Genetic Characterization of the Norwegian Apple Collection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotyping
2.3. SNP-Data Curation
2.4. Diversity Assessment
2.5. Population Structure and Genetic Relatedness
3. Results
3.1. Quality Control
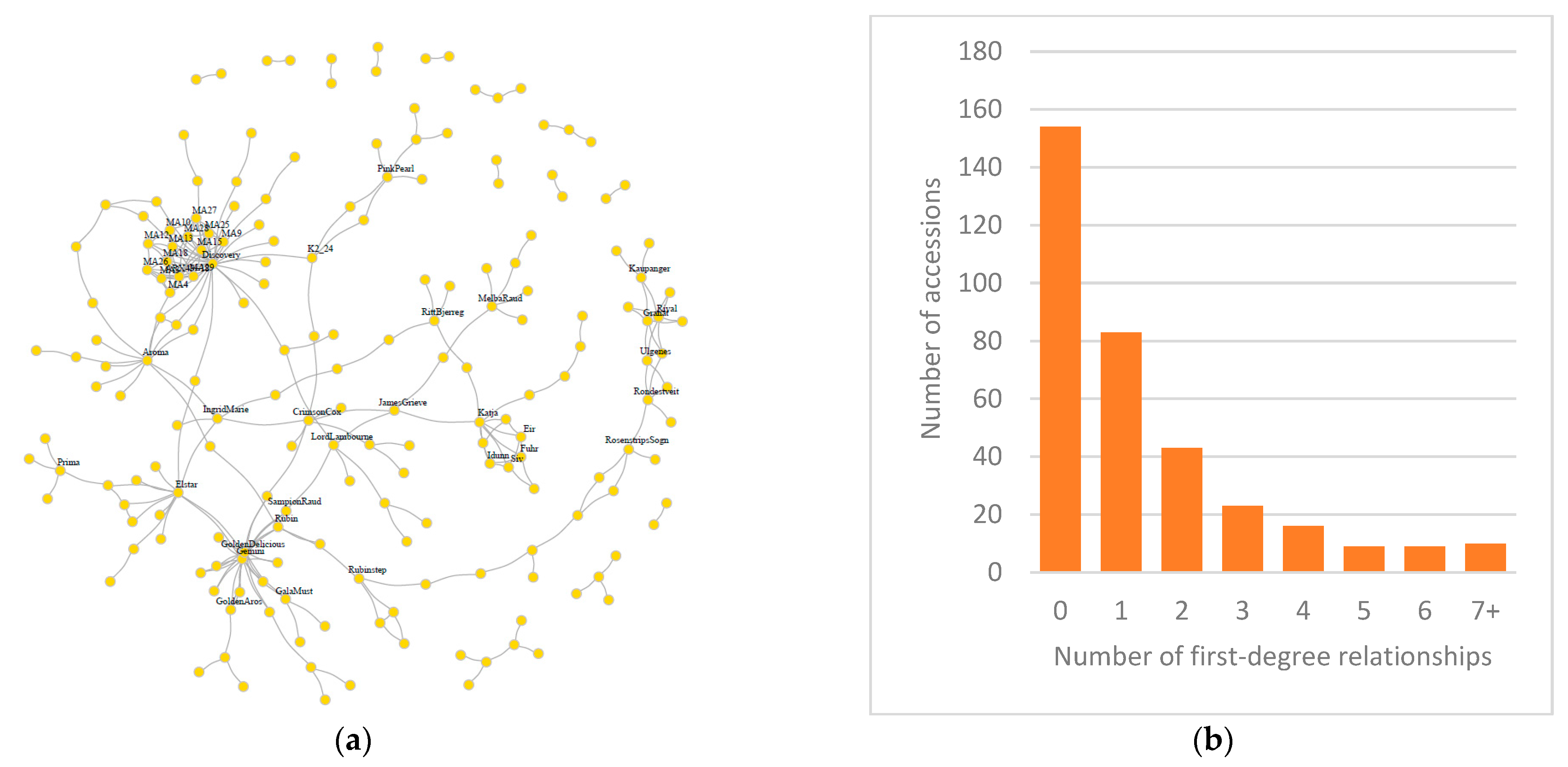
3.2. Relatedness and Ploidy Level
3.3. Possible Parents
3.4. Pedigree
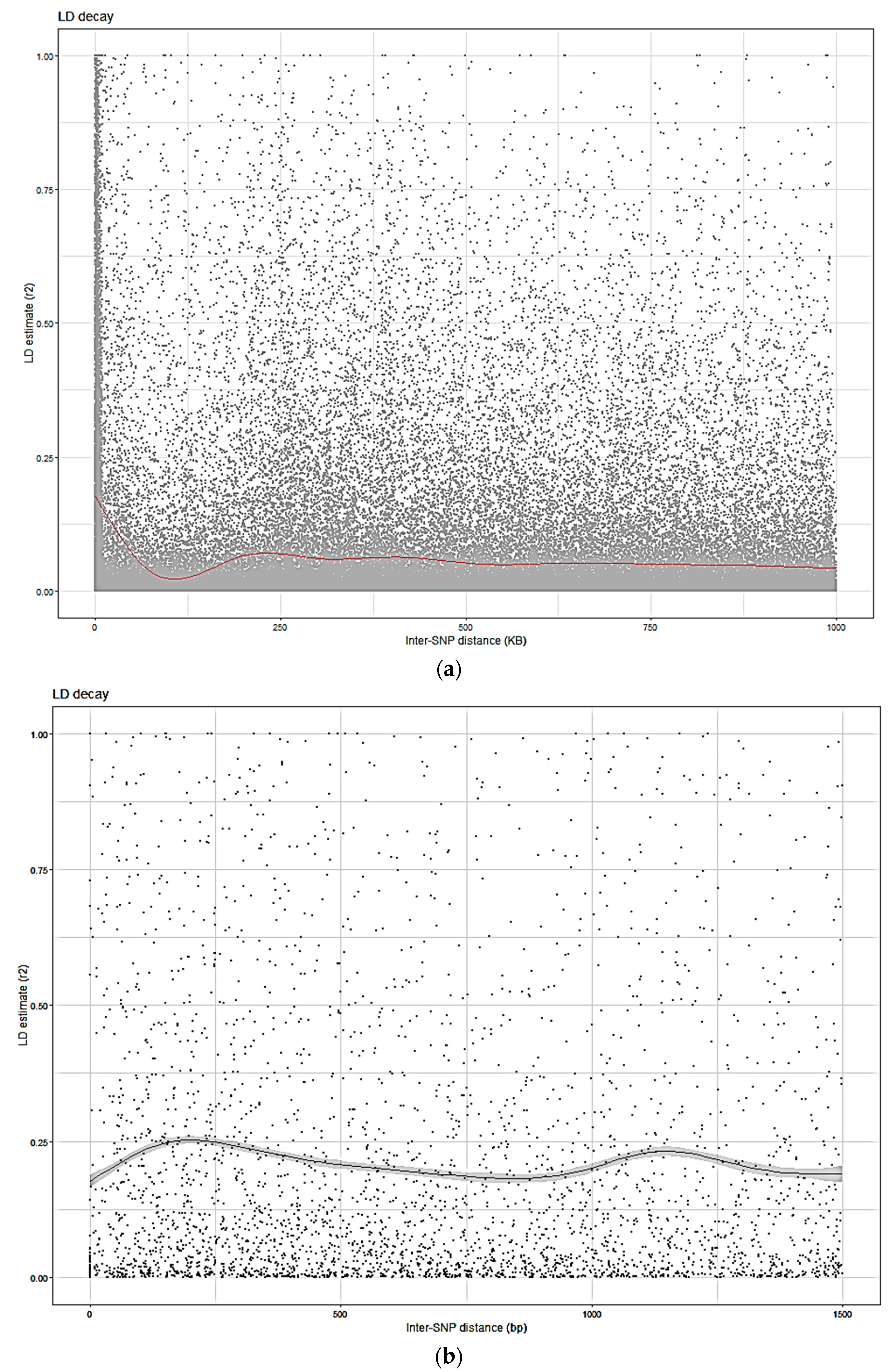
3.5. LD
3.6. Genetic Diversity
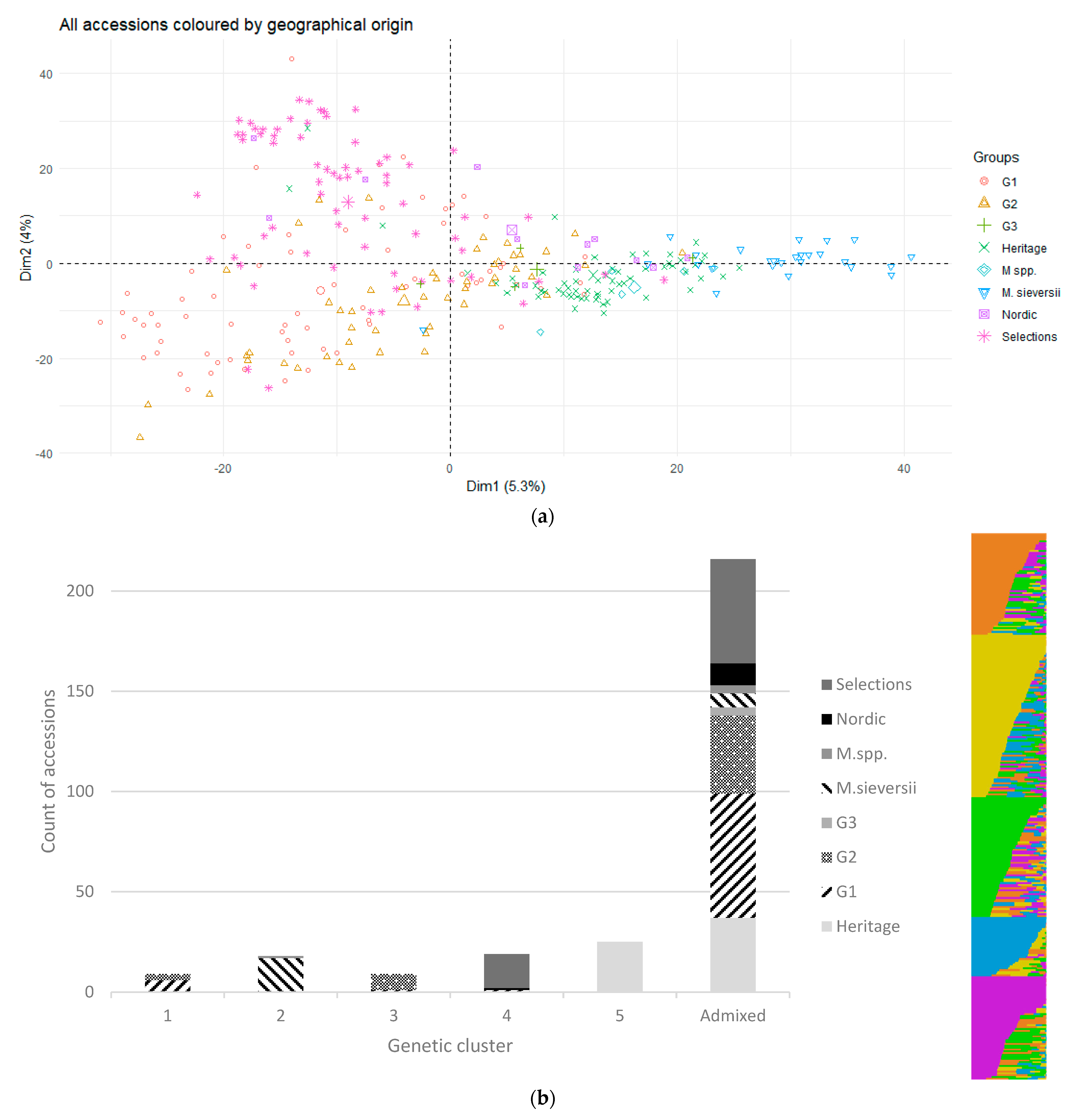
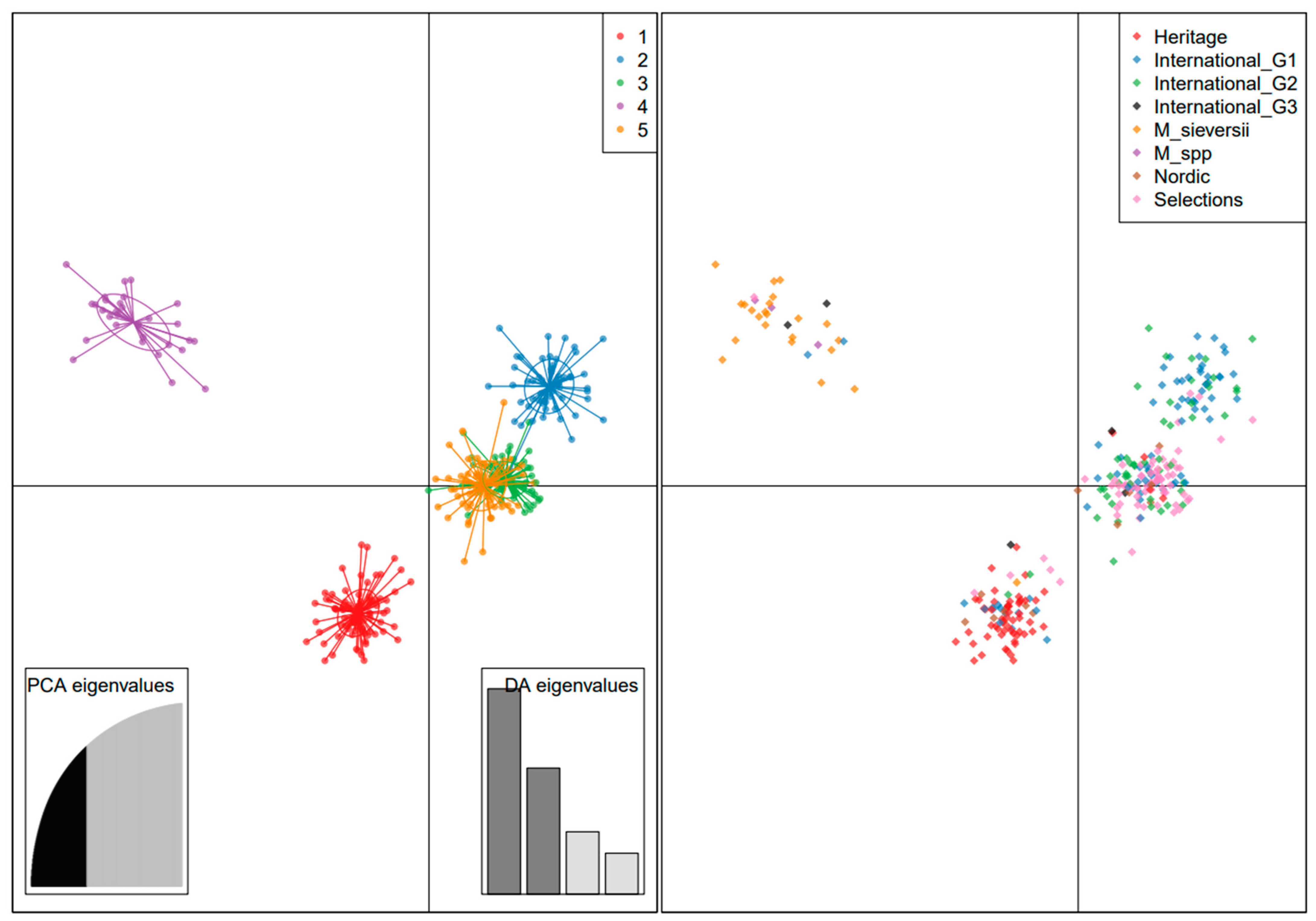
3.7. Genetic Structure
4. Discussion
4.1. Quality Check, Relatedness, and Ploidy Level
4.2. Possible Parents and Pedigree
4.3. LD
4.4. Genetic Structure
4.5. Genetic Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oye, I. Middelalderbyenes Agrare Trekk; Bryggen Museum: Bergen, Norway, 1998; 80p. [Google Scholar]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Juniper, B.E.; Mabberley, D. The Story of the Apple; The Timber Press: Portland, OR, USA, 2006; Volume 47. [Google Scholar]
- Fotirić Akšić, M.; Cerović, R.; Hjeltnes, S.H.; Meland, M. The Effective Pollination Period of European Plum (Prunus domestica L.). Cultiv. West. Norway. Hortic. 2022, 8, 55. [Google Scholar] [CrossRef]
- Ikase, L. Results of fruit breeding in Baltic and Nordic states. In Proceedings of the 25th Congress of the Nordic Association of Agricultural Scientists (NJF) Nordic View to Sustainable Rural Development, Riga, Latvia, 16–18 June 2015. [Google Scholar]
- Hammer, K.; Arrowsmith, N.; Gladis, T. Agrobiodiversity with emphasis on plant genetic resources. Naturwissenschaften 2003, 90, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Hjeltnes, S.H. Norske Klonarkiv i Frukt; Norsk Fruktmuseum og Skog og Landskap: Ås, Norway, 2008; p. 51. [Google Scholar]
- Gasi, F.; Kanlić, K.; Stroil, B.K.; Pojskić, N.; Asdal, Å.; Rasmussen, M.; Kaiser, C.; Meland, M. Redundancies and genetic structure among ex situ apple collections in Norway examined with microsatellite markers. HortScience 2016, 51, 1458–1462. [Google Scholar] [CrossRef]
- Garkava-Gustavsson, L.; Mujaju, C.; Sehic, J.; Zborowska, A.; Backes, G.M.; Hietaranta, T.; Antonius, K. Genetic diversity in Swedish and Finnish heirloom apple cultivars revealed with SSR markers. Sci. Hortic. 2013, 162, 43–48. [Google Scholar] [CrossRef]
- Larsen, B.; Toldam-Andersen, T.B.; Pedersen, C.; Ørgaard, M. Unravelling genetic diversity and cultivar parentage in the Danish apple gene bank collection. Tree Genet. Genomes 2017, 13, 14. [Google Scholar] [CrossRef]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Hibrand-Saint-Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C.-E. Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol. Biol. Rep. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Marconi, G.; Ferradini, N.; Russi, L.; Concezzi, L.; Veronesi, F.; Albertini, E. Genetic characterization of the apple germplasm collection in central Italy: The value of local varieties. Front. Plant Sci. 2018, 9, 1460. [Google Scholar] [CrossRef]
- Patocchi, A.; Fernández-Fernández, F.; Evans, K.; Gobbin, D.; Rezzonico, F.; Boudichevskaia, A.; Dunemann, F.; Stankiewicz-Kosyl, M.; Mathis-Jeanneteau, F.; Durel, C.E. Development and test of 21 multiplex PCRs composed of SSRs spanning most of the apple genome. Tree Genet. Genomes 2009, 5, 211–223. [Google Scholar] [CrossRef]
- Routson, K.J.; Reilley, A.A.; Henk, A.D.; Volk, G.M. Identification of historic apple trees in the Southwestern United States and implications for conservation. HortScience 2009, 44, 589–594. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Poncet, C.; Lateur, M.; Houben, P.; Ordidge, M. Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. 2016, 16, 130. [Google Scholar] [CrossRef] [PubMed]
- Van Treuren, R.; Kemp, H.; Ernsting, G.; Jongejans, B.; Houtman, H.; Visser, L. Microsatellite genotyping of apple (Malus × domestica Borkh.) genetic resources in the Netherlands: Application in collection management and variety identification. Genet. Resour. Crop Evol. 2010, 57, 853–865. [Google Scholar] [CrossRef]
- Larsen, B.; Gardner, K.; Pedersen, C.; Ørgaard, M.; Migicovsky, Z.; Myles, S.; Toldam-Andersen, T.B. Population structure, relatedness and ploidy levels in an apple gene bank revealed through genotyping-by-sequencing. PLoS ONE 2018, 13, e0201889. [Google Scholar] [CrossRef] [PubMed]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.-L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 2020, 20, 2. [Google Scholar] [CrossRef]
- Vanderzande, S.; Micheletti, D.; Troggio, M.; Davey, M.W.; Keulemans, J. Genetic diversity, population structure, and linkage disequilibrium of elite and local apple accessions from Belgium using the IRSC array. Tree Genet. Genomes 2017, 13, 125. [Google Scholar] [CrossRef]
- Chagné, D.; Crowhurst, R.N.; Troggio, M.; Davey, M.W.; Gilmore, B.; Lawley, C.; Vanderzande, S.; Hellens, R.P.; Kumar, S.; Cestaro, A. Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS ONE 2012, 7, e31745. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; Di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Linsmith, G.; Muranty, H.; Denancé, C.; Théron, A.; Poncet, C.; Micheletti, D.; Kerschbamer, E.; Di Pierro, E.A. Development and validation of the Axiom® Apple480K SNP genotyping array. Plant J. 2016, 86, 62–74. [Google Scholar] [CrossRef]
- Peace, C.P.; Bianco, L.; Troggio, M.; Van de Weg, E.; Howard, N.P.; Cornille, A.; Durel, C.-E.; Myles, S.; Migicovsky, Z.; Schaffer, R.J. Apple whole genome sequences: Recent advances and new prospects. Hortic. Res. 2019, 6, 59. [Google Scholar] [CrossRef]
- Vanderzande, S.; Howard, N.P.; Cai, L.; Da Silva Linge, C.; Antanaviciute, L.; Bink, M.C.; Kruisselbrink, J.W.; Bassil, N.; Gasic, K.; Lezzoni, A. High-quality, genome-wide SNP genotypic data for pedigreed germplasm of the diploid outbreeding species apple, peach, and sweet cherry through a common workflow. PLoS ONE 2019, 14, e0210928. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, M.; Micheletti, D.; Bianco, L.; Koehorst-van Putten, H.J.; Longhi, S.; Costa, F.; Aranzana, M.J.; Velasco, R.; Arús, P.; Troggio, M. ASSIsT: An automatic SNP scoring tool for in-and outbreeding species. Bioinformatics 2015, 31, 3873–3874. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Kirk, C.; Whitworth, C.; Erasmuson, S.; Bicknell, R.; Sargent, D.J.; Kumar, S.; Troggio, M. Polyploid and aneuploid detection in apple using a single nucleotide polymorphism array. Tree Genet. Genomes 2015, 11, 94. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- De Beukelaer, H.; Davenport, G.F.; Fack, V. Core Hunter 3: Flexible core subset selection. BMC Bioinform. 2018, 19, 203. [Google Scholar] [CrossRef]
- Boccacci, P.; Aramini, M.; Ordidge, M.; van Hintum, T.J.; Marinoni, D.T.; Valentini, N.; Sarraquigne, J.-P.; Solar, A.; Rovira, M.; Bacchetta, L. Comparison of selection methods for the establishment of a core collection using SSR markers for hazelnut (Corylus avellana L.) accessions from European germplasm repositories. Tree Genet. Genomes 2021, 17, 48. [Google Scholar] [CrossRef]
- Liang, W.; Dondini, L.; De Franceschi, P.; Paris, R.; Sansavini, S.; Tartarini, S. Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol. Biol. Report. 2015, 33, 458–473. [Google Scholar] [CrossRef]
- Odong, T.L.; Jansen, J.; van Eeuwijk, F.A.; van Hintum, T.J.L. Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor. Appl. Genet. 2013, 126, 289–305. [Google Scholar] [CrossRef]
- Rogers, J.S. Deriving phylogenetic trees from allele frequencies: A comparison of nine genetic distances. Syst. Biol. 1986, 35, 297–310. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Breton, C.; Pinatel, C.; Medail, F.; Bonhomme, F.; Berville, A. Comparison between classical and Bayesian methods to investigate the history of olive cultivars using SSR-polymorphisms. Plant Sci. 2008, 175, 524–532. [Google Scholar] [CrossRef]
- Marra, F.; Caruso, T.; Costa, F.; Di Vaio, C.; Mafrica, R.; Marchese, A. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. subsp. europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genet. Genomes 2013, 9, 961–973. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F.; Package ‘Factoextra’. Extract and Visualize the Results of Multivariate Data Analyses. 2017. Available online: https://rpkgs.datanovia.com/factoextra/index.html (accessed on 21 January 2022).
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 50: An environment for modern phylogenetics evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Evans, K.; Patocchi, A.; Rezzonico, F.; Mathis, F.; Durel, C.E.; Fernandez-Fernandez, F.; Boudichevskaia, A.; Dunemann, F.; Stankiewicz-Kosyl, M.; Gianfranceschi, L. Genotyping of pedigreed apple breeding material with a genome-covering set of SSRs: Trueness-to-type of cultivars and their parentages. Mol. Breed. 2011, 28, 535–547. [Google Scholar] [CrossRef]
- Patzak, J.; Paprštein, F.; Henychová, A.; Sedlák, J. Genetic diversity of Czech apple cultivars inferred from microsatellite markers analysis. Hortic. Sci. 2012, 39, 149–157. [Google Scholar] [CrossRef]
- Skytte af Sätra, J.; Troggio, M.; Odilbekov, F.; Sehic, J.; Mattisson, H.; Hjalmarsson, I.; Ingvarsson, P.K.; Garkava-Gustavsson, L. Genetic Status of the Swedish Central collection of heirloom apple cultivars. Sci. Hortic. 2020, 272, 109599. [Google Scholar] [CrossRef]
- Okie, W.R. Register of New Fruit and Nut Varieties Brooks and Olmo List 38. HortScience 1997, 32, 785–805. [Google Scholar] [CrossRef]
- Stedje, P.; Skard, O.M. Norsk Pomologi. 1: Epler ed. Det Norske Hageselskap; Grøndahl og Søns Forlag: Oslo, Norway, 1943; Volume 2, 497p. [Google Scholar]
- Le Scouarnec, S.; Gribble, S.M. Characterising chromosome rearrangements: Recent technical advances in molecular cytogenetics. Heredity 2012, 108, 75–85. [Google Scholar] [CrossRef]
- Howard, N.P.; Troggio, M.; Durel, C.-E.; Muranty, H.; Denancé, C.; Bianco, L.; Tillman, J.; van de Weg, E. Integration of Infinium and Axiom SNP array data in the outcrossing species Malus × domestica and causes for seemingly incompatible calls. BMC Genom. 2021, 22, 246. [Google Scholar] [CrossRef]
- Bonk, S.; Reichelt, M.; Teuscher, F.; Segelke, D.; Reinsch, N. Mendelian sampling covariability of marker effects and genetic values. Genet. Sel. Evol. 2016, 48, 36. [Google Scholar] [CrossRef]
- Harrison, K. Mouldy core in Gravenstein apples. Sci. Agric. 1935, 15, 358–369. [Google Scholar] [CrossRef]
- McClure, K.A.; Gong, Y.; Song, J.; Vinqvist-Tymchuk, M.; Campbell Palmer, L.; Fan, L.; Burgher-MacLellan, K.; Zhang, Z.; Celton, J.-M.; Forney, C.F.; et al. Genome-wide association studies in apple reveal loci of large effect controlling apple polyphenols. Hortic. Res. 2019, 6, 107. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Gardner, K.M.; Money, D.; Sawler, J.; Bloom, J.S.; Moffett, P.; Chao, C.T.; Schwaninger, H.; Fazio, G.; Zhong, G.-Y.; et al. Genome to Phenome Mapping in Apple Using Historical Data. Plant Genome 2016, 9, plantgenome2015-11. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Korban, S.S. Association mapping in forest trees and fruit crops. J. Exp. Bot. 2012, 63, 4045–4060. [Google Scholar] [CrossRef] [PubMed]
- Myles, S.; Chia, J.-M.; Hurwitz, B.; Simon, C.; Zhong, G.Y.; Buckler, E.; Ware, D. Rapid genomic characterization of the genus Vitis. PLoS ONE 2010, 5, e8219. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S., IV. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Meland, M.; Aksic, M.F.; Frøynes, O.; Konjic, A.; Lasic, L.; Pojskic, N.; Gasi, F. Genetic Identity and Diversity of Apple Accessions within a Candidate Collection for the Norwegian National Clonal Germplasm Repository. Horticulturae 2022, 8, 630. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Gardner, K.M.; Richards, C.; Thomas Chao, C.; Schwaninger, H.R.; Fazio, G.; Zhong, G.-Y.; Myles, S. Genomic consequences of apple improvement. Hortic. Res. 2021, 8, 9. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Miranda, C.; Santesteban, L.G.; Royo, J.B. Genetic diversity and structure of local apple cultivars from Northeastern Spain assessed by microsatellite markers. Tree Genet. Genomes 2012, 8, 1163–1180. [Google Scholar] [CrossRef]
- Frichot, E.; Mathieu, F.; Trouillon, T.; Bouchard, G.; François, O. Fast and efficient estimation of individual ancestry coefficients. Genetics 2014, 196, 973–983. [Google Scholar] [CrossRef]
- Zurn, J.D.; Hummer, K.E.; Bassil, N.V. Exploring the diversity and genetic structure of the US National Cultivated Strawberry Collection. Hortic. Res. 2022, 9, uhac125. [Google Scholar] [CrossRef]
- Janes, J.K.; Miller, J.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Cullingham, C.I.; Andrew, R.L. The K = 2 conundrum. Mol. Ecol. 2017, 26, 3594–3602. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef] [PubMed]
- Beukelaer, H.D.; Smýkal, P.; Davenport, G.F.; Fack, V. Core Hunter II: Fast core subset selection based on multiple genetic diversity measures using Mixed Replica search. BMC Bioinform. 2012, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Geibel, J.; Reimer, C.; Weigend, S.; Weigend, A.; Pook, T.; Simianer, H. How array design creates SNP ascertainment bias. PLoS ONE 2021, 16, e0245178. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.F.; Volk, G.M.; Gardner, C.; Gore, M.A.; Simon, P.W.; Smith, S. Sustaining the future of plant breeding: The critical role of the USDA-ARS National Plant Germplasm System. Crop Sci. 2018, 58, 451–468. [Google Scholar] [CrossRef]
- Langridge, P.; Waugh, R. Harnessing the potential of germplasm collections. Nat. Genet. 2019, 51, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsson, P.K.; Street, N.R. Association genetics of complex traits in plants. New Phytol. 2011, 189, 909–922. [Google Scholar] [CrossRef]

| Offspring | PosPar1 | PosPar2 | Age Offspring | Age Parent 1 | Age Parent 2 |
|---|---|---|---|---|---|
| ‘Bøtuneple’ | ‘Rosenstrips Sogn’ | Unknown | 1781 | ||
| ‘Crimson Cox’ | ‘Steinkyrkje’ | 1825 | 1700/1800 | ||
| ‘Ekely’ | ‘Rosenstrips Hard.’ | 1907 | 1781 | ||
| ‘Enestående’ | ‘Høyneseple’ | Unknown | 1800 | ||
| ‘Filippa’ | ‘Flaske-eple’ | 1880 | 1700 | ||
| ‘Rosenstrips Sogn’ | ‘Fuhr’ | ‘Torstein’ | 1781 | 1660 | 1760 |
| ‘Furuholm’ | ‘Sävstaholm’ | Unknown | 1830 | ||
| ‘Hindals Dronning’ | ‘Worcester Pearmain’ | Unknown | 1874 | ||
| ‘Håkonseple’ | ‘Lærdalseple’ | ‘Torstein’ | 1905 | Unknown | 1760 |
| ‘Tolleivseple’ | ‘Kaupanger’ | Unknown | 1700 | ||
| ‘Tormod’ | ‘Kaupanger’ | Unknown | 1700 | ||
| ‘Langballe’ | ‘Franskar’ | 1870 | 1790 | ||
| ‘Leiknes’ | ‘Granat’ | 1890 | Unknown | ||
| ‘Linda’ | ‘Granat’ | ‘Transp. Blanche’ | 1970 | Unknown | 1800 |
| ‘Løeple’ | ‘Høyneseple’ | ‘Lærdalseple’ | 1925 | 1800 | Unknown |
| ‘Martaeple’ | ‘Lærdalseple’ | ‘Torstein’ | 1915 | Unknown | 1760 |
| ‘Mostereple’ | ‘Høyneseple’ | - | 1800 | ||
| ‘Prins’ | ‘Granat’ | 1860 | Unknown | ||
| ‘Sukkereple’ | ‘Høyneseple’ | Unknown | 1800 | ||
| ‘Teigeple’ | ‘Rosenstrips Hard.’ | Unknown | 1781 | ||
| ‘Pederstrup’ | ‘Torstein’ | 1850 | 1760 | ||
| ‘Tveiteple’ | ‘Granat’ | ‘Rosenstrips Hard.’ | 1925 | Unknown | 1781 |
| ‘Ulgenes’ | ‘Granat’ | ‘Rosenstrips Hard.’ | Unknown | 1781 |
| Core Collection Type | ||
|---|---|---|
| Structure Group | CC–I (A–NE) | CC–X (E–NE) |
| Selections | 15 | 25 |
| Heritage | 22 | 12 |
| International_G1 | 24 | 25 |
| International_G2 | 17 | 20 |
| International_G3 | 5 | 4 |
| M. sieversii | 6 | 7 |
| M.spp. | 3 | 2 |
| Nordic | 8 | 5 |
| Whole Core Collection | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilpin, L.; Røen, D.; Schubert, M.; Davik, J.; Rumpunen, K.; Gardli, K.A.; Hjeltnes, S.H.; Alsheikh, M. Genetic Characterization of the Norwegian Apple Collection. Horticulturae 2023, 9, 575. https://doi.org/10.3390/horticulturae9050575
Gilpin L, Røen D, Schubert M, Davik J, Rumpunen K, Gardli KA, Hjeltnes SH, Alsheikh M. Genetic Characterization of the Norwegian Apple Collection. Horticulturae. 2023; 9(5):575. https://doi.org/10.3390/horticulturae9050575
Chicago/Turabian StyleGilpin, Liv, Dag Røen, Marian Schubert, Jahn Davik, Kimmo Rumpunen, Kristina Alme Gardli, Stein Harald Hjeltnes, and Muath Alsheikh. 2023. "Genetic Characterization of the Norwegian Apple Collection" Horticulturae 9, no. 5: 575. https://doi.org/10.3390/horticulturae9050575
APA StyleGilpin, L., Røen, D., Schubert, M., Davik, J., Rumpunen, K., Gardli, K. A., Hjeltnes, S. H., & Alsheikh, M. (2023). Genetic Characterization of the Norwegian Apple Collection. Horticulturae, 9(5), 575. https://doi.org/10.3390/horticulturae9050575





