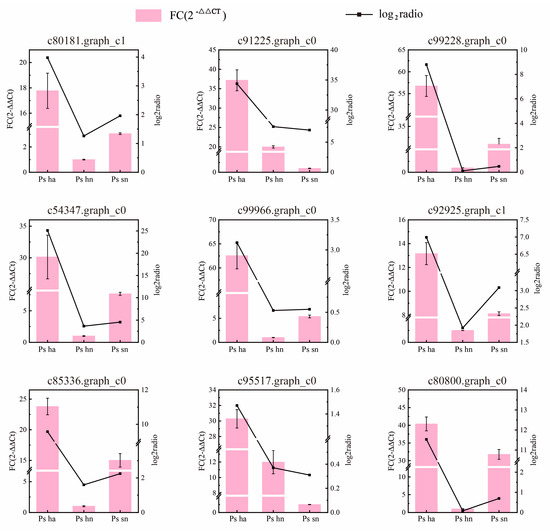
Figure 7.
qRT-PCR validation of the 9 DEGs involved in plant hormone pathways. The ratio of each sample to log2 transformed expression levels in RNA-seq. β-actin served as an internal control. The error bars show the standard error of three biological replicates.
4. Discussion
4.1. Hormones Were Involved in Regulating the Development of the Embryo
Abnormal levels of hormones and imbalances in endogenous hormones during embryonic development can result in embryonic abortion [43]. Transcriptome data revealed that “Plant hormone signal transduction” was the primary difference in gene expression between aborted and normal embryos, especially in the KEGG enrichment map. Additionally, this pathway was absent in the differential enrichment of the two types of normal embryos. Similar findings were reported by Chen et al. on the gene regulatory network of peanut embryo abortion under calcium deficiency [8]. During the development of peony embryos, hormone levels in aborted embryos differ significantly from those in normal embryos. In peony embryos, IAA promotes embryonic cell division and growth, protein synthesis, and embryogenesis [13]. It is confusing that, compared with the normal embryos, the content of IAA was significantly higher in 26 DAP aborted embryos (Figure 1). Upon sensing auxin signals, the AUX/IAA protein family is regulated by the Auxin Response Factor (ARF) family for downstream gene transcription [44,45]. ARF5 is a key transcription factor in response to Auxin [46,47]. In Arabidopsis thaliana, down-regulating the expression of the ARF5 gene leads to defects in embryo sacs, accompanied by seed abortion [48]. However, in our study, the expression of ARF5 (c97786.graph_c1) was up-regulated in aborted embryos compared to normal embryos, consistent with the high levels of IAA in aborted seeds (Figure 5A). GH3 family genes regulate hormone homeostasis and overexpression of GH3 suppresses Auxin’s content [49]. GH3.9 (c89878.graph_c0) is genetically up-regulated in aborted embryos compared to normal embryos (Figure 5A). This suggests that the abnormal increase in IAA levels may disrupt hormonal homeostasis, which may be associated with embryonic abortion.
Similar to the role of IAA, GA facilitates the germination and development of seeds and stimulates the synthesis of amylase, thereby promoting starch accumulation and providing more energy to the embryo [11]. Unexpectedly, the level of GA was also significantly lower in normal embryos compared to the aborted ones (Figure 1). GA is a diterpenoid phytohormone and Gibberellin Insensitive Dwarf 1 (GID1) acts as a GA receptor, interacting with DELLA proteins, which are nuclear transcriptional regulators that repress GA signaling and restrict plant growth [50,51]. It has been suggested that GID1 may be involved in the development of the ovule [52]. Our transcriptome data showed that two GID1 genes (c99466.graph_c0, GID1A; c81061.graph_c0, GID1B) were up-regulated in aborted embryos (Figure 5D). It is speculated that they may play a role in the process of embryogenic abortion. Studies in Arabidopsis thaliana have demonstrated that the GRAS family transcription factor SCL28 may control the mitotic cell cycle and influence cell expansion and differentiation [53]. The down-regulation of SCL28 genes (c99704.graph_c0) in aborted embryos (Figure 5D) may have inhibited cell proliferation, while members of the DELLA proteins family were highly expressed, negatively regulating GA. This abnormal change may be associated with embryonic abortion.
Low concentrations of ABA promote embryogenesis and embryo development and high levels of ABA inhibit amylase synthesis and activity, affect glucose metabolism, and promotes endosperm cellularization [54]. Consistent with previous studies on aborted peanut embryos and Chinese white poplar [55,56], our study found that the content of ABA was abnormally high in aborted peony embryos (Figure 1). The buildup of phosphorylated SnRK2s causes the phosphorylation of bZIP transcription factors known as ABFs/AREBs and induces the activation of ABA-responsive genes [57,58]. In our transcriptome data, all ABF family transcription factors were up-regulated in aborted peony embryos, consistent with the high levels of ABA observed (Figure 5C). The transcription factor ABI5 plays a key role in blocking embryo growth and promoting endosperm cellularization, particularly in the later stages of embryogenesis when the expression is strongest [59,60]. We observed significant up-regulation of ABI5 (c99966.graph_c0) in the peony embryos, suggesting its involvement in the process of embryonic abortion (Figure 5C). Similar to observations in rice hybrid seeds studies, we also found a failure in early endosperm cellularization that resulted in nonviable seeds [61]. Additionally, the ratio of GA+IAA/ABA in peony aborted embryos was lower than in hybrid-normal embryos, similar to findings in peanut and seedless grape embryos [43,62]. We speculate that hormonal imbalances may also contribute to embryonic abortion in peonies.
CTK hormone is a pivotal regulator of plant growth and development and plays a crucial role in determining seed size and enhancing seed set, particularly during the development of endosperm and embryo [14]. Although we did not assess the content of other hormones in the embryos due to difficulties and high costs, we identified numerous genes through transcriptome analysis. CTK signaling is transduced by Arabidopsis histidine kinases (AHK), which recognize CTK and transfer the phosphate group to Arabidopsis histidine-phosphotransfer proteins (AHPs) [63]. Subsequently, phosphorylated AHPs enter the nucleus and transfer the phosphoryl group to B-ARRs, which triggers the immediate induction of target genes such as the A-ARR genes that inhibit CTK [64]. The A-ARR and AHP genes were found to be significantly down-regulated during embryonic development in cucumber [65], whereas, in our study, the ARR4 gene (c100663.graph_c0) and AHP gene (c107303.graph_c0) were found to be significantly up-regulated in aborted embryos (Figure 5B). It suggests that they may be implicated in the development of aborted embryos.
BR hormone plays a crucial role in controlling the size and count of embryonic cells, and the BR-mediated transcription factor, BRASSINAZOLE-RESISTANT1 (BZR1), can affect the development of both the embryo and endosperm [15]. BEH2 (c90256.graph_c0) and BEH4 (c92915.graph_c0) are up-regulated in aborted embryos and they both belong to the same family as BZR1, speculating that they may be involved in the process of embryo development. The plant GSK-3 kinase, Brassinosteroid insensitive 2 (BIN2), acts as a key regulator of BR signaling. Previous research has shown that the up-regulation of BRI1-EMS-suppressor 1 (BES1) and the suppression of BIN2 gene expression can promote somatic embryogenesis [66]. In the aborted peony embryos that we studied, the BIN2 gene (c103346.graph_c0) is highly expressed in aborted embryos (Figure 5E). A similar situation was found in maize, where the BIN2 homolog ZmSK2 was overexpressed, resulting in severe BR phenotypic defects and abnormalities [67]. Hence, the abnormally high expression of BIN2 may be associated with embryo abortion.
During embryonic development, JA has been found to potentially induce cell death or malformation of somatic embryos, as demonstrated in the hybrid studies of tomato and Liriodendron [17,68]. In the JA signaling pathway, inhibition of JASMONATE-ZIM-PROTEIN (JAZ1) and up-regulation of MYC2 can impede embryogenesis [69]. In the aborted peony embryos that we studied, MYC2 (c91429.graph_c0) was found to be significantly up-regulated and JAZ1 (c81087.graph_c0) was down-regulated in the aborted embryos (Figure 5F), suggesting that abnormal expression of these genes may be associated with embryo abortion.
4.2. The Regulatory Relationship between TFs and Pathway Genes
Based on the shared expression patterns, we discovered genes that had a strong correlation with phytohormones to further investigate the function of the six hormones during embryonic development. In the two clusters, we were able to find 119 genes associated with phytohormones, of which 48 TFs were identified in all six hormone signaling pathways (Figure 6A). The GLKs (GOLDEN 2 LIKEs) family of transcription factors primarily controls the growth of chloroplasts in plants, affecting the buildup of starch and sugar in fruit and enhancing plant seed production [70]. During peony hybridization, the GLKs transcription factor family occupies a considerable proportion of the seeds, and it is speculated that they may regulate the development of photosynthetic products in the seeds and that there is some regulatory interaction with phytohormones. Studies have shown that the bZIP transcription factor family, the most widespread and conserved class of eukaryotic transcription factors, primarily controls the accumulation of stored proteins during seed maturation. The bHLH transcription factor family controls the embryo, endosperm, and seed skin, primarily regulating the process of morphological development in the initial stage of seed development [34]. A large number of bZIP and bHLH transcription factors involved in the regulation can be observed (Figure 6A). It is speculated that they may be involved in regulating seed development and controlling storage proteins, which is consistent with our results. The rest of these transcription factors are involved in the phytohormone pathway, and they all play some regulatory role in plant embryo development.
These TFs can stimulate specific pathways through signaling which can induce gene expression and affect embryonic development. In the protein interaction network, ABI5 (c99966.graph_c0), a member of the bZIP transcription factor family, has a high degree of interaction with other genes (Figure 6B). Research on Arabidopsis has revealed that ABA inhibits lipid mobilization in embryos, but not in the endosperm [71,72], suggesting that embryo development is likely regulated by endosperm lipid breakdown under ABA regulation. ABI5, whose expression is restricted to the micropylar region of the endosperm and embryo [73], is involved in ABA-mediated early seed development [32], especially in the process that promotes endosperm cellularization via ABI5-related bZIP transcription factors [16]. It can be speculated that peony aborted embryos may be due to the high expression of ABI5 leading to endosperm cellularization during the developmental stage, which affects the accumulation of seed storage proteins, resulting in the seeds crumpled and failing to provide sufficient energy for normal development. Additionally, transcription factors related to growth hormones such as ARF5 (c97786.graph_c1) and GAI (c95517.graph_c0) are highly expressed in aborted embryos, but the exact mechanism remains to be investigated. Previous studies have found that abnormal balance between plant hormones can also affect seed development [43]. It is presumed that the transcription factors ABI5, which stands in for ABA and ARF5, which stands in for IAA and GAI, which represents GA and has strong interactions with other transcription factors and proteins. According to this, the IAA, GA, and ABA hormone concentration imbalance may also be a contributing factor in abortion.
A previous study demonstrated that ABA treatment can impact the expression levels of genes in the ARF family, specifically augmenting the ubiquitination of ARF6, indicating a potential connection between ABA and ARF-controlled genes [74]. DELLA proteins act in opposition to the nuclear transcriptional regulator ICE1 through direct interaction to control ABA signaling during seed development [75,76,77]. Denay et al. discovered that ICE1 encodes an MYC-like basic helix-loop-helix (bHLH) transcription factor that plays a significant role in regulating the breakdown of the endosperm in Arabidopsis [78]. In the ABA pathways, some studies have shown that ARR4, as a member of A-ARR, can adversely regulate the expression of ABI5 [79]. Additionally, the transcription factor BZR1 can directly interact with the transcription factor ABI5 in the ABA signaling pathway [80,81]. Studies have found that JAZ1 can regulate ABI5 transcription factors and hinder their activity [82]. BRAHMA is believed to be important in plant embryonic and reproductive development in the ABA pathway, regulating the transcription of related target genes and suppressing the expression of ABI5 [83,84]. It has been demonstrated that BRAHMA plays a crucial role in regulating phytohormone signaling pathways such as ABA, IAA, CTK, and GA [85].
Due to ABI5 having a significant inhibitory effect on embryonic development in late embryonic development and it has been found that ABA and other plant hormones can be integrated into signals via ABI5. Studies have shown that CTK, GA, and BR signaling, and metabolic pathways are involved in the regulation of ABI5 or are controlled by ABI5 [33]. We speculate that during embryo abortion, the ABA signaling pathway interacts with other pathways through various mechanisms (Figure 8). Specifically, ABA increases the expression of ABI5, while CTK inhibits ABI5 through ARR4, GA inhibits ABI5 by promoting the expression of DELLA protein and ICE1 in an antagonistic manner, BR inhibits ABI5 by increasing the expression of BZR1, and JA inhibits ABI5 expression through JAZ1. However, the specific relationship between IAA and ABA in the signaling pathway still requires further investigation.
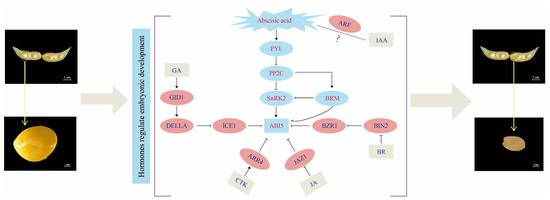
Figure 8.
Map of related hormonal regulatory networks leading to embryo abortion. On the far left of the image is the hybrid-normal seeds (Ps hybrid normal), on the far right is the hybrid-aborted seeds (Ps hybrid aborted), and in the middle is the regulatory network between the six plant hormones.
5. Conclusions
In summary, hormones play an irreplaceable role in embryonic development. We have measured hormone levels and found that they are high in hybrid aborted embryos. Transcriptome analysis and the gene common expressions trends have identified numerous genes and TFs that potentially contribute to embryonic abortion, but more investigation is required to fully understand their mechanisms in peony hybridization. The subsequent validation of key transcription factors can facilitate the breeding of excellent hybrids of peonies and can improve the yield and quality of peony seed crops. This study will provide a fundamental basis for the metabolic regulation of embryo abortion in peony hybridization.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9060694/s1. Table S1. Design of the primer sequences; Table S2. Alignment statistics of the sequencing data and the assembly results; Table S3. The Unigene annotation statistics table; Table S4. TFs and TRs in the common expression trends; Figure S1. GO classification of DEGs; Figure S2. KEGG classification of DEGs; Figure S3. KEGG enrichment of DEGs in (Ps hybrid normal vs. Ps self normal); Figure S4. KEGG enrichment of the common expression trends.
Author Contributions
D.H.: Writing—review and editing, Conceptualization, Project administration. H.G.: Writing—original draft, Investigation and Formal analysis. S.H.: Supervision, Visualization, and Funding acquisition. M.Z.: Investigation and Methodology. Y.C.: Data curation and Formal analysis. Z.W.: Data curation and Resources. Y.L.: Validation and Visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31600568 and 31870698, the Natural Science Foundation of the Henan Province, grant number 232300420006, the Henan Province Science and Technology Research Project, grant number 202102110234, and Key Scientific Research Project of Henan Provincial Colleges and Universities, grant number 23A220001.
Data Availability Statement
The Illumina reads have been deposited in the NCBI Sequence Read Archive (SRA) Database (http://www.ncbi.nlm.nih.gov/sra, accessed on 2 March 2023) and are open for research. Accession number PRJNA940270.
Conflicts of Interest
There are no competing interests and personal relationships in this paper.
References
- De Smet, I.; Lau, S.; Mayer, U.; Jürgens, G. Embryogenesis—The humble beginnings of plant life. Plant J. 2010, 61, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Schon, M.; Baxter, C.; Xu, C.; Enugutti, B.; Nodine, M.D.; Dean, C. Antagonistic activities of cotranscriptional regulators within an early developmental window set FLC expression level. Proc. Natl. Acad. Sci. USA 2021, 118, e2102753118. [Google Scholar] [CrossRef] [PubMed]
- Teng, R.; Shao, Q.; Wu, M.; Wang, H.; Li, M.; Huang, Y. Reproductive barriers to hybridizations between narrow-leaf and broad-leaf Anoectochilus roxburghii. J. Hortic. Sci. Biotech. 2017, 92, 183–191. [Google Scholar] [CrossRef]
- He, D.; Zhang, M.; He, S.; Cao, J.; Hua, C.; Zhang, J.; Liu, Y. Cloning, expression and physiological mechanism of Paeonia suffruticosa distant hybrid seeds abortion PsMTERF2 gene. J. Northwest Sci.-Tech. Univ. Agric. For. Nat. Sci. Ed. 2022, 50, 108–116. [Google Scholar] [CrossRef]
- Xu, S.; Hou, H.; Wu, Z.; Zhao, J.; Zhang, F.; Teng, R.; Chen, F.; Teng, N. Chrysanthemum embryo development is negatively affected by a novel ERF transcription factor, CmERF12. J. Exp. Bot. 2022, 73, 197–212. [Google Scholar] [CrossRef]
- Noguero, M.; Le Signor, C.; Vernoud, V.; Bandyopadhyay, K.; Sanchez, M.; Fu, C.; Torres-Jerez, I.; Wen, J.; Mysore, K.S.; Gallardo, K.; et al. DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 2015, 81, 453–466. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Z.; Dong, W.; Sun, C.; Wang, H.; Song, A.; He, L.; Fang, W.; Chen, F.; Teng, N. Transcriptomic and proteomic analysis reveals mechanisms of embryo abortion during chrysanthemum cross breeding. Sci. Rep. 2014, 4, 6536. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Fu, H.; Chen, K.; Zhao, S.; Zhang, C.; Cai, T.; Wang, L.; Lu, W.; Dang, H.; et al. Identification of Key Gene Networks and Deciphering Transcriptional Regulators Associated With Peanut Embryo Abortion Mediated by Calcium Deficiency. Front. Plant Sci. 2022, 13, 814015. [Google Scholar] [CrossRef]
- Chan, A.; Carianopol, C.; Tsai, A.Y.; Varathanajah, K.; Chiu, R.S.; Gazzarrini, S. SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. Exp. Bot. 2017, 68, 4219–4231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, M.; Li, S. Research progress on mechanism of plant embryo abortion. J. Northeast. Agric. Univ. 2021, 52, 89–96. [Google Scholar] [CrossRef]
- Li, Q.; Yang, A. Comparative studies on seed germination of two rice genotypes with different tolerances to low temperature. Environ. Exp. Bot. 2020, 179, 104216. [Google Scholar] [CrossRef]
- Pang, F.; Ma, X.; Zhang, X.; Li, X.; Ji, W. A study on the factors influencing rescue success of the embryo in stenopermocarpic grape. J. Fruit Sci. 2021, 38, 1699–1707. [Google Scholar] [CrossRef]
- Verma, S.K.; Das, A.K.; Gantait, S.; Gurel, S.; Gurel, E. Influence of auxin and its polar transport inhibitor on the development of somatic embryos in Digitalis trojana. 3 Biotech 2018, 8, 99. [Google Scholar] [CrossRef]
- Brugiere, N.; Humbert, S.; Rizzo, N.; Bohn, J.; Habben, J.E. A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Cytokinin biosynthesis in maize. Plant Mol. Biol. 2008, 67, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, C.; Li, Y.; Yu, H. Mobile TERMINAL FLOWER1 determines seed size inArabidopsis. Nat. Plants 2020, 6, 1146. [Google Scholar] [CrossRef]
- Wasternack, C.; Forner, S.; Strnad, M.; Hause, B. Jasmonates in flower and seed development. Biochimie 2013, 95, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiao, Y.; Zhang, C.; Dou, M.; Weng, K.; Wang, Y.; Xu, Y. VvHDZ28 positively regulate salicylic acid biosynthesis during seed abortion in Thompson Seedless. Plant Biotechnol. J. 2021, 19, 1824–1838. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, Y.; Zhao, A.; Xue, X.; Gong, G.; Du, X.; Li, D.; Du, J. Changes of Endogenous Hormones during Fruit Development and Their Relationship with Embryo Abortion in Ziziphus jujuba ‘Lengbaiyu’. Sci. Silv. Sin. 2020, 56, 55–63. [Google Scholar]
- Li, Z.; Zhang, C.; Guo, Y.; Niu, W.; Wang, Y.; Xu, Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genom. 2017, 18, 744. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, X.; Xuan, X.; Sadeghnezhad, E.; Liu, F.; Dong, T.; Pei, D.; Fang, J.; Wang, C. miR3633a-GA3ox2 Module Conducts Grape Seed-Embryo Abortion in Response to Gibberellin. Int. J. Mol. Sci. 2022, 23, 8767. [Google Scholar] [CrossRef]
- Florez-Rueda, A.M.; Paris, M.; Schmidt, A.; Widmer, A.; Grossniklaus, U.; Stadler, T. Genomic Imprinting in the Endosperm Is Systematically Perturbed in Abortive Hybrid Tomato Seeds. Mol. Biol. Evol. 2016, 33, 2935–2946. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.-J.; Zhang, J.-J.; Xue, H.-W. Genome-Wide Analysis of the Complex Transcriptional Networks of Rice Developing Seeds. PLoS ONE 2012, 7, e31081. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.-P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Angel Perez-Amador, M.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR Receptors Play a Major Role in Quantitative Regulation of Stomatal Aperture and Transcriptional Response to Abscisic Acid. Plant Cell. 2012, 24, 2483–2496. [Google Scholar] [CrossRef]
- Di, F.; Jian, H.; Wang, T.; Chen, X.-Y.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Xin, Q.; Yu, L.; Wang, Z.; Wu, W.; Jiang, L.; Wang, G.; Tian, W.; Deng, Z.; et al. Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism. Structure 2012, 20, 780–790. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Jiang, L.; Xin, Q.; Liu, Y.; Tan, J.X.; Chen, Z.Z. Structural basis and functions of abscisic acid receptors PYLs. Front. Plant Sci. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-J.; Zhu, Y.; Wang, P.; Zhao, Y.; Xie, S.; Batelli, G.; Wang, B.; Duan, C.-G.; Wang, X.; Xing, L.; et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet. 2016, 12, e1005835. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic Acid Regulates Early Seed Development in Arabidopsis by ABI5-Mediated Transcription of Short Hypocotyl under Blue1. Plant Cell. 2014, 26, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants. Cell 2021, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shen, Y.-B.; Shi, F.-H. Research Progress on Transcription Factors Regulating Plant Seed Development. Biotech. Bull. 2019, 35, 150–159. [Google Scholar] [CrossRef]
- Gou, Q.M. Recombination of ABF1,ABF2,and ABF4 Genes of ABI5 Subfamily and Construction of Overexpressed Transgenic Arabidopsis. Master’s Thesis, Lanzhou University of Technology, Lanzhou, China, 2021. [Google Scholar]
- Yang, Y.; Sun, M.; Li, S.; Chen, Q.; Da Silva, J.A.T.; Wang, A.; Yu, X.; Wang, L. Germplasm resources and genetic breeding of Paeonia: A systematic review. Hortic. Res. 2020, 7, 107. [Google Scholar] [CrossRef]
- Yang, Y.; He, C.; Wu, Y.; Yu, X.; Li, S.; Wang, L. Characterization of stilbenes, in vitro antioxidant and cellular anti-photoaging activities of seed coat extracts from 18 Paeonia species. Ind. Crops Prod. 2022, 177, 114530. [Google Scholar] [CrossRef]
- Tong, N.-N.; Peng, L.-P.; Liu, Z.-A.; Li, Y.; Zhou, X.-Y.; Wang, X.-R.; Shu, Q.-Y. Comparative transcriptomic analysis of genes involved in stem lignin biosynthesis in woody and herbaceous Paeonia species. Physiol. Plant. 2021, 173, 961–977. [Google Scholar] [CrossRef]
- Zhao, D.; Xue, Y.; Shi, M.; Tao, J. Rescue and in vitro culture of herbaceous peony immature embryos by organogenesis. Sci. Hortic. 2017, 217, 123–129. [Google Scholar] [CrossRef]
- Dietz, K.J.; Sauter, A.; Wichert, K.; Messdaghi, D.; Hartung, W. Extracellular beta-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000, 51, 937–944. [Google Scholar] [CrossRef]
- Gi, S. Temporal and Spatial Expression of Rooting Gene PsARRO-1 in Tree Peony. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2013. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Xu, J.; Tang, F.-S.; Dong, W.-Z.; Zang, X.-W.; Zhang, Z.-X. Embryonic Development and Changes of Endogenous Hormones in Interspecific Hybrids between Peanut (A. hypogaea L.) and Wild Arachis Species. Acta Agron. Sin. 2013, 39, 1127–1133. [Google Scholar] [CrossRef]
- Kubes, M.; Napier, R. Non-canonical auxin signalling: Fast and curious. J. Exp. Bot. 2019, 70, 2609–2614. [Google Scholar] [CrossRef]
- Luo, P.; Di, D.; Wu, L.; Yang, J.; Lu, Y.; Shi, W. MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling. Int. J. Mol. Sci. 2022, 23, 510. [Google Scholar] [CrossRef] [PubMed]
- Rademacher, E.H.; Lokerse, A.S.; Schlereth, A.; Llavata-Peris, C.I.; Bayer, M.; Kientz, M.; Freire Rios, A.; Borst, J.W.; Lukowitz, W.; Jurgens, G.; et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 2012, 22, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Ai, W.; Liu, L.; Xu, X.; Lu, X. Genome-wide identification of the auxin response factor (ARF) gene family in Magnolia sieboldii and functional analysis of MsARF5. Front. Plant Sci. 2022, 13, 958816. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Miao, L.; Huo, R.; Song, X.; Johnson, C.; Kong, L.; Sundaresan, V.; Yu, X. ARF2-ARF4 and ARF5 are Essential for Female and Male Gametophyte Development in Arabidopsis. Plant Cell Physiol. 2018, 59, 179–189. [Google Scholar] [CrossRef]
- Mellor, N.; Bennett, M.J.; King, J.R. GH3-Mediated Auxin Conjugation Can Result in Either Transient or Oscillatory Transcriptional Auxin Responses. Bull. Math. Biol. 2016, 78, 210–234. [Google Scholar] [CrossRef]
- Harberd, N.P.; Belfield, E.; Yasumura, Y. The Angiosperm Gibberellin-GID1-DELLA Growth Regulatory Mechanism: How an “Inhibitor of an Inhibitor” Enables Flexible Response to Fluctuating Environments. Plant Cell 2009, 21, 1328–1339. [Google Scholar] [CrossRef]
- Sun, T.-P. Gibberellin-GID1-DELLA: A Pivotal Regulatory Module for Plant Growth and Development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef]
- Ferreira, L.G.; De Alencar Dusi, D.M.; Irsigler, A.S.T.; Gomes, A.; Mendes, M.A.; Colombo, L.; De Campos Carneiro, V.T. GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Rep. 2018, 37, 293–306. [Google Scholar] [CrossRef]
- Goldy, C.; Pedroza-Garcia, J.A.; Breakfield, N.; Cools, T.; Vena, R.; Benfey, P.N.; De Veylder, L.; Palatnik, J.; Rodriguez, R.E. The Arabidopsis GRAS-type SCL28 transcription factor controls the mitotic cell cycle and division plane orientation. Proc. Natl. Acad. Sci. USA 2021, 118, e2005256118. [Google Scholar] [CrossRef]
- Li, W.R. Study on the Dormancy and Germination of Paeonia Rockii Seeds. Master’s Thesis, Gansu Agricultural University, Lanzhou, China, 2020. [Google Scholar]
- Ma, K.; Song, Y.; Huang, Z.; Lin, L.; Zhang, Z.; Zhang, D. The low fertility of Chinese white poplar: Dynamic changes in anatomical structure, endogenous hormone concentrations, and key gene expression in the reproduction of a naturally occurring hybrid. Plant Cell Rep. 2013, 32, 401–414. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y.; et al. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.-J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-H.; Qu, L.; Xu, Z.-H.; Zhu, J.-K.; Xue, H.-W. EL1-like Casein Kinases Suppress ABA Signaling and Responses by Phosphorylating and Destabilizing the ABA Receptors PYR/PYLs in Arabidopsis. Mol. Plant 2018, 11, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, B.; Mclachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]
- Wu, L.M. Analysis of Genes Regulating Seed Abortion in Inter-Ploid Crosses of Rice. Master’s Thesis, Sichuan Agricultural University, Chengdu, China, 2016. [Google Scholar]
- Li, S.; Liu, K.; Yu, S.; Jia, S.; Chen, S.; Fu, Y.; Sun, F.; Luo, Q.; Wang, Y. The process of embryo abortion of stenospermocarpic grape and it develops into plantlet in vitro using embryo rescue. Plant Cell Tissue Organ Cult. 2020, 143, 389–409. [Google Scholar] [CrossRef]
- Li, K. Transcriptome Rrofiling of Adventitious Rooting Development and Expression Analysis of Candidate Mdrrs and Mdcrfs Genes in Apple Stock. Master’s Thesis, Northwest A & F University, Xianyang, China, 2018. [Google Scholar]
- Ou, X.; Wang, Y.; Zhang, J.; Xie, Z.; He, B.; Jiang, Z.; Wang, Y.; Su, W.; Song, S.; Hao, Y.; et al. Identification of BcARR Genes and CTK Effects on Stalk Development of Flowering Chinese Cabbage. Int. J. Mol. Sci. 2022, 23, 7412. [Google Scholar] [CrossRef]
- Xue, W.; Liu, N.; Zhang, T.; Li, J.; Chen, P.; Yang, Y.; Chen, S. Substance metabolism, IAA and CTK signaling pathways regulating the origin of embryogenic callus during dedifferentiation and redifferentiation of cucumber cotyledon nodes. Sci. Hortic. 2022, 293, 110680. [Google Scholar] [CrossRef]
- Berenguer, E.; Carneros, E.; Perez-Perez, Y.; Gil, C.; Martinez, A.; Testillano, P.S. Small molecule inhibitors of mammalian GSK-3 beta promote in vitro plant cell reprogramming and somatic embryogenesis in crop and forest species. J. Exp. Bot. 2021, 72, 7808–7825. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Yu, J.; Zhu, D.; Zhao, Q. Maize GSK3-like kinase ZmSK2 is involved in embryonic development. Plant Sci. 2022, 318, 111221. [Google Scholar] [CrossRef]
- Cheng, T.; Meng, Y.; Chen, J.; Shi, J. Effects of methyl jasmonic acid on somatic embryogenesis of Liriodendron hybrid. J. Nanjing For. Univ. Nat. Sci. Edn. 2017, 60, 41. [Google Scholar]
- Mira, M.M.; Wally, O.S.D.; Elhiti, M.; El-Shanshory, A.; Reddy, D.S.; Hill, R.D.; Stasolla, C. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. J. Exp. Bot. 2016, 67, 2231–2246. [Google Scholar] [CrossRef]
- Shen, S.; Yuan, J.; Xu, Y.; Ma, B.; Chen, X. Biological function and molecular mechanism of the transcription factor GLKs in plants: A review. Chin. J. Biotechnol. 2022, 38, 2700–2712. [Google Scholar] [CrossRef]
- Penfield, S.; Li, Y.; Gilday, A.D.; Graham, S.; Graham, I.A. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 2006, 18, 1887–1899. [Google Scholar] [CrossRef]
- Lee, K.P.; Piskurewicz, U.; Tureckova, V.; Carat, S.; Chappuis, R.; Strnad, M.; Fankhauser, C.; Lopez-Molina, L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012, 26, 1984–1996. [Google Scholar] [CrossRef] [PubMed]
- Barros-Galvao, T.; Vaistij, F.E.; Graham, I.A. Control of seed coat rupture by ABA-INSENSITIVE 5 in Arabidopsis thaliana. Seed Sci. Res. 2019, 29, 143–148. [Google Scholar] [CrossRef]
- Li, K.; Wang, S.; Wu, H.; Wang, H. Protein Levels of Several Arabidopsis Auxin Response Factors Are Regulated by Multiple Factors and ABA Promotes ARF6 Protein Ubiquitination. Int. J. Mol. Sci. 2020, 21, 9437. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-H.; Yang, C.-C. Identification of ICE1 as a negative regulator of ABA-dependent pathways in seeds and seedlings of Arabidopsis. Plant Mol. Biol. 2015, 88, 459–470. [Google Scholar] [CrossRef]
- Macgregor, D.R.; Zhang, N.; Iwasaki, M.; Chen, M.; Dave, A.; Lopez-Molina, L.; Penfield, S. ICE1 and ZOU determine the depth of primary seed dormancy in Arabidopsis independently of their role in endosperm development. Plant J. 2019, 98, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The Transcription Factor INDUCER OF CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef]
- Denay, G.; Creff, A.; Moussu, S.; Wagnon, P.; Thevenin, J.; Gerentes, M.-F.; Chambrier, P.; Dubreucq, B.; Ingram, G. Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix-loop-helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development 2014, 141, 1222–1227. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, T.; Zhao, S.; Liu, Z.; Feng, Y.-Q.; Wu, Y. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 2011, 68, 249–261. [Google Scholar] [CrossRef]
- Yang, X.; Bai, Y.; Shang, J.; Xin, R.; Tang, W. The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 2016, 39, 1994–2003. [Google Scholar] [CrossRef]
- Xiong, M. Study on the Molecular Mechanism of Coordinated Regulation of Brassinosteroid and Gibberellin on Rice Seed Germination. Ph.D. Thesis, Yangzhou University, Yangzhou, China, 2021. [Google Scholar]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular Mechanism Underlying the Synergetic Effect of Jasmonate on Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef] [PubMed]
- Peirats-Llobet, M.; Han, S.-K.; Gonzalez-Guzman, M.; Jeong, C.W.; Rodriguez, L.; Belda-Palazon, B.; Wagner, D.; Rodriguez, P.L. A Direct Link between Abscisic Acid Sensing and the Chromatin-Remodeling ATPase BRAHMA via Core ABA Signaling Pathway Components. Mol. Plant. 2016, 9, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, S.; Sakamoto, T.; Matsunaga, S. Roles of BRAHMA and Its Interacting Partners in Plant Chromatin Remodeling. Cytologia 2020, 85, 263–267. [Google Scholar] [CrossRef]
- Thouly, C.; Le Masson, M.; Lai, X.; Carles, C.C.; Vachon, G. Unwinding BRAHMA Functions in Plants. Genes 2020, 11, 90. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).