Abstract
Light-harvesting chlorophyll a/b binding (Lhc) proteins play a key role in the efficiency of the photosynthetic system of plants and participate in the regulation of plant growth, development, and the response to abiotic stress. In this study, 28 PbrLhc genes of the family were identified in the pear Pyrus bretschneideri genome database, which were distributed on 25 of the 27 chromosomes. Phylogenetic relationship analysis demonstrated that PbrLhc proteins could be divided into five subfamilies. Using MEME Suite, PlantRegMap, and Softberry, the conserved motifs, transcription binding sites, and predicated location of all PbrLhc proteins, respectively, were determined. A collinearity relationship using MCScanX software showed that there was a collinearity relationship between different PbrLhc genes. Using the hierarchical clustering method to develop a heat map of gene expression, five genes were found to be down-regulated under irrigation treatment and up-regulated under drought treatment; seven genes were up-regulated under irrigation treatment and down-regulated under drought treatment. Finally, the results of these analyses will provide theoretical basis for future study on the function and differential expression of PbrLhc genes in response to drought stress.
1. Introduction
Light energy can be converted into chemical energy by green plants for cellular processes via photosynthesis [1]. In this process, light energy is captured and transferred by chlorophyll [2]. Light-harvesting chlorophyll a/b-binding (Lhc) protein is one of the most abundant thylakoid membrane proteins encoded by the nuclear genome. Lhc is one of the earliest cloned genes. It plays a significant role in the efficiency of the photosynthetic system and crop yield. The Lhc family is mainly divided into two groups, Lhca and Lhcb, which are related to antenna proteins of photosystem I and photosystem II. They are composed of three transmembrane helices and can combine chlorophylls (e.g., a, b or c), carotenoids, and lipids [3]. Lhc proteins can also combine with these pigment molecules to form a protein complex that participates in the collection and transportation of solar energy during photosynthesis [4].
In plants, as a type of membrane protein with high abundance in photosystem II, Lhcb acts as an antenna protein and participates in regulating plant growth and development [4]. For example, AtLhcb1 regulates seed germination and plant growth in Arabidopsis thaliana by responding to the hormone abscisic acid (ABA) [5]. Additionally, five single nucleotide polymorphisms (SNPs) in barley Lhcb1 were significantly associated with agronomic traits, such as plant height, leaf color, etc. [6]. Finally, overexpression of Sedum alfredii SaLhcb2 can confer increased the biomass of roots and branches [7].
LHC is also involved in the response of plants to abiotic stress. Some studies have demonstrated that Lhcb1-6 can affect the response of stomata to abscisic acid inflow, thus improving the sensitivity of plants to drought stress when they are down-regulated [8]. In addition, the down-regulation of Lhcb results in an ABA-insensitive phenotype in germination and growth of seed. In celery, the expression level of Lhcb1 can be up-regulated by abiotic stress, such as cold, salt, and drought stress. In rice seedlings, the level of Lhca1-4 subunit decreases during iron deficiency, weakening the light-harvesting ability of plants and leading to a decline in photosynthetic efficiency. AtLhcb1-6, which responds to stomatal movement and participates in ABA signal transduction by affecting reactive oxygen species homeostasis, plays a significant role in plant resistance to stress [9]. LeLhcb2 overexpression enhances tolerance of tobacco to cold stress and reduces the photooxidation of photosystem II [10]. In addition, formaldehyde stress can affect the expression of photosynthetic genes in Arabidopsis, including Lhca4, Lhcb2.1, and Lhcb3 [11]. In previous studies, the Malus domestica Lhc gene MdLhc was reported to show different expression patterns under drought stress, and transgenic Arabidopsis and apple calli expressing MdLhcb4.3 confer resistance to osmotic and drought stress [12]. These results demonstrate that Lhc proteins play a significant role in regulating plant resistance to various stressors.
Pear (Pyrus bretschneideri) is one of the fruits with the largest cultivation area and yield in northern China. However, drought stress is an important obstacle limiting the economic cultivation of fruit trees. When fruit trees are subjected to abiotic stresses, such as drought, their cultivation is vulnerable, resulting in serious production losses. Completion of the P. brestschneideri genome sequencing database has provided the possibility of mining key candidate genes associated with the drought stress response. In view of the potential application value of PbrLhc genes in regulating plant growth and development and the genetic improvement of drought-resistant fruit trees, the comprehensive bioinformatic analysis of chromosome location, physical and chemical properties, gene structure, predicted subcellular location, collinearity, and expression profiles of the PbrLhc genes in response to drought stress have been carried out in this study. This information will provide a theoretical basis for future research concerning the function and molecular mechanism of PbrLhc genes in response to drought stress, as well as the genetic improvement of drought-resistant germplasm.
2. Materials and Methods
2.1. Genome Wide Identification of PbrLhc in P. bretschneideri
To identify the Lhc proteins of P. bretschneideri, AtLhc protein sequences were obtained from the Arabidopsis database [13], which were taken as the construction of a hidden Markov model using HMMER 3.0 (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer accessed on 10 December 2022). Using this model, all potential Lhc protein sequences of the P. bretschneideri genome database (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/315/295/GCF_000315295.1_Pbr_v1.0/ accessed on 16 December 2022) were identified. Next, blastp (version: ncbi blast 2.10.1+) was used to obtain the Lhc family reference sequence by aligning the sequences of these proteins (the e-value was set to 1 × 10−5). The alignment sequence was used as the candidate Lhc family sequence. These sequences were combined as candidate Lhc protein sequences. Pfamscan (version: v1.6) and the Pfam A database (version: v33.1) were used to annotate the sequence domain obtained [14] and to determine the sequences containing the PF00504 domain as the final Lhc sequences.
2.2. Gene Sequence Alignment and Phylogenetic Relationships Analysis
Compute pI/MW in the ProtParam tool [15] were used for the analysis of physical and chemical characteristics of PbrLhc potential proteins, including the theoretical molecular weight, instability index, coded amino acid number, isoelectric point, and aliphatic index.
ClustalW was used for multiple alignments of amino acid sequences [16]. The neighbor-joining method with 1000 bootstrap replicates was used for phylogenetic tree construction of the PbrLhc protein sequences in P. bretschneideri and the AtLhc protein sequences in Arabidopsis, and the resulting figure was visualized by MEGA7.0 (Arizona State University, Tempe, AZ, USA).
2.3. Prediction Analysis of Chromosome Distribution, Subcellular Localization, and Gene Structure
The distribution of PbrLhc genes on chromosomes was identified using MapInspect software and visualized using Tbtools [17]. The subcellular localization of PbrLhc proteins was predicted using the SoftBerry database (Mount Kisco, NY, USA).
Intron/exon structure of PbrLhc genes was analyzed using the GSDS [18]. The conserved motifs of the PbrLhc amino acid sequences were identified using MEME Suite [19]. The sequences of the conserved motifs were listed in Supplemental Table S1.
2.4. Cis-Element Analysis of the Promoter Region and Signal Peptide Prediction
The 2 kb promoter regulatory region sequences upstream of the PbrLhc starting site were used for the analysis of potential TF binding sites by the PlantRegMap database [20]. Pyrus bretschneideri was selected as the species, and the e-value was set to 1 × 10−4. In the physical map of the gene promoter, only the top 12 TF families were displayed by labeling and displaying their binding sites. The signal peptide prediction software SignalP 3.0 [21] was used to predict their positions hidden and the signal peptide cutting sites in the given amino acid sequences of PbrLhc proteins. This prediction method is based on various artificial neural network algorithms.
2.5. Collinearity Analysis and Expression Pattern Analysis
Collinearity analysis of PbrLhc genes was performed using MCScanX software [22], and the data obtained were visualized using Circos software.
The RNA-seq data of PbrLhc genes under different irrigation and drought conditions were obtained from the pear drought transcriptome database (https://peerj.com/articles/12921/#supp-4 accessed on 14 January 2023) [23], and the data were analyzed using MAV software, and a clustering heat map of expression was generated using the hierarchical clustering (HCL) method.
3. Results
3.1. Identification of PbLhc Genes
To identify potential Lhc proteins in the P. bretschneideri genome, 21 acquired AtLhc protein sequences were used as query sequences. Finally, 28 PbrLhc sequences were obtained, and some identification and comparison results are shown in Table 1.

Table 1.
Statistical table of family member information.
To determine the distribution of these PbrLhc genes on chromosomes, the physical positions of the 28 PbrLhc sequences on the 27 total P. bretschneideri chromosomes were studied using MapInspect. As shown in Figure 1, these sequences were distributed on 25 chromosomes with a relatively uniform distribution. However, three genes, PbrLhcb1.6, PbrLhcb1.7, and PbrLhcb1.8, were located in close proximity on chromosome NW_008988048.1, and two genes, PbrLhcb1.4 and PbrLhcb1.5, were located on chromosome NW_008988041.1. The other 23 chromosomes possessed only one gene.
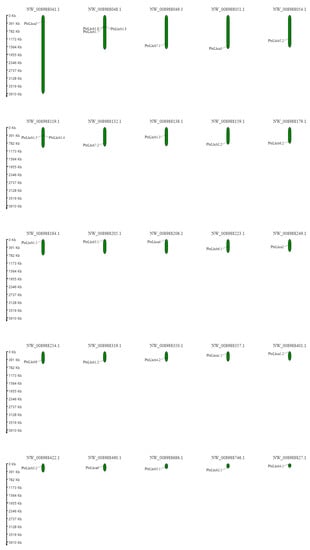
Figure 1.
Distribution of PbrLhc genes on 25 of the 27 chromosomes in the Pyrus bretschneideri genome. The physical location map was drawn based on the location of the PbrLhc genes on the chromosomes by TBtools.
3.2. Characteristics of PbrLhc Genes
The physical and chemical properties of PbrLhc genes were also analyzed, as shown in Table 2. The prediction results showed that these genes potentially encode polypeptides of 246 (PbrLhcb5.1) to 347 (PbrLhcb8). The predicted molecular weights range from 26.68 kDa (PbrLhca1.2) to 37.83 kDa (PbrLhcb8). The protein isoelectric point range is from 4.81 (PbrLhcb1.5) to 8.52 (PbrLhcb8).

Table 2.
Physical and chemical characteristics of partial PbrLhc proteins.
3.3. Phylogenetic Analysis of PbrLhc Proteins
To analyze the phylogenetic relationship between different PbrLhc proteins, the phylogenetic tree containing the Lhc amino acid sequences of Arabidopsis and P. bretschneideri was constructed. As shown in Figure 2, PbrLhc proteins can be divided into five subfamilies (CP24, CP26, CP29, Lhca, and Lhcb). The distribution of PbrLhc proteins between the subgroups was uneven. Among them, Lhcb subfamily had 13 members, followed by the Lhca subfamily with seven, while the CP24 and CP26 subfamilies had the fewest members, each with only two genes.
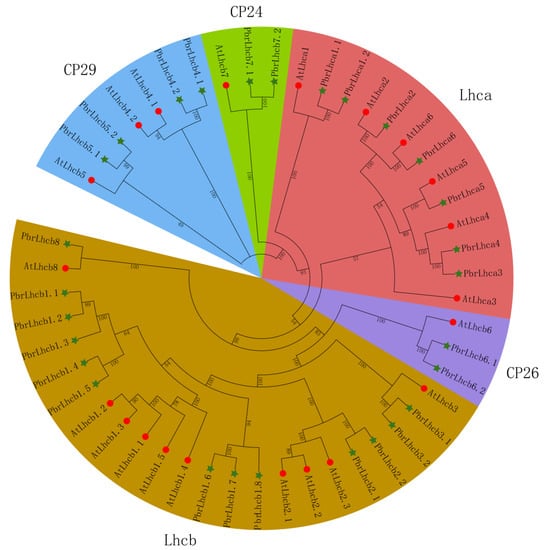
Figure 2.
Phylogenetic relationship of Lhc proteins in Arabidopsis and P. bretschneideri, divided into five subfamilies, CP24, CP26, CP29, Lhca, and Lhcb. AtLhc proteins are marked with red circles, and PbrLhc proteins are marked with green stars.
3.4. Gene Structure Analysis of PbrLhc Genes
To analyze the distribution of the introns and exons of PbrLhc genes, their structural characteristics were analyzed with the GSDS. The results revealed that differences in the exon/intron structures of the different PbrLhc genes were apparent (Figure 3). Among these, PbrLhcb1.1–1.8 each have only one exon. Additionally, there were two exons in seven genes: PbrLhcb2.1, PbrLhcb2.2, PbrLhcb4.1, PbrLhcb4.2, PbrLhcb5.2, PbrLhcb7.1, and PbrLhcb7.2. However, the number of exons in PbrLhca5 is only six.
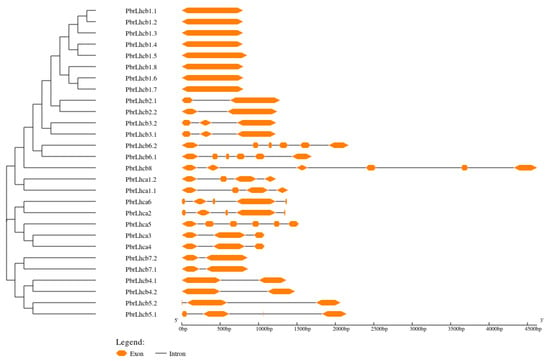
Figure 3.
Gene structures of PbrLhc genes. Intron/exon structure of PbrLhc genes was analyzed by GSDS. Orange boxes present exons, and single lines present introns. Gene models are drawn to scale, as shown in the bar at the bottom.
3.5. Promoter Elements and Conserved Motifs Analysis of PbrLhc Genes
To study the similarity and diversity of motifs among different proteins, the 15 conserved motifs found in the PbrLhc genes were analyzed (Figure 4). Most of the identified PbrLhc genes contained motifs 1, 2, 3, 9, and 10. Different members had different conserved motifs. However, members of the same subfamily had similar ones. For instance, all members of the Lhcb subgroup contain the same motif.
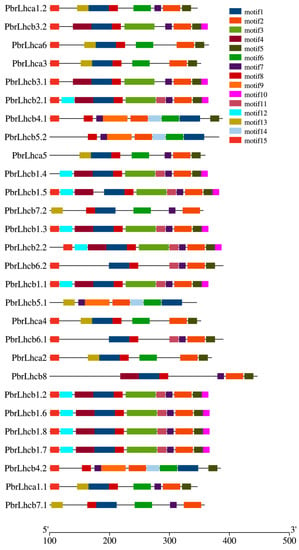
Figure 4.
Conserved motifs of the PbrLhc genes. The MEME Suite program was used for the motifs analysis of the 28 PbrLhc protein sequences [19]. Colors of boxes represent motifs at the corresponding position in each PbrLhc protein.
To further understand the transcriptional regulation of the PbrLhc genes, functional elements 2000 bp upstream of their coding regions were predicted using the PlantRegMap database. The results indicated that several TF-binding sites were found, such as TCP, MYB, ERF, NAC, etc., in the promoter region of PbrLhc genes (Figure 5), demonstrating that the expression of given genes might be regulated by these TFs.
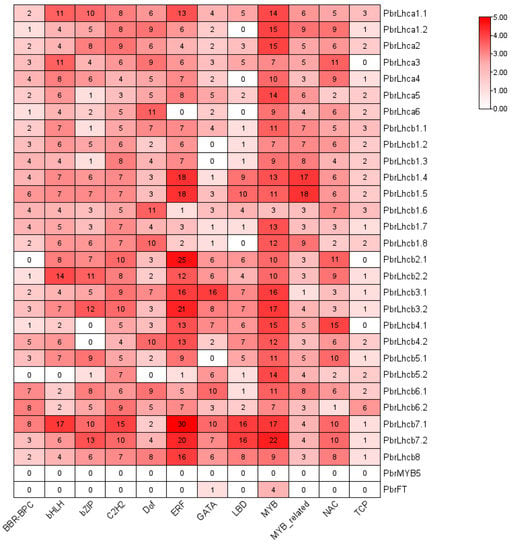
Figure 5.
Promoter element prediction analysis of the PbrLhc genes. Transcription factor binding sites within a region 2000 bp upstream of the PbrLhcs start site was predicted. Only the top 12 transcription factor families were shown. The number of putative TF sites in the promoters of the PbrLhc genes were marked.
3.6. Subcellular Localization and Signal Peptide Prediction of PbrLhc Proteins
Using the SoftBerry database, an analysis of the subcellular localization of PbrLhc family members indicated that all identified PbrLhc proteins were predicted to be in membrane-bound chloroplasts (Table 3).

Table 3.
Predicted subcellular location of the PbrLhc family members.
The amino acid sequences encoded by the identified 28 PbrLhc genes were also analyzed using SignalP software to predict whether potential signal peptide cutting sites and positions were observed. The predicted scores of each PbrLhc protein were less than 0.5, and no significant peak turning was found (Supplemental Figure S1). Therefore, no signal peptide splicing site was identified in any PbrLhc protein.
3.7. Expression Pattern Analysis of PbrLhc Genes
To explore the potential function of PbrLhc proteins, based on previous drought transcriptome data on P. bretschneideri [21], an expression heat map of 18 PbrLhc genes under simulated irrigation and drought treatments at different times was constructed. The results demonstrated that the gene expression levels of PbrLhca3, PbrLhca4, PbrLhca5, PbrLhcb1.5, and PbrLhca1.6 were down-regulated during irrigation treatment but were up-regulated following drought treatment (Figure 6). The gene expression levels of PbrLhcb6.2, PbrLhcb8, PbrLhcb1.8, PbrLhcb3.2, PbrLhcb2.2, PbrLhcb6.1, and PbrLhca6 were up-regulated during irrigation treatment but were down-regulated following drought treatment. However, there was no significant change in the expression of PbrLhcb1.7 under two treatments. The expression levels of the other genes followed no distinct trend under either treatment. Therefore, given the correlation between gene activity and irrigation/drought conditions, it is possible that some of these PbrLhc genes could possibly participate in regulating the response of pear to drought stress.
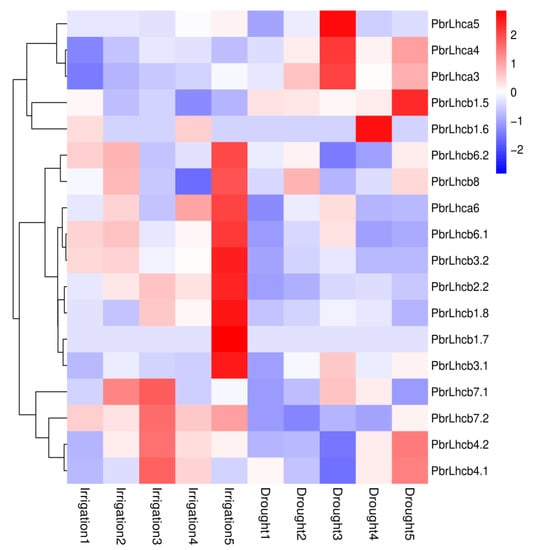
Figure 6.
Expression pattern analysis of PbrLhc genes in response to irrigation or drought conditions. All raw reads were deposited in the NCBI SRA database under the accession numbers of SRR12424088–SRR12424107 (Bioproject PRJNA655255). The FPKM values of PbrLhc genes in different tissues were obtained, and the log2 based on FPKM values were taken as generation of the heat map by MeV4 software.
3.8. Collinearity Analysis of PbrLhc Genes
A collinearity analysis of the 28 identified PbrLhc family members was identified between different PbrLhc genes, and the results demonstrated that it had a collinearity relationship between some PbrLhc genes, such as between PbrLhca3 and PbrLhca4 and between PbrLhcb5.1 and PbrLhcb5.2 (Figure 7). In addition, a collinearity analysis was conducted between P. bretschneideri and Arabidopsis, which revealed collinearity between family members of both plants, such as between PbrLhcb4.2, AtLhcb4.1, PbrLhca5, and AtLhca5 and between PbrLhcb3.1/3.2 and AtLhcb3, indicating that members of the same subtribe often have similarities and that some similarities might exist between members who are close to each other in different species (Supplemental Figure S2).
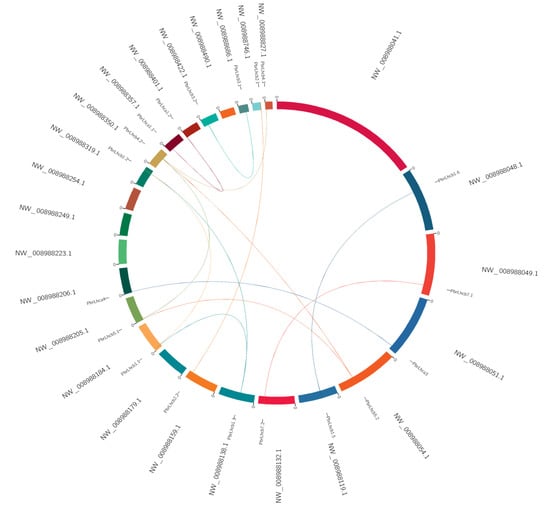
Figure 7.
Collinearity analysis of the 28 identified PbrLhc genes. Collinearity analysis was performed using the MCScanX software.
4. Discussion
Currently, the Lhc gene family has been identified in Arabidopsis [24], rice [24], Populus [25], Zostera marina [26], Ricinus communis [27], Hordeum vulgare, and apple [12]. However, no relevant studies on pear have been previously reported. In this study, 28 PbrLhc genes were identified, and the diversity for which is higher than that for Arabidopsis (21) and rice (17). In general, members of the same subgroup often have similar domain conserved motifs.
In view of the important role of Lhc proteins in regulating plant growth and abiotic stress resistance, 27 MdLhc genes identified in previous studies were randomly distributed on 17 chromosomes in apples. The MdLhc proteins could be divided into Lhca, CP24-CP29, and Lhcb-CP26 subfamilies, all of which contained typical Lhc motifs, indicating that the sequence of Lhc family proteins is highly conserved in plants. In addition, an analysis of MdLhc gene structure showed that there were structural differences among different subgroups, while genes of the same subgroup had similar structures [12]. Among these, six genes had no introns and seven genes contained only one intron [12]. In the present study, 28 PrLhc genes were identified. Phylogenetic analysis demonstrated that the PbrLhc proteins were divided into five subfamilies (CP24, CP26, CP29, Lhca, and Lhcb) and that the distribution of the PbrLhc genes in different subgroups was uneven. The gene structure and conserved motif of the same branch in the evolutionary tree were similar and large differences between different subfamilies were apparent, similar to what is observed in apples. For example, compared with PbrLhcb, the structure of PbrLhca is more complex and contains more introns.
Some studies have shown that the expression of Lhc genes associate with a variety of abiotic stressors. For example, previous studies have found that the expression of Lhcb1 in the celery cultivars could be up-regulated under several abiotic stresses. Heterologous expression of LeLhcb2 in transgenic tobacco can improve its tolerance to low temperatures [10]. The down-regulation of Lhcb1-Lhcb6 in Arabidopsis affects the response of stomata to ABA, thus reducing the tolerance of plants to drought stress [8]. In this study, numerous bZIP, MYB, NAC, and other TF-binding sites were predicted to present in the promoter region of the PbrLhc genes, and these transcription factors are reported to participate in light response, flavonoid biosynthesis, and drought induction, which are associated with the stress response of plants. Combined with the analysis of PbrLhc gene expression under drought stress and irrigation conditions, PbrLhc proteins might play an important role in the response to abiotic stress or hormone regulation, with regards to plant growth and development.
In conclusion, these results provide a foundation for future research on the function of PbrLhc genes regarding the growth and development of P. bretschneideri, as well as its response to drought stress and the molecular mechanisms underlying its drought resistance. These results also provide a theoretical basis for the molecular breeding of new drought-resistant pear varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9050522/s1, Figure S1: Signal peptide prediction of PbrLhc proteins. The prediction of potential signal peptide locations and cleavage sites in PbrLhc protein sequences was determined by SignalP software based on various artificial neural network algorithms. Figure S2: Similarity analysis of members of the PbrLhc family in P. bretschneideri and the AtLhc family in Arabidopsis. Table S1: The sequences of the conserved motifs.
Author Contributions
Conceptualization, K.R.; methodology, K.R.; software, S.Z.; validation, R.W. and K.R.; formal analysis, R.W.; investigation, R.W.; resources, R.W.; data curation, R.W.; writing—original draft preparation, R.W.; writing—review and editing, F.C.; visualization, F.C.; supervision, R.W.; project administration, R.W.; funding acquisition, F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Earmarked Fund of Industrial Innovation Team Construction of Hebei Modern Agricultural Industrial Technology System (HBCT2021210203), the Handan Science and Technology Research and Development Plan Project (21422012321), the General Project of the Natural Science Foundation of Hebei Province (C2022402006), and the Innovation and Entrepreneurship Training Program for College Students (X202210076061).
Data Availability Statement
The following information was supplied regarding data availability: The sequence data for expression pattern analysis of PbrLhc genes in response to irrigation or drought conditions is available at NCBI: PRJNA655255. The raw data are available in the Supplemental Files.
Acknowledgments
The logical presentation of ideas and the structure of the manuscript was checked and performed by professional editors at Editage, a division of Cactus communications.
Conflicts of Interest
The authors declare there is no competing interest.
References
- Kirilovsky, D.; Büchel, C. Chapter Nine. Evolution and function of light-harvesting antenna in oxygenic photosynthesis. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2019; Volume 91, pp. 247–293. [Google Scholar]
- Wang, P.; Grimm, B. Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Yang, J. Genomics analysis of the light-harvesting chlorophyll a/b-binding (Lhc) superfamily in cassava (Manihot esculenta Crantz). Gene 2019, 702, 171–181. [Google Scholar] [CrossRef]
- Jansson, S. A guide to the Lhc genes and their relatives in Arabidopsis/IT>. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell. 2014, 26, 3646–3660. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Ning, Z.; Bai, G.; Li, R.; Yan, G.; Siddique, K.H.; Baum, M.; Guo, P. Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLoS ONE 2012, 7, e37573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Senoura, T.; Yang, X.; Chao, Y.; Nishizawa, N.K. Lhcb2 gene expression analysis in two ecotypes of Sedum alfredii subjected to Zn/Cd treatments with functional analysis of SaLhcb2 isolated from a Zn/Cd hyperaccumulator. Biotechnol. Lett. 2011, 33, 1865–1871. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Ma, J.; Wu, N.; Su, Y.Q.; Zhang, Z.W.; Yuan, M.; Zhang, H.Y.; Zeng, X.Y.; Yuan, S. The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 2018, 130, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.S.; Kong, F.Y.; Zhou, B.; Zhang, S.; Yue, M.M.; Meng, Q.W. Heterology expression of the tomato LeLhcb2 gene confers elevated tolerance to chilling stress in transgenic tobacco. Plant Physiol. Biochem. 2014, 80, 318–327. [Google Scholar] [CrossRef]
- Wang, S.; Song, Z.; Sun, Z.; Zhang, J.; Mei, Y.; Nian, H.; Li, K.; Chen, L. Effects of formaldehyde stress on physiological characteristics and gene expression associated with photosynthesis in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2012, 30, 1291–1302. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Luo, J.; Wang, H.; Dong, Q.; Wang, Y.; Yang, K.; Mao, K.; Ma, F. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress. Plant Physiol. Biochem. 2020, 154, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.H.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the Expasy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; Volume 52, pp. 571–607. ISSN 1064-3745. [Google Scholar]
- Hung, J.H.; Weng, Z.P. Sequence alignment and homology search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot093088. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 9. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan = Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MeMe: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. Plantregmap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S.S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debary, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Yang, S.; Bai, M.; Hao, G.; Guo, H.; Fu, B. Transcriptomics analysis of field-droughted pear (Pyrus spp.) reveals potential drought stress genes and metabolic pathways. PeerJ 2022, 10, e12921. [Google Scholar] [CrossRef] [PubMed]
- Umate, P. Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal. Behav. 2010, 5, 1537–1542. [Google Scholar] [CrossRef] [PubMed]
- Klimmek, F.; Sjödin, A.; Noutsos, C.; Leister, D.; Jansson, S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 2006, 140, 793–804. [Google Scholar] [CrossRef]
- Kong, F.; Zhou, Y.; Sun, P.; Cao, M.; Li, H.; Mao, Y. Identification of light-harvesting chlorophyll a/b-binding protein genes of Zostera marina L. and their expression under different environmental conditions. J. Ocean Univ. China 2016, 15, 152–162. [Google Scholar] [CrossRef]
- Zou, Z.; Huang, Q.; An, F. Genome-wide identification, classification and expression analysis of Lhc supergene family in castor bean (Ricinus communis L.). Agric. Biotechnol. 2013, 2, 44. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).