Identification of ABA Signaling Pathway Genes and Their Differential Regulation in Response to Suboptimal Light Stress in Grape (Vitis vinifera L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Analysis of APGs in Grapevine
2.2. Phylogenetic Analysis and Gene Conserved Motif Analysis
2.3. Chromosomal Location, Gene Duplication, Synteny Analysis and Cis-Acting Regulatory Element Analysis
2.4. Plant Materials and Treatments
2.5. Measurement of Leaf Gas Exchange Parameters
2.6. Measurement of Berry Quality
2.7. RNA Extraction and qRT-PCR
2.8. Statistical Analysis
3. Results
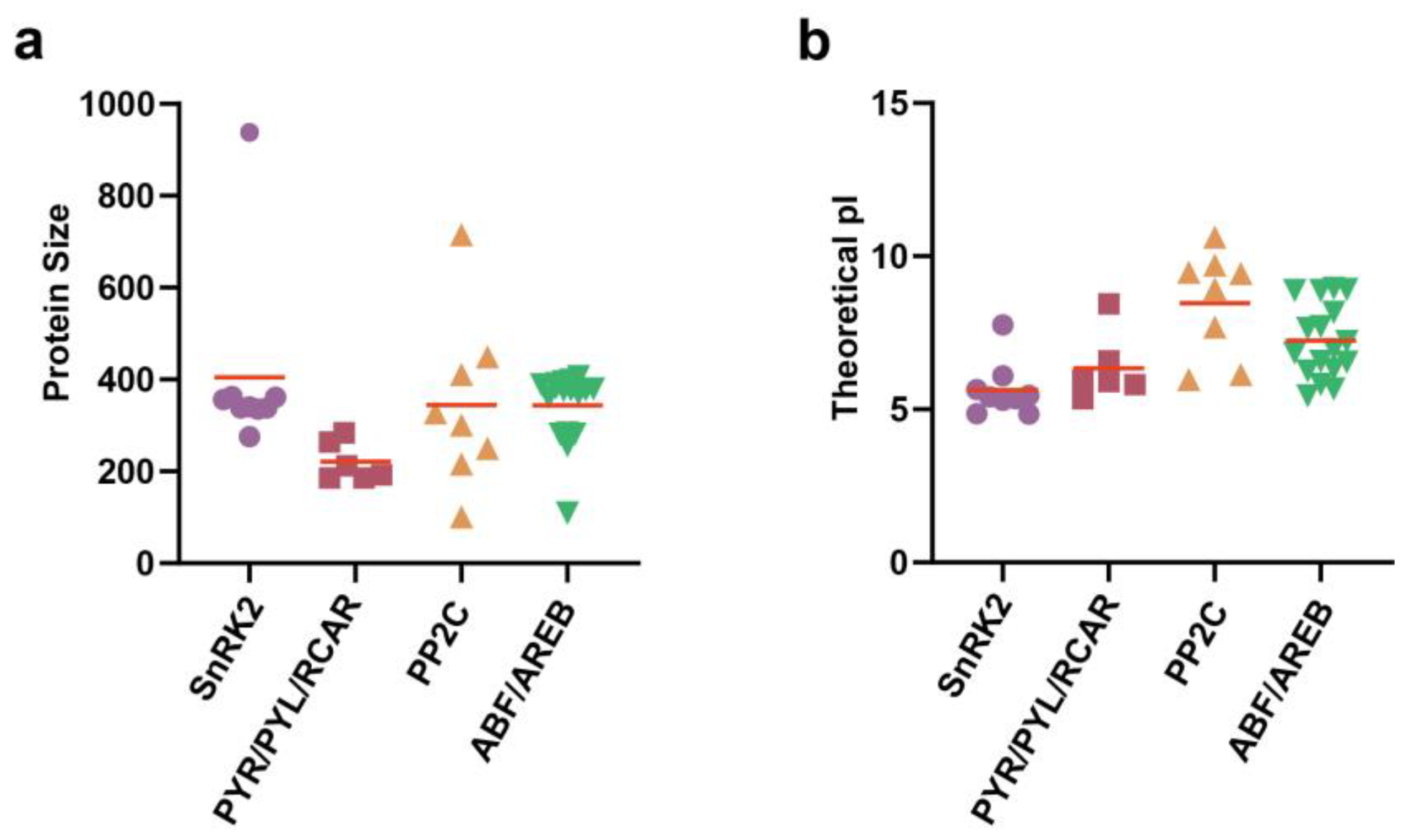
3.1. Identification and Analysis of APGs in Grapevine
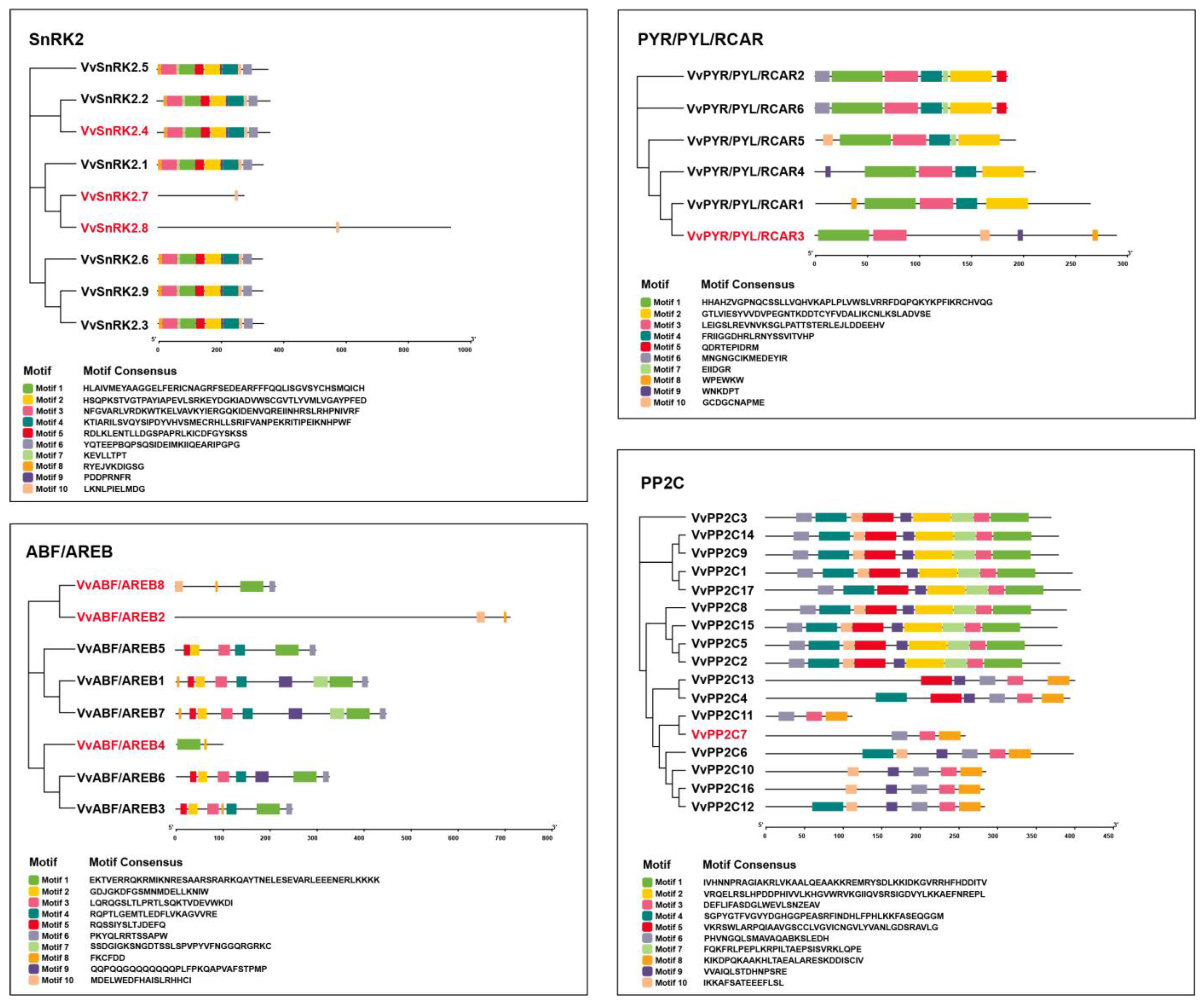
3.2. Phylogeny and Conserved Motifs of APGs in Grapevine
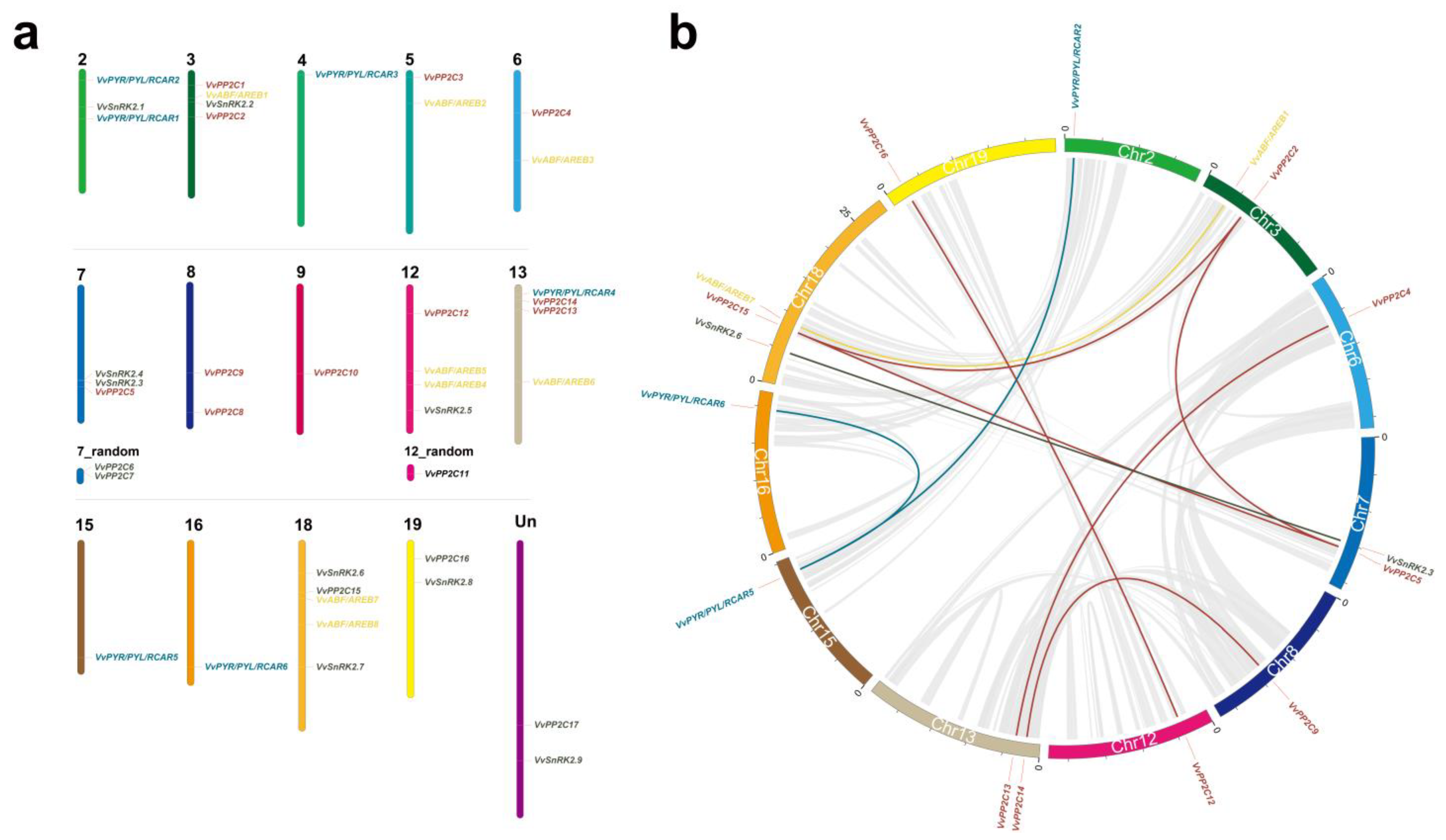
3.3. Chromosomal Location and Collinearity Analysis
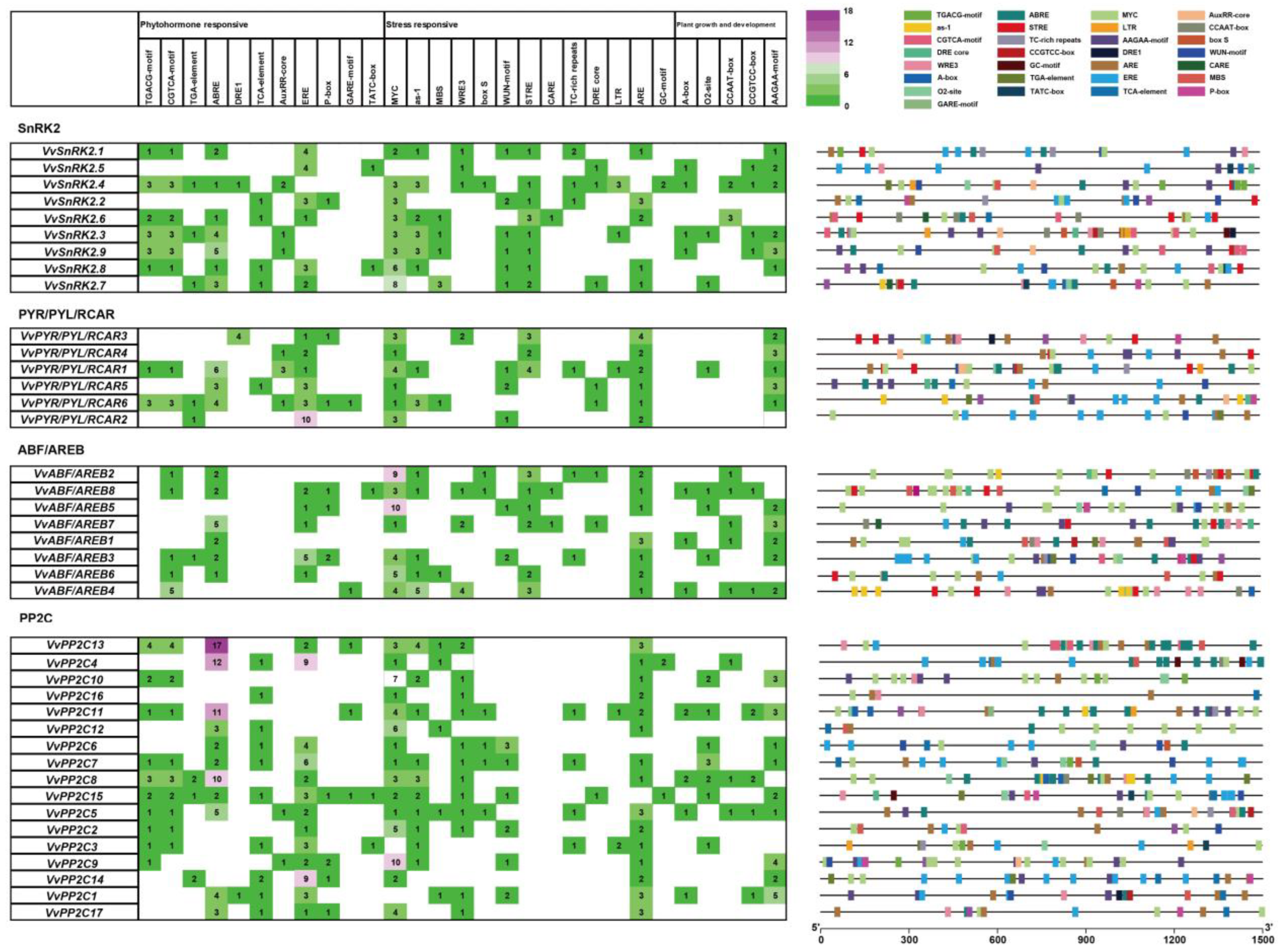
3.4. Cis-Acting Elements Present in the Promoter Regions
3.5. Growth of Leaves and Berry under Weak Light Stress
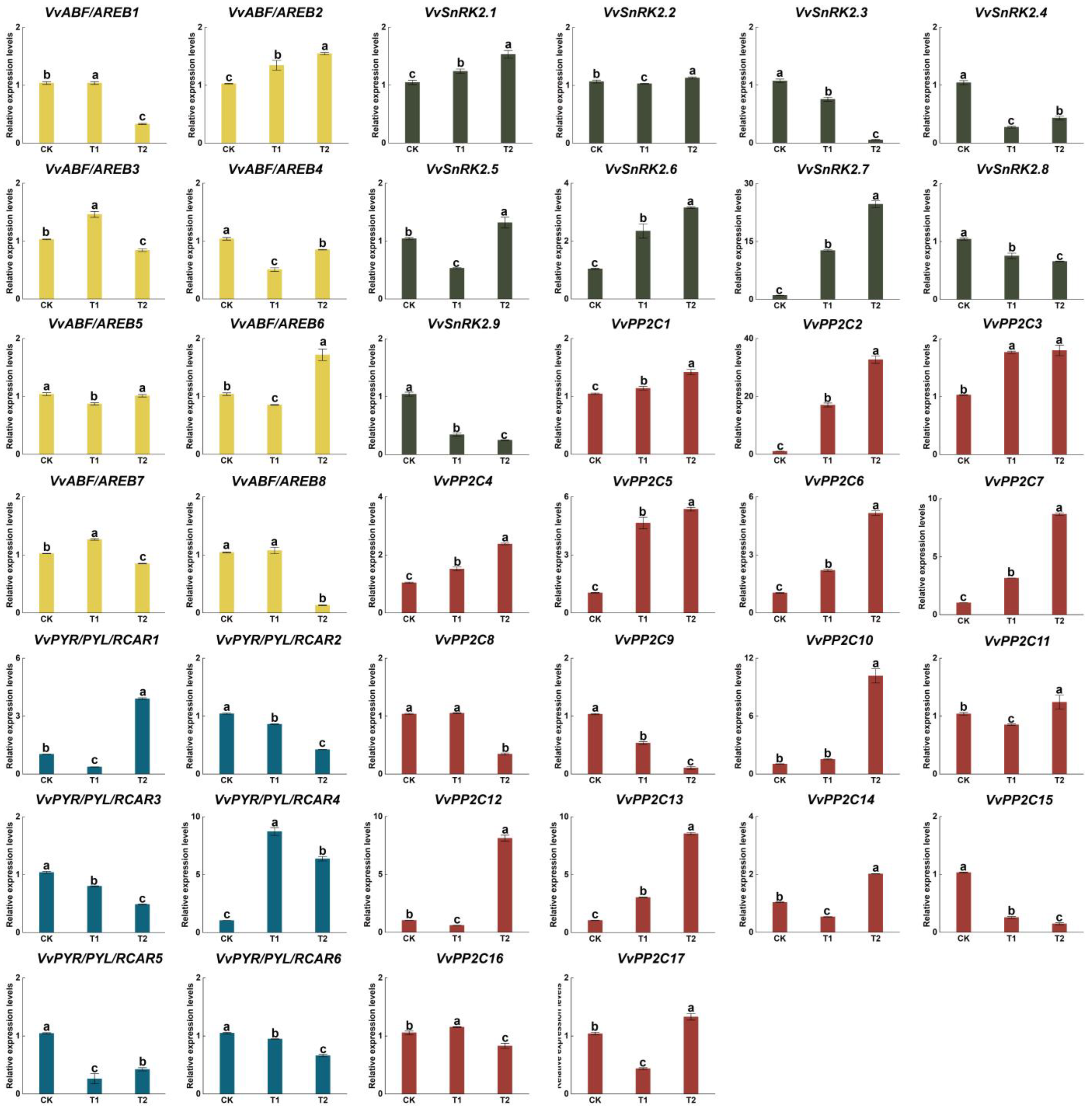
3.6. Expression Pattern of APGs under Low Light Treatments
3.7. Expression Pattern of APGs under ABA Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, B.; Zhang, H.; Jing, Q.; Wang, J. Light pollution on the growth, physiology and chlorophyll fluorescence response of landscape plant perennial ryegrass (Lolium perenne L.). Ecol. Indic. 2020, 115, 106448. [Google Scholar] [CrossRef]
- Velez-Ramirez, A.I.; van Ieperen, W.; Vreugdenhil, D.; Millenaar, F.F. Plants under continuous light. Trends Plant Sci. 2011, 16, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Y.; Qiao, Y.; Sun, H.; Liu, W.; Qiao, W.; Li, W.; Liu, M.; Dong, B. Low light stress promotes new tiller regeneration by changing source-sink relationship and activating expression of expansin genes in wheat. Plant Cell Environ. 2023, 46, 1562–1581. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Gu, X.; Lu, W.; Lu, D. Effects of weak-light stress during grain filling on the physicochemical properties of normal maize starch. Carbohydr. Polym. 2018, 202, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Li, N.J.; Shahid, M.; Zong, X.F.; Lv, J.; Wang, D.B.; Saleem, A.; Anjum, S.A.; Wang, S.G. Exogenously applied 5-aminolevulinic acid mediated physiochemical regulations ameliorate weak light stress in tobacco seedlings. Bangladesh J. Bot. 2019, 48, 353–358. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, W.; Yu, H. Effects of Exogenous Epibrassinolide on Photosynthetic Characteristics in Tomato (Lycopersicon esculentum Mill) Seedlings under Weak Light Stress. J. Agric. Food Chem. 2010, 58, 3642–3645. [Google Scholar] [CrossRef] [PubMed]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Shi, Y.; Ke, X.; Yang, X.; Liu, Y.; Hou, X. Plants response to light stress. J. Genet. Genom. 2022, 49, 735–747. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Jia, H.; Lu, C. Effects of abscisic acid on photoinhibition in maize plants. Plant Sci. 2003, 165, 1403–1410. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, C.; Yan, Y.; Tie, W.; Ding, Z.; Liu, G.; Yan, W.; Li, Y.; Wang, W.; Peng, M.; et al. Genomic analysis of the core components of ABA signaling reveals their possible role in abiotic stress response in cassava. Environ. Exp. Bot. 2019, 167, 103855. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins Struct. Funct. Bioinform. 2001, 43, 134–144. [Google Scholar] [CrossRef]
- Schweighofer, A.; Meskiene, I. Phosphatases in plants. Methods Mol. Biol. 2015, 1306, 25–46. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007, 12, 343–351. [Google Scholar] [CrossRef]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The Regulatory Domain of SRK2E/OST1/SnRK2.6 Interacts with ABI1 and Integrates Abscisic Acid (ABA) and Osmotic Stress Signals Controlling Stomatal Closure in Arabidopsis*. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3D structure analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.-H.; Hu, S.; Li, X.-B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, M.; Zhou, S.; Xu, B.; Chen, P.; Ma, F.; Mao, K. The ABA receptor gene MdPYL9 confers tolerance to drought stress in transgenic apple (Malus domestica). Environ. Exp. Bot. 2022, 194, 104695. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, T.; Lian, M.; Liu, T.; Hou, J.; Ijaz, R.; Song, B. Genome-Wide Identification and Characterization of the AREB/ABF/ABI5 Subfamily Members from Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 311. [Google Scholar] [CrossRef] [PubMed]
- Yong, X.; Zheng, T.; Zhuo, X.; Ahmad, S.; Li, L.; Li, P.; Yu, J.; Wang, J.; Cheng, T.; Zhang, Q. Genome-wide identification, characterisation, and evolution of ABF/AREB subfamily in nine Rosaceae species and expression analysis in mei (Prunus mume). PeerJ 2021, 9, e10785. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Li, S.; Yang, L.; Liang, Z. ABA signaling pathway genes and function during abiotic stress and berry ripening in Vitis vinifera. Gene 2021, 769, 145226. [Google Scholar] [CrossRef]
- Ren, C.; Kuang, Y.; Lin, Y.; Guo, Y.; Li, H.; Fan, P.; Li, S.; Liang, Z. Overexpression of grape ABA receptor gene VaPYL4 enhances tolerance to multiple abiotic stresses in Arabidopsis. BMC Plant Biol. 2022, 22, 271. [Google Scholar] [CrossRef]
- Nai, G.; Liang, G.; Ma, W.; Lu, S.; Li, Y.; Gou, H.; Guo, L.; Chen, B.; Mao, J. Overexpression VaPYL9 improves cold tolerance in tomato by regulating key genes in hormone signaling and antioxidant enzyme. BMC Plant Biol. 2022, 22, 344. [Google Scholar] [CrossRef]
- Xu, T.; Chen, T.C.; Zhang, T.Y.; Shen, L.Y.; Chen, Z.; Xu, Y.; Wu, Y.Y.; Yang, J. Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.). Horticulturae 2022, 8, 1024. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative Evolutionary Analysis of Chalcone Synthase and Alcohol Dehydrogenase Loci in Arabidopsis, Arabis, and Related Genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xu, T.; Shen, L.; Zhang, T.; Wang, L.; Chen, Z.; Wu, Y.; Yang, J. Effects of Girdling and Foliar Fertilization with K on Physicochemical Parameters, Phenolic and Volatile Composition in ‘Hanxiangmi’ Table Grape. Horticulturae 2022, 8, 388. [Google Scholar] [CrossRef]
- Gao, J. Experimental Guidance for Plant Physiology; Higer Education Press: Beijing, China, 2006. [Google Scholar]
- Xu, C.; Yagiz, Y.; Zhao, L.; Simonne, A.; Lu, J.; Marshall, M.R. Fruit quality, nutraceutical and antimicrobial properties of 58 muscadine grape varieties (Vitis rotundifolia Michx.) grown in United States. Food Chem. 2017, 215, 149–156. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guo, B.-B.; Li, J.-M.; Liu, X.; Qiao, X.; Musana, R.F.; Wang, P.; Zhang, S.-l.; Wu, J.-Y. Identification and expression analysis of the PbrMLO gene family in pear, and functional verification of PbrMLO23. J. Integr. Agric. 2021, 20, 2410–2423. [Google Scholar] [CrossRef]
- Xu, D.; Li, J.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; An, F.; Sun, Y.; Guo, R.; Pan, L.; Wan, T.; Hao, Y.; Cai, Y. Genome-wide identification of the ABA receptor PYL gene family and expression analysis in Prunus avium L. Sci. Hortic. 2023, 313, 111919. [Google Scholar] [CrossRef]
- Tao, X.; Chengjie, C.; Chuhao, L.; Jiarou, L.; Chaoyang, L.; Yehua, H.J.B.G. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar]
- Li, H.; Yue, M.; Jiang, L.; Liu, Y.; Zhang, N.; Liu, X.; Ye, Y.; Lin, X.; Zhang, Y.; Lin, Y.; et al. Genome-Wide Identification of Strawberry C2H2-ZFP C1-2i Subclass and the Potential Function of FaZAT10 in Abiotic Stress. Int. J. Mol. Sci. 2022, 23, 13079. [Google Scholar] [CrossRef]
- Xu, H.; Li, D.; Hao, Y.; Guo, X.; Lu, J.; Zhang, T. Genome-wide analysis of DGAT gene family in Perilla frutescens and functional characterization of PfDGAT2-2 and PfDGAT3-1 in Arabidopsis. Plant Sci. 2022, 324, 111426. [Google Scholar] [CrossRef]
- Safder, I.; Shao, G.; Sheng, Z.; Hu, P.; Tang, S. Identification and analysis of the structure, expression and nucleotide polymorphism of the GPAT gene family in rice. Plant Gene 2021, 26, 100290. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Jin, Z.; Yang, Z.; Li, Y. Leaf anatomy, photosynthesis, and chloroplast ultrastructure of Heptacodium miconioides seedlings reveal adaptation to light environment. Environ. Exp. Bot. 2022, 195, 104780. [Google Scholar] [CrossRef]
- Van Heerden, P.D.R.; KrÜGer, G.H.J.; Loveland, J.E.; Parry, M.A.J.; Foyer, C.H. Dark chilling imposes metabolic restrictions on photosynthesis in soybean: Dark chilling imposes restrictions on photosynthesis. Plant Cell Environ. 2003, 26, 323–337. [Google Scholar] [CrossRef]
- Li, H.; Bai, Y.; Yang, Y.; Zheng, H.; Xu, X.; Li, H.; Wang, W.; Tao, J. Transcriptomic analyses reveal light-regulated anthocyanin accumulation in ‘ZhongShan-HongYu’ grape berries. Sci. Hortic. 2023, 309, 111669. [Google Scholar] [CrossRef]
- Song, H.; Duan, Z.; Wang, Z.; Li, Y.; Wang, Y.; Li, C.; Mao, W.; Que, Q.; Chen, X.; Li, P. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crops Prod. 2022, 178, 114582. [Google Scholar] [CrossRef]
- Hu, L.; Wang, P.; Hao, Z.; Lu, Y.; Xue, G.; Cao, Z.; Qu, H.; Cheng, T.; Shi, J.; Chen, J. Gibberellin Oxidase Gene Family in L. chinense: Genome-Wide Identification and Gene Expression Analysis. Int. J. Mol. Sci. 2021, 22, 7167. [Google Scholar] [CrossRef] [PubMed]


| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplicated Type | Divergence-Time (Mya) |
|---|---|---|---|---|---|
| VvABF/AREB1/VvABF/AREB7 | 0.25 | 3.12 | 0.08 | Segmental | 104.12 |
| VvPP2C2/VvPP2C5 | 0.13 | 2.11 | 0.06 | Segmental | 70.34 |
| VvPP2C4/VvPP2C13 | 0.40 | 0.44 | 0.92 | Segmental | 14.55 |
| VvPP2C5/VvPP2C15 | 0.73 | 1.81 | 0.40 | Segmental | 60.39 |
| VvPP2C9/VvPP2C14 | 0.18 | 2.00 | 0.09 | Segmental | 66.75 |
| VvPP2C12/VvPP2C16 | 0.15 | 1.50 | 0.10 | Segmental | 50.19 |
| VvPP2C2/VvPP2C15 | 0.16 | 1.54 | 0.11 | Segmental | 51.41 |
| VvPYR/PYL/RCAR5/VvPYR/PYL/RCAR6 | 0.18 | 1.91 | 0.09 | Segmental | 63.75 |
| VvPYR/PYL/RCAR5/VvPYR/PYL/RCAR2 | 0.17 | 1.96 | 0.09 | Segmental | 65.27 |
| VvSnRK2.6/VvSnRK2.3 | 0.12 | 1.05 | 0.12 | Segmental | 34.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Zhang, M.; Chen, T.; Gong, L.; Hu, L.; Yang, J.; Si, H.; Wu, Y. Identification of ABA Signaling Pathway Genes and Their Differential Regulation in Response to Suboptimal Light Stress in Grape (Vitis vinifera L.). Horticulturae 2023, 9, 789. https://doi.org/10.3390/horticulturae9070789
Xu T, Zhang M, Chen T, Gong L, Hu L, Yang J, Si H, Wu Y. Identification of ABA Signaling Pathway Genes and Their Differential Regulation in Response to Suboptimal Light Stress in Grape (Vitis vinifera L.). Horticulturae. 2023; 9(7):789. https://doi.org/10.3390/horticulturae9070789
Chicago/Turabian StyleXu, Tao, Min Zhang, Tianchi Chen, Lili Gong, Lingling Hu, Jie Yang, Haoxuan Si, and Yueyan Wu. 2023. "Identification of ABA Signaling Pathway Genes and Their Differential Regulation in Response to Suboptimal Light Stress in Grape (Vitis vinifera L.)" Horticulturae 9, no. 7: 789. https://doi.org/10.3390/horticulturae9070789
APA StyleXu, T., Zhang, M., Chen, T., Gong, L., Hu, L., Yang, J., Si, H., & Wu, Y. (2023). Identification of ABA Signaling Pathway Genes and Their Differential Regulation in Response to Suboptimal Light Stress in Grape (Vitis vinifera L.). Horticulturae, 9(7), 789. https://doi.org/10.3390/horticulturae9070789





