Abstract
Apple leaf characteristics are the most important vegetative growth and development traits. The apple rhizosphere is also indirectly responsible for apple growth and development. It provides roots with elements and compounds that improve roots’ and shoots’ growth and development. The application of exogenous plant growth regulators such as strigolactones (SLs) has become one of the main trends for improving vegetative growth and enhancing the rhizosphere microbiome. This study aimed to investigate the effect of the exogenous SLs on some leaf characteristics, such as leaf area, angle, nitrogen content, chlorophyll content, and apical area diameter, in addition to the composition of the rhizosphere microbiome of apple M9 rootstock. The apple rootstocks were treated with various concentrations of GR24, an analog of SLs; the concentrations were 0, 1, 5, and 10 µM. The study found that the treatments of 5 µM increased the leaf–stem angles and leaf length while decreasing the apical diameter. The treatments of 1 and 5 µM increased leaf nitrogen content; however, this effect was not observed when using the higher concentration of 10 µM. The lower concentration (1 µM) led to a different abundance and diversity of microorganisms compared to the higher concentration (10 µm).
1. Introduction
Apples are among the most consumed fruits worldwide [1]. According to USDA [2], apples are consumers’ top U.S. fruit choice. China is the biggest producer of apples in terms of the harvested area and production quantity [3]. Apple fruits can be used fresh or processed; thus, they are a valuable commodity for many consumers. Due to consumers’ high demand for apples, optimizing apple tree health and production is necessary.
Phytohormones regulate plants’ growth, development, and environmental interactions [4]. Moreover, they stimulate the interaction between plants and surrounding microorganisms [5]. Plant hormones have various physiological effects inside plant tissues, subsequently affecting plant organs. For instance, Auxin is the primary precursor of cell division [6] and elongation [7]. Cytokinin participates in these processes and other reactions, such as cell division [6]. Other effects of Auxin, such as bud initiation, apical dominance, and regulation of other hormones, have been studied and reported [8]. Plant hormones can be up-regulated or down-regulated depending on the stimulation circumstances; therefore, they can negatively and positively affect plant growth and development. Changes in phytohormones can impact leaves, stems, buds, and other shoot organs [9,10]. In the same trend, strigolactone could be used as a plant growth regulator for plant growth improvement.
Strigolactones (SLs) are carlactonic compounds identified as phytohormones, which enhance plant growth directly and indirectly [11,12,13,14,15]. They improve vegetative plant growth, attract rhizosphere microorganisms [8,16,17,18,19], and play a key role in the relationship between plants and their rhizosphere microorganisms [20]. There are many studies on the effects of SLs on different plant species. However, there is still a knowledge gap in SLs’ role in fruit tree health and production. Therefore, further experimental studies are needed to examine the effect of SLs on fruit tree growth and their relation to the rhizosphere microbiome.
Generally, shoot architecture is the main element for plant adaptation, competition, and survival [21]. Apical area, leaf angle, length, and width are significant factors in forming canopy structures and are considered specific architectural characteristics of the plant [22]. The leaf area is the most important leaf characteristic because it regulates light absorption and photosynthesis and modifies carbon outputs such as carbohydrates and other compounds [22,23]. These carbon outputs form biomass accumulation in different parts of the plant, such as roots and leaves [23]. Leaf angle and apical leaf area have also demonstrated an important role in light absorption and photosynthesis with similar output products [22]. In addition, these plant characteristics may support the plants’ response to environmental factors [8,15]. For instance, shoot architecture modifications support plants against drought stress [24], light stress, salinity, and other surrounding factors [22,25]. These physiological changes impact all organs of the plant, especially fruits.
In the same manner, plant architectural characteristics can be affected by many factors, such as nutrition composition [26], hormonal and metabolic compound reactions, and environmental factors. For example, there is a significant correlation between leaf nitrogen (N) content and chlorophyll (Ch) content [27]. This relation subsequently affects the leaf’s appearance or dimensions [28]. These combinations strongly affect the photosynthesis rate and many other physiological and morphological characteristics [23,29]. In addition, the surrounding microorganisms provide plants with nitrogen and other nutrients and enhance growth [30].
The rhizosphere microbiome is considered an essential factor in the growth and development of apple trees because it provides trees with accumulated nutrients. Nutrient accumulation promotes tree health and high production [15]. Vegetative growth and root rhizosphere health are vital factors in the apple’s survival. Plants can secrete hormones or metabolic compounds to communicate with surrounding microorganisms, especially the rhizosphere microbiome. This relation may lead to plant growth improvements, signaled by the rhizosphere microbiome. Many studies have explored the trend of examining the effect of plant microbiomes on improving vegetative growth [31,32]. The rhizosphere microbiome plays many roles in plants’ responses to environmental factors such as nutrient uptake [33]. Furthermore, the plant itself can change its rhizosphere microbiome to improve its nutrient uptake and proportions [34,35]. Microbial communities play a vital role in enhancing plant growth by influencing plant physiology and development. The symbiotic relationship between the root rhizosphere and its microbiome is essential for plant health, particularly in soil rich with organic matter and microorganisms. Furthermore, the role of strigolactone as a phytohormone on vegetative apple growth and its rhizosphere microbiome needs more investigation and understanding.
Roots and the rhizosphere microbiome can be affected by hormonal changes [36,37]. In addition, the roots can secrete many chemical compounds as secondary metabolites, which attract microorganisms that react with the root biochemical changes [6]. As a result of these reactions between rhizosphere microorganisms and roots, the plant and its growth characteristics are supported. For example, Strigolactone enables plants to modify growth to compete with neighboring organisms for limited resources. In addition, SLs have other roles in improving vegetative growth directly and indirectly. For instance, they improve the absorption of nutrients such as nitrogen and phosphorus, increasing chlorophyll levels and, subsequently, vegetative growth [11,38].
The under- and above-ground environments of plants have a complicated interaction. The synthetic analog GR24 was used to investigate SLs’ regulation in integrating plants with the subsurface ecosystem. Furthermore, the microorganisms can modify the soil pH, secrete organic matter, and act as osmo-regulators in the soil. Osmo-regulators buffer and condition the soil [39], which improves roots’ absorption and provides plants with nutrients such as nitrogen that enhance vegetative growth. Plants and microbiomes are separate communities with harmonious relationships; SLs are moderators between plants and microbes [40,41,42].
In this article, we investigate the role of SLs in improving some leaf characteristics of the apple rootstock and identify rhizosphere microbiome changes that may enhance plant growth by providing substances or nutrients such as nitrogen.
2. Materials and Methods
2.1. Plant Material and Experimental Design and Conditions
The study aimed to detect the effect of the exogenous application of SLs on vegetative growth characteristics and the rhizosphere microbiome composition of apples. Seedlings of one-year-old apple rootstock seedlings v M9 (Malus × domestica Borkh) were used in the study. The experiment was conducted in a greenhouse belonging to the China Agricultural University, Beijing, China. The average greenhouse temperature was 18–30 °C, the relative humidity was 40–70%, and normal light and darkness interval periods were used. Transplants of the experiment were cultivated in soil and then transferred to a mixed peat moss with vermiculite propagation-growth media and were planted in plastic pots without any sterilization of the growth media. The plants were treated with different concentrations of GR24 as a Strigolactone analog. Each treatment had four replicates, and each replicate included three transplants. The transplants were typically irrigated and fertilized before starting experimental treatments.
2.2. GR24 Preparation, Concentrations, and Application
The 1 mg GR24 powder (C17H14O5, produced by PhytoTechnology Laboratories® (Lenexa, KS, USA), purity 100% mol. Weight 298.29 g/mol) was dissolved in 3% acetone and 0.1% Tween 20 and mixed to prepare the stock solution [12]. The stock solution was diluted with distilled water into levels of concentration. All the plants’ topsoil was covered with plastic covers to prevent the cross-contamination of soil by the GR24 and to ensure the absorption of GR24 was only by leaves through a foliar application. Hand sprayers with different concentrations were used to spray the leaves until they trickled droplets from their surfaces [12].
2.3. Leaf Characteristics
Leaf dimensions (length and width) were measured by a digital caliper. Mature leaves midway up the shoot were used to measure the leaf length and width. Leaf area was calculated according to Boyacı and Küçükönder [43].
2.4. Apical Diameter and Leaf Angle
The apical area diameter was measured by using a protractor and ruler. The approximate circle formed by the group of leaves in the apical part was detected, and the diameter was measured. The angles of the leaves were determined by measuring the angle degree at the attachment point between the leaves and the stem using a ruler and protractor.
2.5. Leaves Chlorophyll and Nitrogen Content
Chlorophyll content was recorded on the mature leaves using a chlorophyll meter (Konica Minolta SPAD 502 Plus Chlorophyll Meter, INC, Osaka, Japan). Five random leaves were chosen per replicate to determine the chlorophyll content index. Five mature leaves at the middle part of the stem were collected and dried, and 5 mg of dry ashes were weighed and wet digested in the microwave [44]. The Kjeldahl Distillation method instrument (Kjeltec™ 8400, Foss, Hilleroed, Denmark) was then used for measuring the total nitrogen content in the leaves.
2.6. Rhizosphere Sample Collection
For the first experiment, the plants were removed from the soil carefully. Then, the rhizosphere media was collected gently using fine brushes. The samples were collected from specific concentrations of 0, 1, and 10 µM for 16S rRNA analysis for microbiome analysis. The samples were immediately stored in sterile bags on dry ice and then transferred to −80 °C until DNA extraction [45].
2.7. DNA Extraction
DNA was extracted from 250 mg of rhizosphere soil using MO BIO PowerSoil™ DNA Isolation Kits (Mo Bio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. All extractions were quantified and qualified on a Nanodrop ND-2000 UV-VIS Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) [19]. After the extraction of genomic DNA was completed, the extracted genomic DNA was detected by 1% agarose gel electrophoresis.
2.8. PCR Amplification and Illumina MiSeq Sequencing
Nine 16S rRNA gene libraries (three treatments × three replicates) were generated and compared. The V4 region of the 16S rRNA genes was amplified using the primer pair 515F and 806R [46,47,48]. Amplification of the bacterial 16S rRNA sequences was performed using the TransStart® FastPfu DNA Polymerase (TransGen Biotech Co., Ltd., Beijing, China). All amplifications were performed under the following conditions. The total reaction volume was 50 μL, including 1μM for the forward primer and 1 μM for the reverse primer. A 1 μM volume of FastPfu DNA Polymerase was also used, and the final concentration was 2.5 units. The PCR thermal cycling conditions were one cycle of 95 °C for 2 min, followed by 30 cycles of 95 °C for 20 s, TM 5 °C for 20 s, and 72 °C for 4 min (4 kb/min), with a final one cycle of 5 min at 72 °C. The PCR products of the same sample were mixed and detected by 2% agarose gel electrophoresis. The AxyPrep DNA Gel Recovery Kit (AXYGEN®, CORNING company, Glendale, CA, USA) was used to cut the gel and recover the PCR products. Sequencing was performed using the Illumina MiSeq sequencing platform at Allwegene Technology Co., Beijing, China.
2.9. Bioinformatics Processing and Analyses
Raw data were stored as a FASTQ format file. Sequencing was allocated to each sample according to the unique barcode of each sample. QIIME was used for Sequencing analysis [49] in addition to the software package (Quantitative Insights Into Microbial Ecology) and UPARSE pipeline [50]. Additionally, QIIME was used for filtering the reads by quality filter [51]. UPARSE pipeline was used to pick up operational taxonomic units (OTUs) by making an OTU table. After filtering the reads, the retained sequences were assigned to OTUs at 97% similarity, while chimeras were filtered. RDP classifier was used to assign the taxonomic data to each representative sequence. OTUs were clustered using QIIME (v1.8.0) and VSEARCH (v2.7.1) [52], which were performed at 97% similarity levels. Clean tags were used for clustering to generate OTUs. The clustering was made by using UPRISE [50], UCLUST [53], and Cd-hit [54]. R language tools for statistics and charting were used [48] to compare the overlap between the OTU number groups in the Venn diagram. Alpha diversity indices as the richness index of the Chao estimator (Chao1), Shannon, PD_whole_tree, observed-species, and good-coverage were calculated using QIIME [55,56,57,58].
The databases Silva (http://www.arb-silva.de, accessed on 13 August 2019), RDP (http://rdp.cme.msu.edu, accessed on 13 August 2019), Greengene (http://greengenes.secondgenome.com, accessed on 13 August 2019), and NCBI Nucleotide 13 a (https://www.ncbi.nlm.nih.gov, accessed on 13 August 2019) were used. Multivariant statistical and beta diversity analyses were analyzed using R language packages and QIIME software Version 1.8.
2.10. Statistical Analysis
Alpha diversity, multivariant statistical analysis, and other molecular statistical analyses were calculated using QIIME and R package software. Data from the vegetative measurements were statistically analyzed with CoStat software. One-way analysis of variance (ANOVA) and Duncan’s multiple range test were used to determine the significance of the differences among samples (p < 0.05).
3. Results
3.1. Effect of SLs on Leaf Nitrogen Content and Chlorophyll Index (SPAD)
SLs at 1 and 5 µM significantly increased the leaf nitrogen content. Using the concentration of 10 µM, GR24 increased the nitrogen content in the leaf compared to the control. However, the difference was not statistically significant (Figure 1A). Data of the chlorophyll index showed no significant difference between 1 µM and control; however, using 5 and 10 µM significantly decreased the chlorophyll content (Figure 1B).
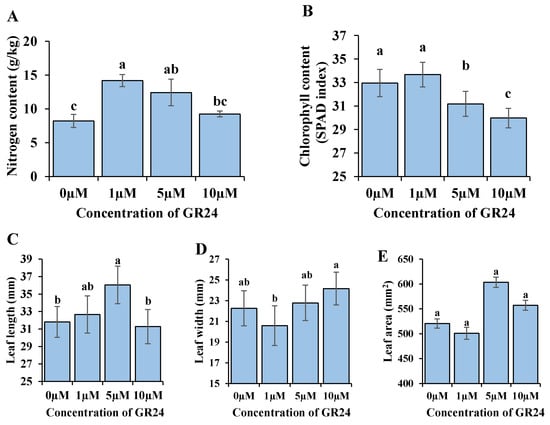
Figure 1.
The effect of SLs on the nitrogen and chlorophyll content (SPAD index) and the leaf dimensions (leaf length, width, and area) in the M9 apple rootstocks. (A,B) The leaf nitrogen content per gram and the chlorophyll SPAD index, respectively, according to the different concentrations of GR24. (C–E) The leaf length, width, and area, respectively, according to the different concentrations of GR24. Bars labeled with a different letter are significantly different (p ≤ 0.05) according to Duncan’s test. The bars show standard errors.
3.2. Effect of SLs on Leaf Length, Width, and Area
SLs at moderate concentrations (5 µM) significantly increased leaf length. However, the lowest and the highest levels of SLs (1 and 10 µM) showed no significant increments. The effect of the lowest level of SLs (1 µM) on leaf length was closer to 5 µM than 10 µM (Figure 1C). The lowest leaf width occurred on the plant treated with 1 µM, and the highest occurred on a plant treated with 10 µM, with a significant difference between the two measurements (Figure 1D). Thus, the relative lowest level of SLs (1 µM) decreased leaf width, and the concentration of 5 µM increased leaf width. The highest level of SLs (10 µM) showed the highest values of leaf width but did not show a clear difference in leaf length. Consequently, the highest leaf area was in plants treated with 5 and 10 µM (Figure 1E).
3.3. Effect of SLs on Leaf Angle and Apical Radius
SLs at 5 and 10 µM significantly increased the angle between the stem and leaf. However, there was no significant difference between the 0 µM and 1 µM concentrations. On the contrary, SLs significantly decreased the diameter of the apical area, and there was no significant difference between 0 µM and 1 µM. However, 1 µM showed slightly lower diameter values than 0 µM (Figure 2). The leaf–stem angles of plants treated with 5 and 10 µM ranged from 80° to 90°.
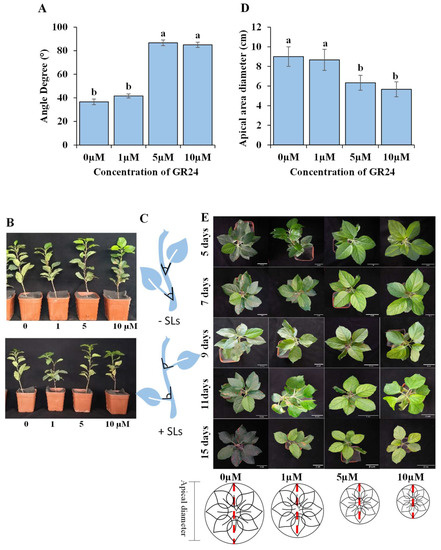
Figure 2.
The effect of SLs on the apical area diameter. (A) Bar chart describes the angle degrees of the leaves and stem of the M9 apple rootstocks according to different concentrations of GR24. (B) Photographs demonstrate the changes in the angles between the leaves and stem. (C): an illustration of the changes in leaf-stem angles as an effect of SLs presence. (D) Bar chart showing the apical diameter (length of the apical area) according to the different concentrations of GR24. (E) Changing apical diameter during the time intervals. Each white scale bar is 2 cm in length and illustration of the different changes in apical diameter according to the different concentrations of GR24 at the end of the experiment. Bars with different letters are significantly different at p < 0.05 (ANOVA, Duncan correction). The bars show standard errors.
3.4. Effect of SLs on Rhizosphere Microbiome Composition
After a quality protocol was implemented for paired-end alignments, filtering, and deletion of chimeric and singletons sequences through sequencing of the V3–V4 region of the 16S rRNA, 293,215 quality reads were recovered. A total of 2555 OTUs at 97% sequence similarity were generated, and the remaining 2535 after level processing. In the rhizosphere microbial communities for bacteria and archaea, there were differences in taxa relative abundance between samples and groups. Bacteria were found in the rhizosphere of all groups under study, while archaea were not found in the rhizosphere microbiome.
The Venn and petal core diagrams showed differences in the attracted microorganisms (Figure 3). Of note, the rhizosphere of the plants treated with GR24 attracted a higher relative abundance of Fibrobacteres, Planctomycetes, Microgenomates, Gemmatimonadetes, Chloroflexi, FCPU426, WS6, WS2, Chlamydiae, Nitrospirae, TM6_Dependentiae, BRC1, Hydrogenedentes, and Peregrinibacteria Phyla compared to the non-treated plants (0 µM). There are variations in different taxa abundance in the rhizosphere among different groups of samples.
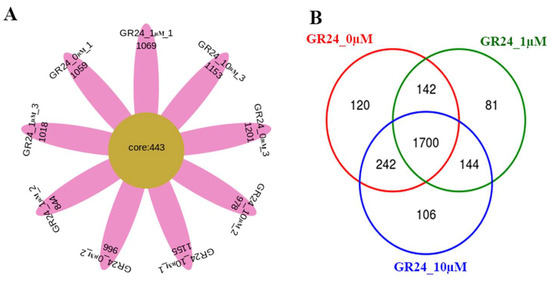
Figure 3.
OTUs Cluster Analysis. (A) The petal map. Each petal represents a sample. The core number in the middle represents the number of OTUs shared by all samples in the rhizosphere of the M9 apple rootstocks. The number on the petal represents the unique OTU number of the sample. (B) Venn diagram of OTUs for 3 groups of samples. Different colors represent a different group of samples. The overlapping areas of the colored circles are intersections that express shared OTUs. The non-overlapping sections mean the OTU is unique to the specific samples.
The relative abundance of bacterial phyla, orders, classes, and families was analyzed across all samples, as shown in Figure 4. All the samples showed a dominance of the Proteobacteria, ranging from 50 to 65% of the community. Actinobacteria were ranked the second dominant phylum with a maximum of 20% of the relative abundance. Chloroflexi, Bacteroidetes, Gemmatimonadetes, Acidobacteria, and Firmicutes followed Proteobacteria and Actinobacteria in rank, respectively. The plants treated with 1 µM attracted more microbes from the Actinobacteria phylum. The main classes were Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria, respectively. Alphaproteobacteria and Gammaproteobacteria classes were within the Proteobacteria phylum, while the Actinobacteria class was within the Actinobacteria phylum. In addition, 1 µM showed a noticeable increase in the relative abundance of the Actinobacteria class. The main orders were Rhizobiales, which were within Alphaproteobacteria and unidentified classes of all treatments. The 1 µM treatment showed a slightly higher relative abundance in Gemmatimonadales, Sphingomonadales, and Streptomycetales. Furthermore, 10 µM showed higher relative abundance in different orders, including unidentified, Nitrosomonadales, Cytophagales, Bacteroidales, and Solibacterales. In addition, the two concentrations showed higher values in Acidimicrobiales and Anaerolineales orders. The main two families were unidentified and Hyphomicrobiaceae, which were within the Rhizobiales order. Moreover, 10 μM showed higher abundance in the unidentified family than in the control. The 1 µM treatment showed an increase in the family Gemmatimonadaceae within the phylum Gemmatimonadetes and the family Streptomycetaceae within the phylum Actinobacteria. Finally, 10 μM showed an increase in the family Nitrosomonadaceae within the phylum proteobacteria (Figure 4).
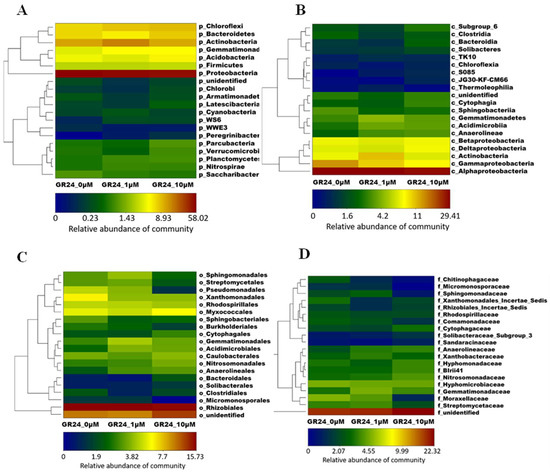
Figure 4.
OTUs and their taxonomy level heatmaps and hierarchical clustering analysis between different taxa. Each heat map represents the relative abundance of the community taxa in the different applied GR24 concentrations in the rhizosphere of the M9 apple rootstocks. (A) phylum-level, (B) Class-level, (C) Order-level, and (D) Family-level. The difference in colors represents the relative microbial abundance of the community.
3.5. Effect of SLs on Microbiome Diversity
Diversity at different indices of the community bacterial species was studied. There were some differences in microbial diversity in the soil rhizosphere across treatments. The α diversity Good’s coverage values indicated that, on average, 96.4 of the total species richness was accounted for in bacterial communities. Chao1 diversity values ranged from 1666.9 to 2104.5, while Shannon diversity ranged from 8.7 to 9.5. Bacterial diversity, as well as richness and evenness within the microbial community, differed between rhizosphere samples when comparing the 0 and 10 µM treatments. For example, 0 µM decreased the bacterial diversity in the rhizosphere bacterial community, while 10 µM increased the community’s diversity. While the 10 µM treatment increased the diversity (Shannon index) in the rhizosphere bacteria, the richness of the community was close to that of the control (Chao1, observed species) (Figure 5).
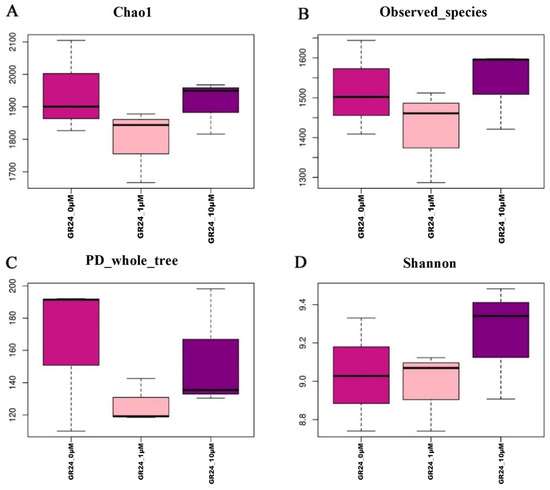
Figure 5.
Alpha diversity metrics (Chao1 index, observed species, PD_whole_tree, and Shannon index) of OTU-level rhizosphere communities of studied M9 apple rootstocks. The Box diagrams of alpha diversity metrics show the relationship among the three groups. The box chart contains 4 main data node indexes. The data are arranged sequentially, and the upper edge, upper quartile, median, lower quartile, and lower edge are calculated. The abscissa is the group name, and the ordinate is the Alpha index. (A,B) Box plots for comparison of species richness (Chao1 index; observed species) between the two study groups. (C) Boxplots for comparison of phylogenetic diversity (PD_whole_tree). (D) Boxplots for comparison of species diversity (Shannon index).
Distance metric indices were used to measure the β diversity between the bacterial communities with the influence of 0, 1, and 10 µM of GR24 and to use the OTU phylogenetic level to analyze the diversity in the different communities (OTUs defined at a 97% similarity cut-off). To compare the bacterial composition of identified microbial community members of the rhizosphere UniFrac and Bray–Curtis dissimilarity, matrices were calculated on normalized. Similarities and dissimilarities in microbial community structures among samples were displayed using principal component analysis (PCA), principal coordinates analysis (PCoA), partial least squares discrimination analysis (PLs-DA), and the non-metric multidimensional scaling method (NMDs). The analysis was constructed using UniFrac weighted and unweighted distance metric indices to analyze the quantitative and qualitative diversity between samples. It was demonstrated that there was a difference between samples in the community based on the UniFrac distances between the rhizosphere samples and qualitative variation (presence and absence of organism) between the control and the treatments of 0 and10 µM. Treatment of 1 µM revealed apparent quantitative variation with 0 µM compared to the variation between 0 and 10 µM (Figure 6). When using PCA, PLS-DA, and PCoA analysis at the OTU level, PC1 revealed 23.52%, 15%, and 27%, respectively, of the total variation; PC2 revealed 20.12%, 13.19%, and 49%, respectively. The beta diversity metric analysis showed a distinction between the microbial communities of 0, 1, and 10 µM treatment concentrations. However, there was relative similarity in the rhizosphere bacterial community between 0 and 10 µM (Figure 7).
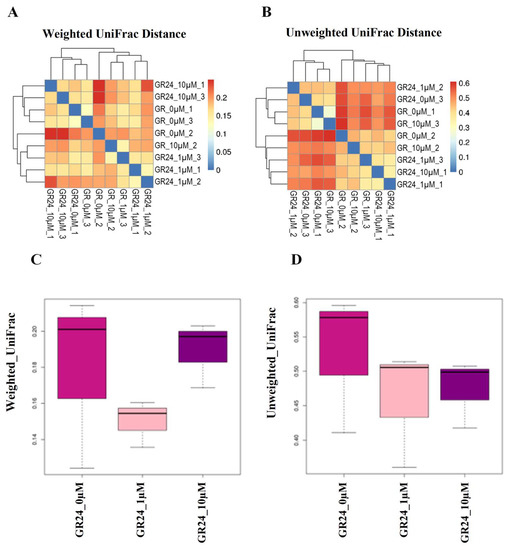
Figure 6.
An illustration of microbial beta diversity indices in the M9 apple rootstock rhizosphere. (A,B) Based on UniFrac heatmaps, the left and upper axes are based on the UniFrac calculation of the sample-to-sample distance relationship. The horizontal and vertical coordinates are the names of the samples, and the color depth represents the level of similarity between the samples. (C,D) Box diagrams based on UniFrac distance.
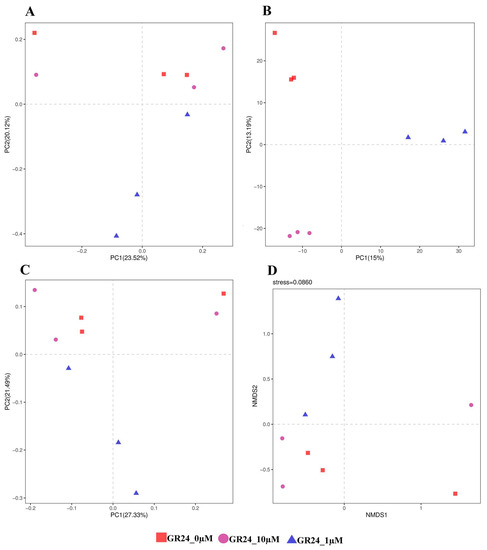
Figure 7.
Beta diversity analysis in the M9 apple rootstock rhizosphere between the sample microbial community composition per group. (A) PCA analysis (Principal Component Analysis) based on OTU level. Points of different colors or shapes represent different sample groupings, and the scales of the horizontal and ordinate axes are relative distances. (B) PLS-DA (discriminant analysis of partial least squares) based on OTU level. This method uses PLS-DA to establish a relationship model between microbial content and sample class to predict the sample category. (C) PCoA (principal coordinates analysis) based on Bray–Curtis. The dots of different colors or shapes in the figure represent different grouping situations, and the scales of the horizontal and vertical axes are relative distances. PC1 and PC2 represent the suspected influencing factors for the deviation of the microbial composition of the two groups of samples. (D) NMDS analysis (Non-metric multidimensional scale method) based on OTU level. The samples of the same group are in a circle, indicating that the difference between groups is not apparent, while the absence of circles between groups indicates that there is a certain difference between groups. The four analysis illustrations demonstrate the similarity or difference in sample community composition depending on the distance between sample groups.
4. Discussion
Leaf characteristics are based on several factors, such as plant nutrient content, phenotype, and physiological processes. Therefore, this study investigated the effect of SLs on leaf characteristics such as length, width, area, nitrogen, and chlorophyll content. On the other hand, the study tried to connect leaf characteristics to the rhizosphere microbiome of apple rootstock.
The SLs were sprayed on the leaves, which may have had multiple effects. First, SLs might play a role in changing the leaf shape and its position by altering the leaf and stem attachment angles and the apical leaves’ diameters. Second, SLs may be transported to the roots to attract rhizosphere microorganisms and enhance nitrogen absorption. Finally, the nitrogen might be translocated to the leaves, affecting the chlorophyll content and, thus, the leaf characteristics (Figure 8).
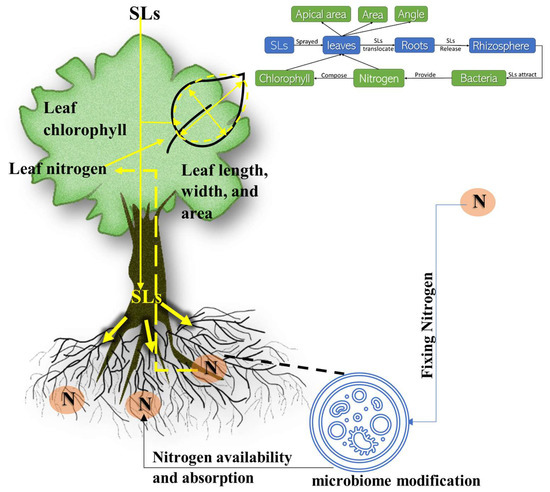
Figure 8.
Diagram of the signaling pathways of SLs and their effects on leaf length, width, leaf–stem angle, and apical area of the M9 apple rootstocks. The diagram further illustrates the effect of SLs on leaf nitrogen and chlorophyll content. It shows the impact of SLs in attracting microorganisms to the rhizosphere, changing its composition.
Many studies reported and reviewed various effects of Strigolactone on plant branching [8,59,60,61], leaf features, and vegetative growth and development improvements [62,63]. Previous studies support these researchers’ investigation of SLs’ impact on leaf shape modification and positioning. SLs may elongate the leaf length when applied at a mild concentration. In this study, SLs increased the leaf length when the application concentration rose from the lower level (1 µM) to the mild levels (5 µM) and vice versa for the leaf width. This agrees with Hu et al. [64], who found the same scenario; SLs elongated cells and consequently increased leaf length. This may have occurred due to the crosstalk between the SLs and other hormones, such as Auxin, responsible for cell elongation. Due to SLs’ influence, these leaf changes may support plants against abiotic stress. The changes in the leaf morphological features slightly affected the leaf area, although this difference was not significant when compared to the control treatment.
SLs increased the angle between the leaf and stem, which led to changes in the position of leaves. In the same trend, Zhao et al. [65] found that endogenous SLs affect the leaf angle. At low concentrations (1 µM), SLs did not affect apical diameter. On the contrary, the angles between the leaves and stem increased with the increasing treatment concentrations (5 and 10 µM). Sang et al. (2014) [66] demonstrated that tiller angles changed by applying strigolactones in an experiment on rice. Leaf area and angle were explained as important factors in changing plant architecture and providing support against gravitropism [66]. On the contrary, Shindo et al. (2020) [67] found that SLs play a different role under nutrient deficiency by decreasing the leaf angle.
The SLs have different pathways for providing plants with nitrogen. Some are based on enhancing absorption, attracting nitrogen-fixing microorganisms, or facilitating nitrogen translocation through the tissues [16,68,69]. The lowest concentration of SLs increased leaf nitrogen content and slightly decreased as the concentration increased. The increasing SL concentration caused decreases in the chlorophyll index. However, the lower concentration did not show a reduction in chlorophyll content.
The rhizosphere microbiome was impacted by the application of SLs [15,70,71,72]. Both high and low concentrations had a similar effect on nitrogen content, chlorophyll content, and the rhizosphere microbiome. This association may demonstrate that Strigolactone promotes the attraction of bacteria that provide nitrogen to plants. There were differences in the abundance, richness, and diversity of the different rhizospheres. The rhizosphere of the plants treated with the lowest concentration (1 µM) differed from the control and the relatively higher concentration (10 µM). Furthermore, the higher concentration may attract different taxa of microorganisms or have less relative abundance than the lower concentration of applied SLs (GR24) [71,72] (Figure 8).
It was noticed that plants treated with 1 µM attracted more microbes from the Actinobacteria and Proteobacteria phyla. The main classes were Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria, respectively. Furthermore, the concentration of 1 µM showed a noticeable increase in the relative abundance of the Rhizobiales, unidentified, Gemmatimonadales, Sphingomonadales, and Streptomycetales orders. On the other hand, 10 µM showed higher relative abundance in different orders, including unidentified, Nitrosomonadales, Cytophagales, Bacteroidales, and Solibacterales.
The 1 µM showed increases while 0 µM showed decreases in the bacterial α-diversity in the rhizosphere bacterial community. Although the 10 µM treatment increased the diversity (Shannon index) in the rhizosphere bacteria, the richness of the community was close to the control (Chao1, observed species)
According to the β-diversity metric analysis of the apple rootstock rhizosphere. There appeared to be a difference between the microbial communities of control, 1, and 10 µM treatments. However, there was relative similarity in the rhizosphere bacterial community between 0 and 10 µM. On the contrary, the concentration of 10 µM showed different β-diversity than the control.
5. Conclusions
SLs affect various aspects of vegetative growth, such as leaf length, width, stem angles, and the diameter of the leaf apical area. Moreover, SLs affect leaf nitrogen and chlorophyll contents. The treatments of 5 and 10 µM increased leaf–stem angles and decreased the apical diameter. The treatment with 5 µM significantly increased leaf length but did not show a significant increase in leaf width. Nitrogen content increased significantly at concentrations of 1 and 5 µM. Furthermore, compared to the control, 1 µM values had higher concentrations. However, there was no significant difference between them. The lower concentration (1 µM) attracted different microorganisms compared to the higher concentration (10 µM). The relation between the concentration of SLs and its effects can be explained by the bell curve graph.
Author Contributions
Conceptualization, S.S., Y.W. and Z.H.; methodology, S.S., Y.W. and Z.H.; formal analysis, S.S. and Y.W.; investigation, S.S., Y.W. and Z.H.; resources, S.S., Y.W., Z.H. and A.E.-k.; writing—original draft preparation, S.S., Y.W., Z.H. and A.E.-k.; writing—review and editing. Y.W. and Z.H.; supervision, Y.W. and Z.H.; project administration, Y.W. and Z.H.; funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by China Agricultural Research System (CARS-27).
Data Availability Statement
Data generated or analyzed during this study are provided in full within the published article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wassermann, B.; Müller, H.; Berg, G. An Apple a Day: Which Bacteria Do We Eat with Organic and Conventional Apples? Front. Microbiol. 2019, 10, 1629. [Google Scholar] [CrossRef]
- USDA. Apples and Oranges Are the Top U.S. Fruit Choices. 2021. Available online: https://www.ers.usda.gov/data-products/chart-gallery/gallery/chart-detail/?chartId=58322 (accessed on 24 May 2022).
- FAO. FAO STAT. Available online: https://www.fao.org/faostat/en/#home (accessed on 24 May 2022).
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Foo, E.; Plett, J.M.; Lopez-Raez, J.A.; Reid, D. Editorial: The Role of Plant Hormones in Plant-Microbe Symbioses. Front. Plant Sci. 2019, 10, 1391. [Google Scholar] [CrossRef]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2017, 114, E7641–E7649. [Google Scholar] [CrossRef]
- Masuda, Y. Auxin-induced cell elongation and cell wall changes. Bot. Mag. Shokubutsu Gaku Zasshi 1990, 103, 345–370. [Google Scholar] [CrossRef]
- Ferguson, B.; Beveridge, C.A. Roles for Auxin, Cytokinin, and Strigolactone in Regulating Shoot Branching. Plant Physiol. 2009, 149, 1929–1944. [Google Scholar] [CrossRef]
- Van de Poel, B.; Smet, D.; Van Der Straeten, D. Ethylene and Hormonal Cross Talk in Vegetative Growth and Development. Plant Physiol. 2015, 169, 61–72. [Google Scholar] [CrossRef]
- Zhao, B.; Liu, Q.; Wang, B.; Yuan, F. Roles of Phytohormones and Their Signaling Pathways in Leaf Development and Stress Responses. J. Agric. Food Chem. 2021, 69, 3566–3584. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Y.; Li, J. 10—Strigolactones. In Hormone Metabolism and Signaling in Plants; Li, J., Li, C., Smith, S.M., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 327–359. [Google Scholar]
- Min, Z.; Li, R.; Chen, L.; Zhang, Y.; Li, Z.; Liu, M.; Ju, Y.; Fang, Y. Alleviation of drought stress in grapevine by foliar-applied strigolactones. Plant Physiol. Biochem. 2019, 135, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Halouzka, R.; Zeljković, S.; Klejdus, B.; Tarkowski, P. Analytical methods in strigolactone research. Plant Methods 2020, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Seto, Y.; Yamaguchi, S. Strigolactone biosynthesis, transport and perception. Plant J. 2021, 105, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.; Wang, Y.; Han, Z.; Pervaiz, T.; El-Kereamy, A. Strigolactones in Plants and Their Interaction with the Ecological Microbiome in Response to Abiotic Stress. Plants 2022, 11, 3499. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Davies, N.W. Strigolactones promote nodulation in pea. Planta 2011, 234, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of Strigolactones to the Inhibition of Tiller Bud Outgrowth under Phosphate Deficiency in Rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Duna, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Dynamics of Strigolactone Function and Shoot Branching Responses in Pisum sativum. Mol. Plant 2013, 6, 128–140. [Google Scholar] [CrossRef]
- Zheng, W.; Gong, Q.; Zhao, Z.; Liu, J.; Zhai, B.; Wang, Z.; Li, Z. Changes in the soil bacterial community structure and enzyme activities after intercrop mulch with cover crop for eight years in an orchard. Eur. J. Soil Biol. 2018, 86, 34–41. [Google Scholar] [CrossRef]
- Stassen, M.J.J.; Hsu, S.-H.; Pieterse, C.M.J.; Stringlis, I.A. Coumarin Communication along the Microbiome–Root–Shoot Axis. Trends Plant Sci. 2021, 26, 169–183. [Google Scholar] [CrossRef]
- Guo, W.; Chen, L.; Herrera-Estrella, L.; Cao, D.; Tran, L.-S.P. Altering Plant Architecture to Improve Performance and Resistance. Trends Plant Sci. 2020, 25, 1154–1170. [Google Scholar] [CrossRef]
- Sarlikioti, V.; De Visser, P.H.B.; Buck-Sorlin, G.H.; Marcelis, L.F.M. How plant architecture affects light absorption and photosynthesis in tomato: Towards an ideotype for plant architecture using a functional–structural plant model. Ann. Bot. 2011, 108, 1065–1073. [Google Scholar] [CrossRef]
- Weraduwage, S.M.; Chen, J.; Anozie, F.C.; Morales, A.; Weise, S.E.; Sharkey, T.D. The relationship between leaf area growth and biomass accumulation in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 167. [Google Scholar] [CrossRef]
- Durigon, A.; Evers, J.; Metselaar, K.; Lier, Q.D.J.V. Water Stress Permanently Alters Shoot Architecture in Common Bean Plants. Agronomy 2019, 9, 160. [Google Scholar] [CrossRef]
- Li, Y.L.; Stanghellini, C. Analysis of the effect of EC and potential transpiration on vegetative growth of tomato. Sci. Hortic. 2001, 89, 9–21. [Google Scholar] [CrossRef]
- Gatti, M.; Squeri, C.; Garavani, A.; Vercesi, A.; Dosso, P.; Diti, I.; Poni, S. Effects of Variable Rate Nitrogen Application on cv. Barbera Performance: Vegetative Growth and Leaf Nutritional Status. Am. J. Enol. Vitic. 2018, 69, 196–209. [Google Scholar] [CrossRef]
- Jifon, J.L.; Syvertsen, J.P.; Whaley, E. Growth Environment and Leaf Anatomy Affect Nondestructive Estimates of Chlorophyll and Nitrogen in Citrus sp. Leaves. J. Am. Soc. Hortic. Sci. 2005, 130, 152–158. [Google Scholar] [CrossRef]
- Vos, J.; van der Putten, P.; Birch, C. Effect of nitrogen supply on leaf appearance, leaf growth, leaf nitrogen economy and photosynthetic capacity in maize (Zea mays L.). Field Crops Res. 2005, 93, 64–73. [Google Scholar] [CrossRef]
- Heuvelink, E.; Bakker, M.J.; Elings, A.; Kaarsemaker, R.C.; Marcelis, L.F.M. Effect of leaf area on tomato yield. Acta Hortic. 2005, 691, 43–50. [Google Scholar] [CrossRef]
- Ingham, R.E.; Trofymow, J.A.; Ingham, E.R.; Coleman, D.C. Interactions of Bacteria, Fungi, and their Nematode Grazers: Effects on Nutrient Cycling and Plant Growth. Ecol. Monogr. 1985, 55, 119–140. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621, Correction in Nat. Rev. Microbiol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Qu, Q.; Zhang, Z.; Peijnenburg, W.J.G.M.; Liu, W.; Lu, T.; Hu, B.; Chen, J.-M.; Chen, J.; Lin, Z.; Qian, H. Rhizosphere Microbiome Assembly and Its Impact on Plant Growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Liang, W.; Liu, S. Rhizosphere Microbiome: The Emerging Barrier in Plant-Pathogen Interactions. Front. Microbiol. 2021, 12, 772420. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, D.; Rangappa, K.; Das, A.; Layek, J.; Basavaraj, S.; Kandpal, B.K.; Shouche, Y.; Rahi, P. Pea (Pisum sativum L.) Plant Shapes Its Rhizosphere Microbiome for Nutrient Uptake and Stress Amelioration in Acidic Soils of the North-East Region of India. Front. Microbiol. 2020, 11, 968. [Google Scholar] [CrossRef]
- Sayer, E.J.; Crawford, J.A.; Edgerley, J.; Askew, A.P.; Hahn, C.Z.; Whitlock, R.; Dodd, I.C. Adaptation to chronic drought modifies soil microbial community responses to phytohormones. Commun. Biol. 2021, 4, 516. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition—Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [PubMed]
- Andreo-Jimenez, B.; Ruyter-Spira, C.; Bouwmeester, H.J.; Lopez-Raez, J.A. Ecological relevance of strigolactones in nutrient uptake and other abiotic stresses, and in plant-microbe interactions below-ground. Plant Soil 2015, 394, 19. [Google Scholar] [CrossRef]
- Smith, S.M. Q&A: What are strigolactones and why are they important to plants and soil microbes? BMC Biol. 2014, 12, 19. [Google Scholar] [CrossRef]
- Pineda, A.; Soler, R.; Pozo, M.J.; Rasmann, S.; Turlings, T.C.J. Editorial: Above-belowground interactions involving plants, microbes and insects. Front. Plant Sci. 2015, 6, 318. [Google Scholar] [CrossRef]
- Boyacı, S.; Küçükönder, H. A research on Non-Destructive Leaf Area Estimation Modeling for some Apple Cultivars. Erwerbs-Obstbau 2022, 64, 1–7. [Google Scholar] [CrossRef]
- Mateo, M.; Sabaté, S. Wet digestion of vegetable tissue using a domestic microwave oven. Anal. Chim. Acta 1993, 279, 273–279. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. Mbio 2015, 6, e02527-14. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Fouts, D.E.; Szpakowski, S.; Purushe, J.; Torralba, M.; Waterman, R.C.; MacNeil, M.D.; Alexander, L.J.; Nelson, K.E. Next Generation Sequencing to Define Prokaryotic and Fungal Diversity in the Bovine Rumen. PLoS ONE 2012, 7, e48289. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Q.; Zhou, J.; Wei, Q. Illumina Amplicon Sequencing of 16S rRNA Tag Reveals Bacterial Community Development in the Rhizosphere of Apple Nurseries at a Replant Disease Site and a New Planting Site. PLoS ONE 2014, 9, e111744. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Washington, H. Diversity, biotic and similarity indices: A review with special relevance to aquatic ecosystems. Water Res. 1984, 18, 653–694. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Bunge, J.; Willis, A.; Walsh, F. Estimating the Number of Species in Microbial Diversity Studies. Annu. Rev. Stat. Its Appl. 2014, 1, 427–445. [Google Scholar] [CrossRef]
- Soundappan, I.; Bennett, T.; Morffy, N.; Liang, Y.; Stanga, J.P.; Abbas, A.; Leyser, O.; Nelson, D.C. SMAX1-LIKE/D53 Family Members Enable Distinct MAX2-Dependent Responses to Strigolactones and Karrikins in Arabidopsis. Plant Cell 2015, 27, 3143–3159. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Rameau, C. Strigolactones, a novel class of plant hormone controlling shoot branching. Comptes Rendus Biol. 2010, 333, 344–349. [Google Scholar] [CrossRef]
- Ma, N.; Hu, C.; Wan, L.; Hu, Q.; Xiong, J.; Zhang, C. Strigolactones Improve Plant Growth, Photosynthesis, and Alleviate Oxidative Stress under Salinity in Rapeseed (Brassica napus L.) by Regulating Gene Expression. Front. Plant Sci. 2017, 8, 1671. [Google Scholar] [CrossRef]
- Zheng, Y.; Kumar, N.; Gonzalez, P.; Etxeberria, E. Strigolactones restore vegetative and reproductive developments in Huanglongbing (HLB) affected, greenhouse-grown citrus trees by modulating carbohydrate distribution. Sci. Hortic. 2018, 237, 89–95. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, S.; Huang, B. Strigolactones Promote Leaf Elongation in Tall Fescue through Upregulation of Cell Cycle Genes and Downregulation of Auxin Transport Genes in Tall Fescue under Different Temperature Regimes. Int. J. Mol. Sci. 2019, 20, 1836. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-Q.; Xiang, J.-J.; Xue, H.-W. Studies on the Rice LEAF INCLINATION1 (LC1), an IAA–amido Synthetase, Reveal the Effects of Auxin in Leaf Inclination Control. Mol. Plant 2013, 6, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.; Meng, X.; Chu, J.; Sun, X.; Dong, G.; Yuan, Y.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef]
- Shindo, M.; Yamamoto, S.; Shimomura, K.; Umehara, M. Strigolactones Decrease Leaf Angle in Response to Nutrient Deficiencies in Rice. Front. Plant Sci. 2020, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Marro, N.; Lidoy, J.; Chico, M.; Rial, C.; García, J.; Varela, R.M.; Macías, F.A.; Pozo, M.J.; Janoušková, M.; López-Ráez, J.A. Strigolactones: New players in the nitrogen–phosphorus signalling interplay. Plant Cell Environ. 2022, 45, 512–527. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.; Kusumoto, D.; Sekimoto, H.; Sugimoto, Y.; Takeuchi, Y.; Yoneyama, K. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 2007, 227, 125–132. [Google Scholar] [CrossRef]
- Kim, B.; Westerhuis, J.A.; Smilde, A.K.; Floková, K.; Suleiman, A.K.A.; Kuramae, E.E.; Bouwmeester, H.J.; Zancarini, A. Effect of strigolactones on recruitment of the rice root-associated microbiome. FEMS Microbiol. Ecol. 2022, 98, fiac010. [Google Scholar] [CrossRef]
- Rochange, S.; Goormachtig, S.; Lopez-Raez, J.A.; Gutjahr, C. The Role of Strigolactones in Plant–Microbe Interactions. In Strigolactones—Biology and Applications; Koltai, H., Prandi, C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 121–142. [Google Scholar] [CrossRef]
- López-Ráez, J.A.; Shirasu, K.; Foo, E. Strigolactones in Plant Interactions with Beneficial and Detrimental Organisms: The Yin and Yang. Trends Plant Sci. 2017, 22, 527–537. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).