Red-TE Homozygous Alleles of MdMYB10 Confer Full-Red Apple Fruit Skin in a High-Temperature Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genome and Transcriptome Sequencing and Analyses
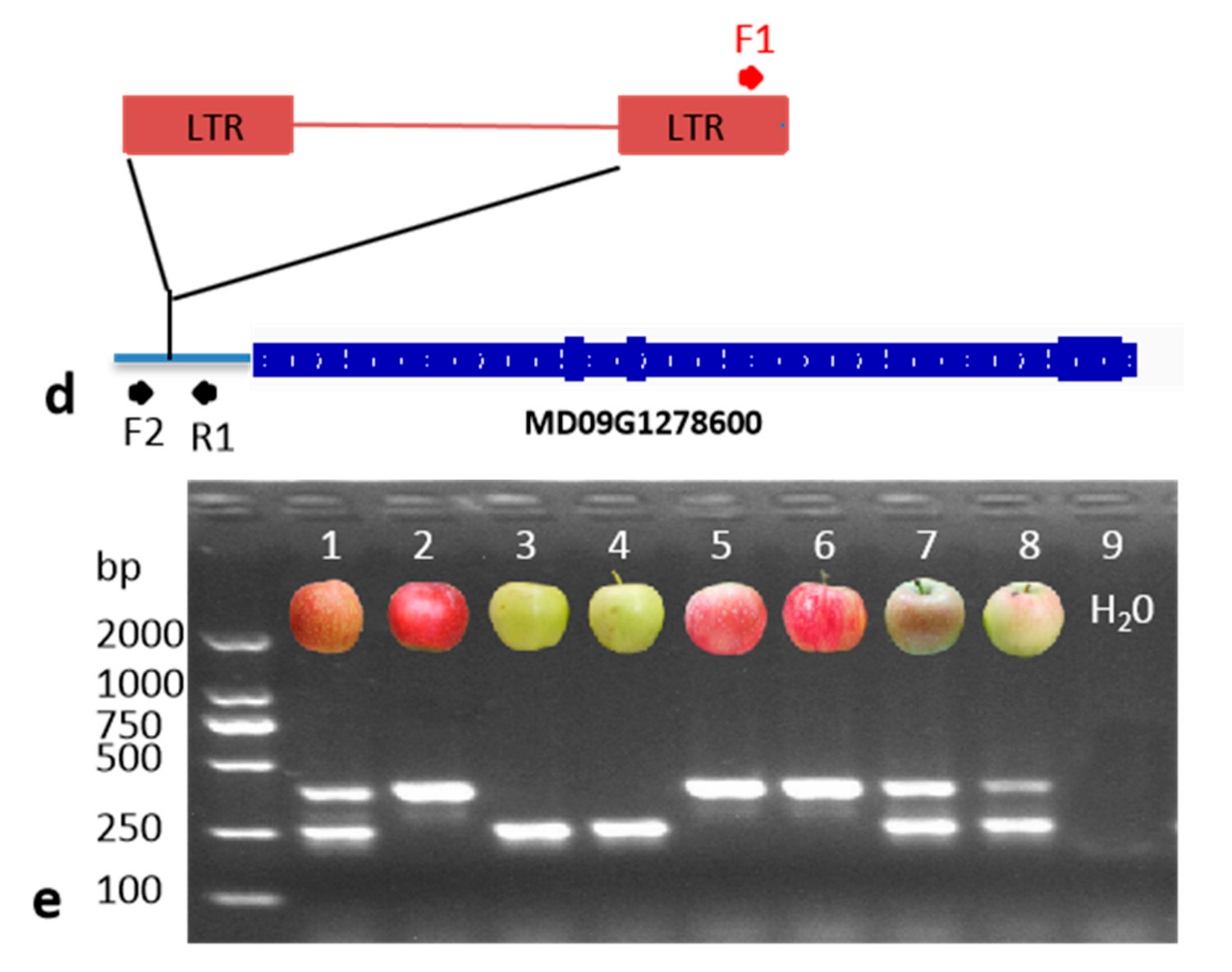
2.3. DNA Extraction and PCR Analysis
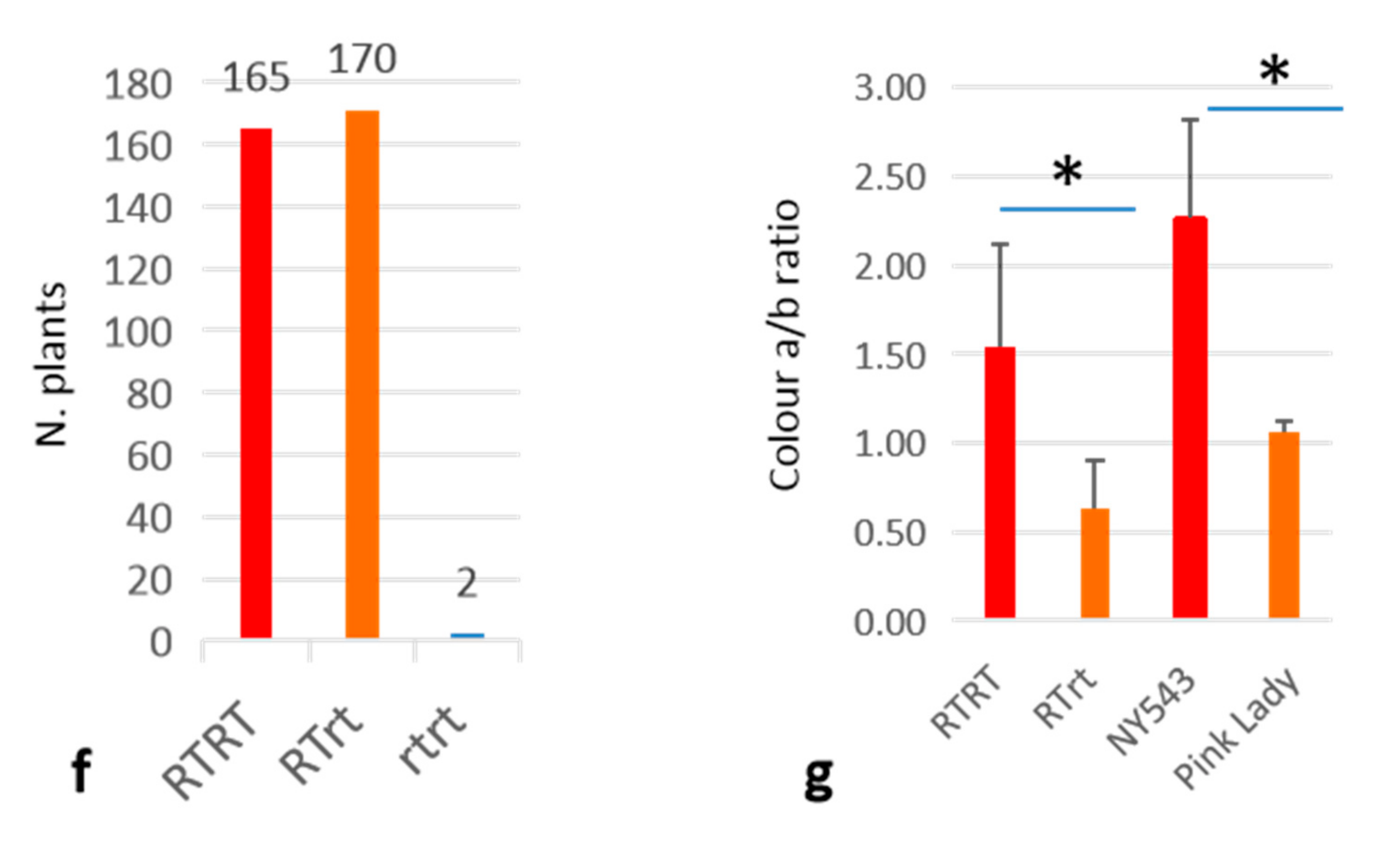
2.4. Measuring Apple Fruit Skin Color
3. Results
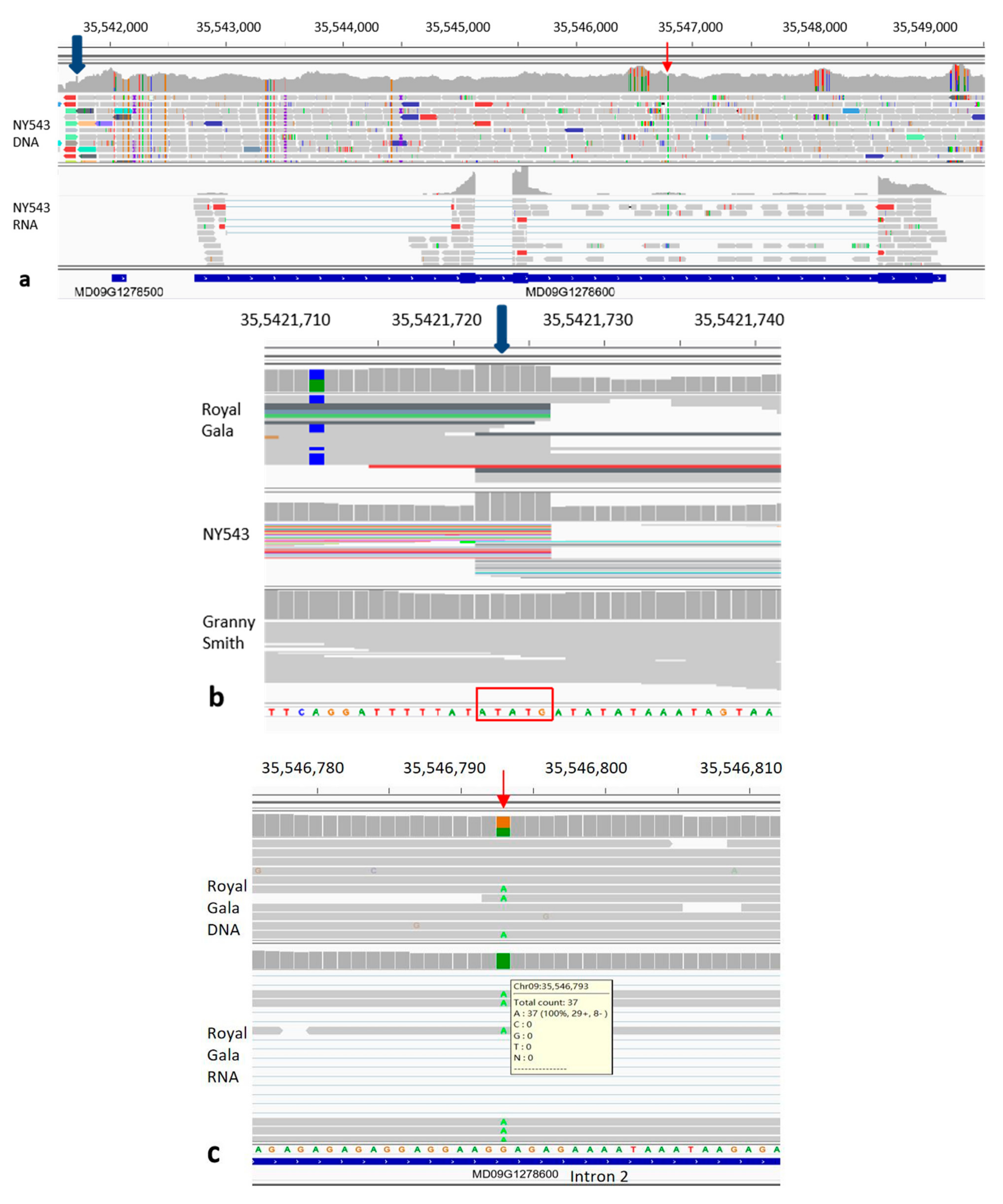
3.1. Allele-Specific Expression of MdMYB10 in Apple Fruit Skin
3.2. The Distribution of MdMYB10_Red-TE Allele in Apple Cultivars
3.3. Homozygous MYB10_Red-TE Allele Conferring Improved Red Fruit Skin Colour in High-Temperature Regions
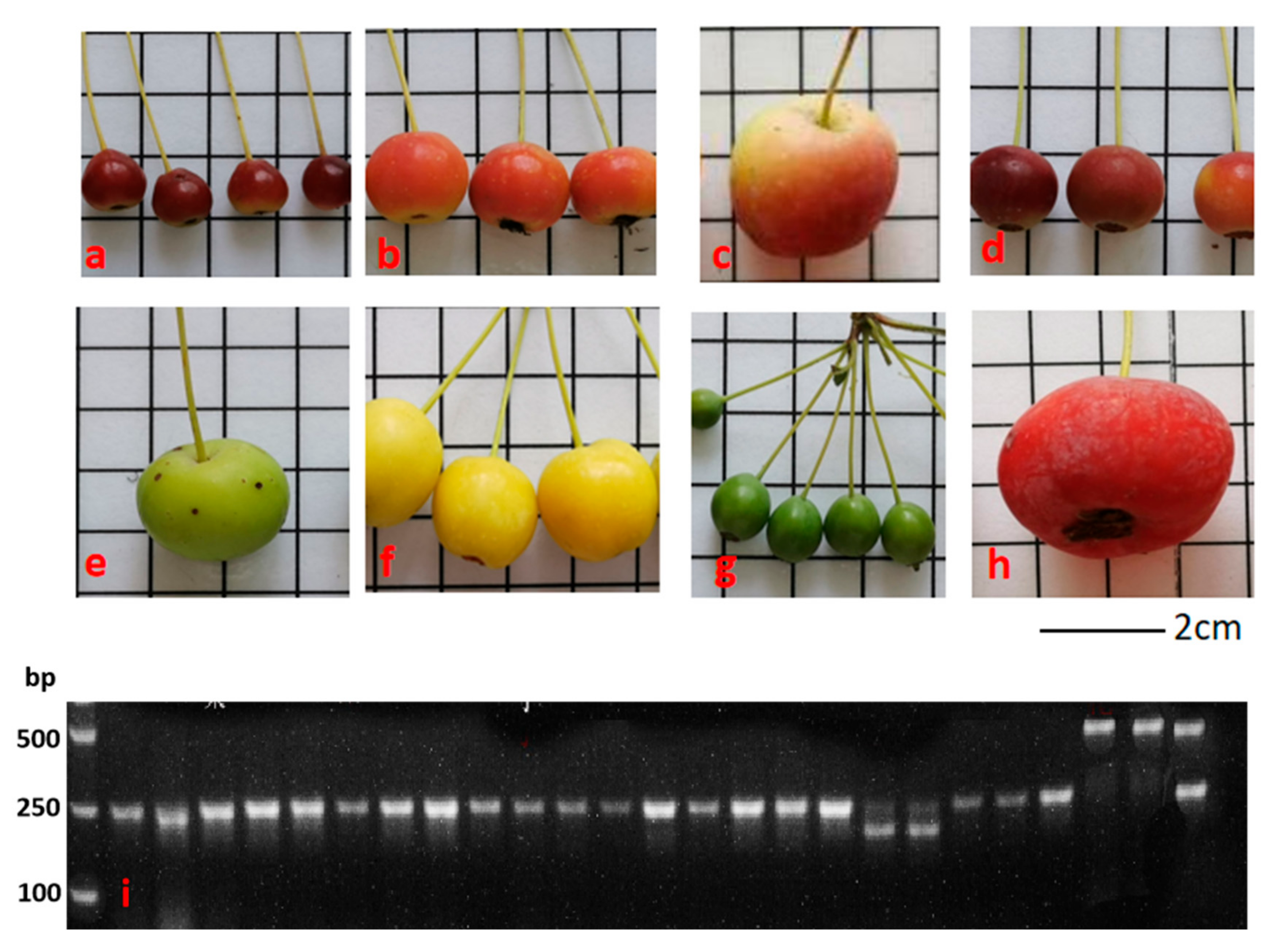
3.4. MYB10_Red-TE Allele Is Not Responsible for Red Fruit Skin in Small Wild Apples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dong, Z.; Song, S.; Song, C.; Zheng, X.; Jiao, J.; Wang, M.; Yan, Z.; Zhang, R.; Bai, T. Pedigree analysis and breeding inspiration of apple cultivars in China. Sci. Agric. Sin. 2020, 53, 4485–4496. [Google Scholar]
- de Pascual-Teresa, S.; Sanchez-Ballesta, M.T. Anthocyanins: From plant to health. Phytochem. Rev. 2008, 7, 281–299. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Espley, R.V. MYBs Drive Novel Consumer Traits in Fruits and Vegetables. Trends Plant Sci. 2018, 23, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 Ubiquitin E3 Ligases Interact with MdMYB1 to Regulate Light-Induced Anthocyanin Biosynthesis and Red Fruit Coloration in Apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Jing, C.; Chang, B.; Yan, J.; Liang, B.; Liu, L.; Yang, Y.; Zhao, Z. The effect of promoter methylation on MdMYB1 expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol. 2018, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Brendolise, C.; Chagne, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P.; et al. Multiple Repeats of a Promoter Segment Causes Transcription Factor Autoregulation in Red Apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Hu, J.; Han, X.L.; Li, J.J.; Gao, Y.; Richards, C.M.; Zhang, C.X.; Tian, Y.; Liu, G.M.; Gul, E.A.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Thrimawithana, A.; Ding, T.Y.; Guo, J.; Gleave, A.; Chagne, D.; Ampomah-Dwamena, C.; Ireland, H.S.; Schaffer, R.J.; Luo, Z.W.; et al. Transposon insertions regulate genome-wide allele-specific expression and underpin flower colour variations in apple (Malus spp.). Plant Biotechnol. J. 2022, 20, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Zhang, R.; Zhang, H.; Zhou, Z.; Liu, C.; Wu, M.; Wang, H.; Dong, H.; Liu, J.; Yao, J.-L.; et al. Identification of gene co-expression networks and key genes regulating flavonoid accumulation in apple (Malus × domestica) fruit skin. Plant Sci. 2021, 304, 110747. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Che, F.; Wang, L.X.; Meng, R.; Zhang, X.J.; Zhao, Z.Y. Fruit Coloration and Anthocyanin Biosynthesis after Bag Removal in Non-Red and Red Apples (Malus × domestica Borkh.). Molecules 2013, 18, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, X.-H.; Tong, L.; Liu, M.-Z.; Zhou, X.-K.; Tahir, M.M.; Xing, L.-B.; Ma, J.-J.; An, N.; Zhao, C.-P.; et al. Multi-omics analyses reveal MdMYB10 hypermethylation being responsible for a bud sport of apple fruit color. Hortic. Res. 2022, 9, uhac179. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.W.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.P.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.M.; Duan, N.B.; Khan, A.; Ban, S.; Xu, K.N.; Cheng, L.L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Cornille, A.; Gladieux, P.; Smulders, M.J.M.; Roldan-Ruiz, I.; Laurens, F.; Le Cam, B.; Nersesyan, A.; Clavel, J.; Olonova, M.; Feugey, L.; et al. New insight into the history of domesticated apple: Secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet. 2012, 8, e1002703. [Google Scholar] [CrossRef] [PubMed]
- Cornille, A.; Gladieux, P.; Giraud, T. Crop-to-wild gene flow and spatial genetic structure in the closest wild relatives of the cultivated apple. Evol. Appl. 2013, 6, 737–748. [Google Scholar] [CrossRef] [PubMed]

| Cultivar Name | Fruit Skin Colour | MdMYB10 Genotype | a/b Ratio | |
|---|---|---|---|---|
| Mean | SD | |||
| Jingqing | Yellow | rtrt | −0.114 | 0.424 |
| Ozark Gold | Yellow | rtrt | −0.210 | 0.111 |
| Gold Spur | Yellow | rtrt | −0.228 | 0.042 |
| Cox’s Orange Pippin 363 | Yellow | rtrt | −0.263 | 0.045 |
| Beidou | Yellow | rtrt | −0.308 | 0.085 |
| Winter Banana | Yellow | rtrt | −0.341 | 0.063 |
| Yuguan | Yellow | rtrt | −0.422 | 0.051 |
| Jingyuan | Yellow | rtrt | −0.427 | 0.006 |
| Mosiketouming | Yellow | rtrt | −0.456 | 0.068 |
| Granny Smith | Green | rtrt | ||
| Indo | Green | rtrt | −0.600 | 0.029 |
| Regent | Red | RTrt | 1.296 | 0.500 |
| Royal Gala | Red | RTrt | ||
| Red 210 | Red | RTrt | 1.274 | 0.211 |
| Pink Lady | Red | RTrt | 1.102 | 0.114 |
| Summerland | Red | RTrt | 0.985 | 0.100 |
| Close | Red | RTrt | 0.968 | 0.395 |
| Fuli | Red | RTrt | 0.871 | 0.298 |
| De No.6 | Red | RTrt | 0.861 | 0.327 |
| Starking Delicious | Red | RTrt | 0.850 | 0.228 |
| Monroe | Red | RTrt | 0.785 | 0.135 |
| Huamei | Red | RTrt | 0.718 | 0.265 |
| Black Ben Davis | Red | RTrt | 0.709 | 0.168 |
| Fuji Spur | Red | RTrt | 0.695 | 0.123 |
| Yuejin | Red | RTrt | 0.680 | 0.259 |
| Starkrimson | Red | RTrt | 0.646 | 0.103 |
| Hardi Brite | Red | RTrt | 0.638 | 0.300 |
| Gongqiduanfu | Red | RTrt | 0.610 | 0.170 |
| Rome Beauty | Red | RTrt | 0.577 | 0.275 |
| Ningqiu | Red | RTrt | 0.556 | 0.417 |
| Senshu | Red | RTrt | 0.470 | 0.370 |
| Kogetsu | Red | RTrt | 0.468 | 0.069 |
| Braeburn | Red | RTrt | 0.453 | 0.230 |
| Fameuse | Red | RTrt | 0.394 | 0.370 |
| Red Jonagold | Red | RTrt | 0.300 | 0.130 |
| Ace | Red | RTrt | 0.215 | 0.210 |
| Anglin | Red | RTrt | 0.194 | 0.125 |
| Discovery | Red | RTrt | 0.172 | 0.003 |
| Yanfeng | Red | RTrt | 0.164 | 0.097 |
| Hongxi Jinqing | Red | RTrt | 0.049 | 0.002 |
| GS-58 | Red | RTRT | 1.854 | 0.177 |
| NY543 | Red | RTRT | 2.240 | 0.479 |
| Liberty | Red | RTRT | 1.815 | 0.679 |
| Mollie’s | Red | RTRT | 1.423 | 0.726 |
| Ralls | Red | RTRT | 1.192 | 0.298 |
| Bella | Red | RTRT | 1.055 | 0.343 |
| Luxiang | Red | RTRT | 0.996 | 0.467 |
| Cox’s Orange Pippin 312 | Red | RTRT | 0.980 | 0.311 |
| Meize | Red | RTRT | 0.948 | 0.111 |
| Hongbaoshi | Red | RTRT | 0.888 | 0.388 |
| Idared | Red | RTRT | 0.857 | 0.585 |
| Changfu 2 | Red | RTRT | 0.780 | 0.403 |
| Longjinmi | Red | RTRT | 0.754 | 0.195 |
| Huadan | Red | RTRT | 0.690 | 0.426 |
| Xinshijie | Red | RTRT | 0.480 | 0.131 |
| Huayu | Red | RTRT | 0.240 | 0.287 |
| Zhongqiu | Red | RTRT | 0.112 | 0.207 |
| Gloster | Red | rtrt | 1.224 | 0.276 |
| Summer Champion | Red | rtrt | 1.109 | 0.419 |
| Xinlimei | Red | rtrt | 0.880 | 0.553 |
| Hongjin | Red | rtrt | 0.805 | 0.458 |
| Kuihua | Red | rtrt | 0.570 | 0.097 |
| Iowa Beauty | Yellow | RTrt | 0.025 | 0.199 |
| Tianyu | Yellow | RTrt | −0.262 | 0.003 |
| Grimes Golden | Yellow | RTrt | −0.398 | 0.070 |
| Accession Name | Species Name | Fruit Skin Colour | MdMYB10 Genotype | a/b Ratio | |
|---|---|---|---|---|---|
| Mean | SD | ||||
| Lijiangshandingzi | M. rockii | Red | rtrt | 1.534 | 0.529 |
| Yingyehaitang-1 | M. prunifolia | Red | rtrt | 1.024 | 0.326 |
| Shanxihaitanghua(zz) | M. robusta | Red | RTRT | 2.192 | 0.135 |
| Hongsanyehaitang | M. sieboldii | Red | rtrt | 1.382 | 0.602 |
| Sanyehaitang | M. sieboldii | green | rtrt | −0.522 | 0.032 |
| Dongbeihuanghaitang | M. honanensis | Yellow | rtrt | −0.064 | 0.067 |
| Lushihongguo | M. honanensis | Green | rtrt | −0.629 | 0.039 |
| Baodehaihong(zz) | M. micromalus | Red | rtrt | 3.832 | 0.376 |
| Shandingzi | M. baccata | Red | rtrt | 2.830 | 0.690 |
| Maoshandingzi | M. manshurica | Red | rtrt | 2.680 | 0.455 |
| Jinxibeishandingzi-1 | M. baccata | Red | rtrt | 2.466 | 0.627 |
| Jixibeishandingzi-2 | M. baccata | Red | rtrt | 2.197 | 0.497 |
| Wushanbianyehaitang | M. toringoides | Red | rtrt | 1.914 | 0.521 |
| Maoshandingzi-1(zz) | M. manshurica | Red | rtrt | 1.903 | 0.537 |
| Hongshijie(zz) | unknown | Red | rtrt | 1.864 | 0.080 |
| Hongxunyihao(zz) | M. sieversii | Red | rtrt | 1.627 | 0.276 |
| Yingyehaitang-2(zz) | M. prunifolia | Red | RTRT | 0.843 | 0.075 |
| Binzi | M. asiatica | Red | RTrt | 0.833 | 0.230 |
| Gansuhaitang | M. kansuensis | Red | rtrt | 0.761 | 0.271 |
| Xiaojinhaitang | M. xiaojinensis | Red | rtrt | 0.491 | 0.658 |
| Zhaai-76 | M. baccata | Red | rtrt | 0.415 | 0.231 |
| Maoshandingzi-2(zz) | M. manshurica | Orange | rtrt | 0.357 | 0.165 |
| Duohuahaitang | M. floribunda | Red | rtrt | −0.043 | 0.207 |
| pingyitiancha | M. hupehensis | Yellow | rtrt | −0.156 | 0.174 |
| Hubeihaitang | M. hupehensis | Yellow | rtrt | −0.338 | 0.287 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wang, Y.; Ding, T.; Yan, Z.; Zhou, Z.; Li, C.; Yao, J.-L.; Zhang, H. Red-TE Homozygous Alleles of MdMYB10 Confer Full-Red Apple Fruit Skin in a High-Temperature Region. Horticulturae 2023, 9, 270. https://doi.org/10.3390/horticulturae9020270
Wang M, Wang Y, Ding T, Yan Z, Zhou Z, Li C, Yao J-L, Zhang H. Red-TE Homozygous Alleles of MdMYB10 Confer Full-Red Apple Fruit Skin in a High-Temperature Region. Horticulturae. 2023; 9(2):270. https://doi.org/10.3390/horticulturae9020270
Chicago/Turabian StyleWang, Meili, Yarong Wang, Tiyu Ding, Zhenli Yan, Zhe Zhou, Cuiying Li, Jia-Long Yao, and Hengtao Zhang. 2023. "Red-TE Homozygous Alleles of MdMYB10 Confer Full-Red Apple Fruit Skin in a High-Temperature Region" Horticulturae 9, no. 2: 270. https://doi.org/10.3390/horticulturae9020270
APA StyleWang, M., Wang, Y., Ding, T., Yan, Z., Zhou, Z., Li, C., Yao, J.-L., & Zhang, H. (2023). Red-TE Homozygous Alleles of MdMYB10 Confer Full-Red Apple Fruit Skin in a High-Temperature Region. Horticulturae, 9(2), 270. https://doi.org/10.3390/horticulturae9020270








