Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Growth Conditions
2.3. Analysis of Anthocyanin Content
2.3.1. Analysis of Anthocyanin Content by Colorimetric Method
2.3.2. HPLC–MS/MS Analyses
2.4. Fluorescence In Situ Hybridization (FISH)
2.4.1. Chromosome Preparation
2.4.2. DNA Probe Preparation
2.4.3. FISH
2.4.4. Image Processing
2.5. Real-Time Quantitative PCR (RT-qPCR)
2.6. Genotype Identification
2.7. Yeast-One Hybridization
3. Results
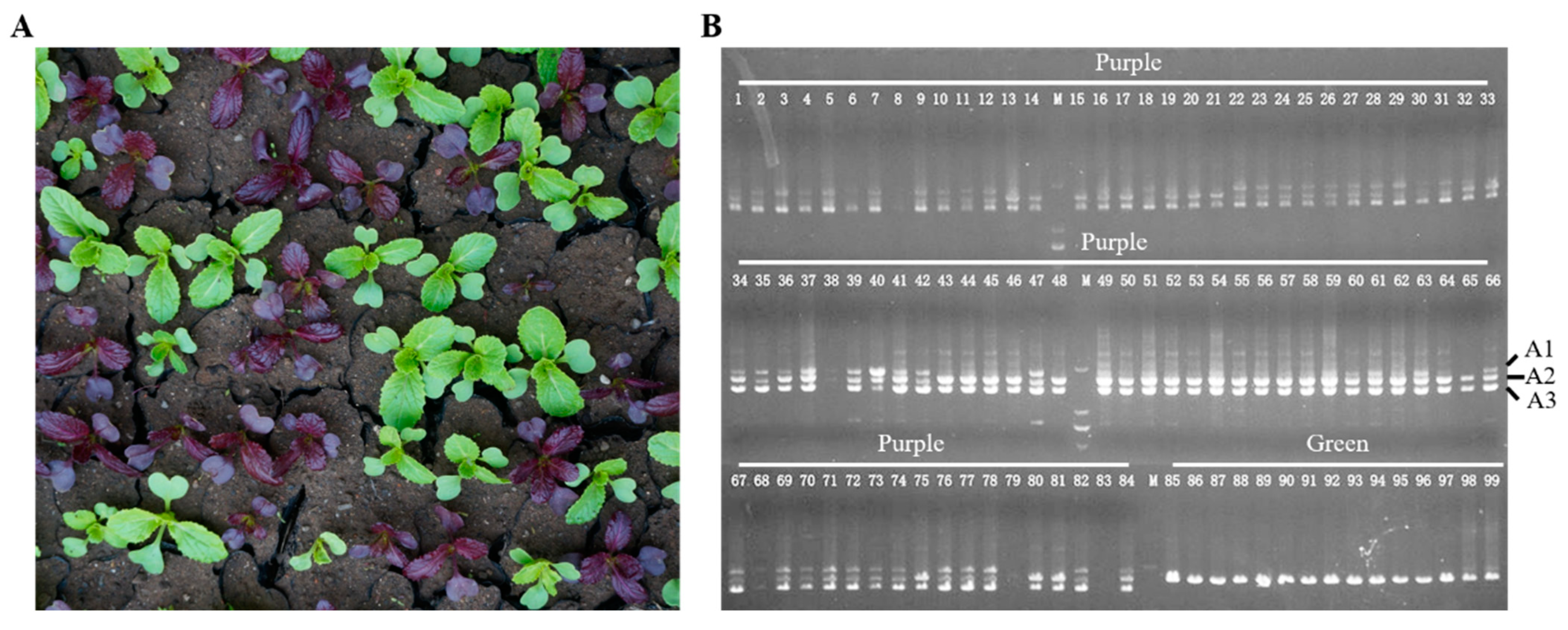
3.1. Generation of a New Purple Chinese Cabbage Germplasm
3.2. Anthocyanin Content in the 18M-245 Leaves
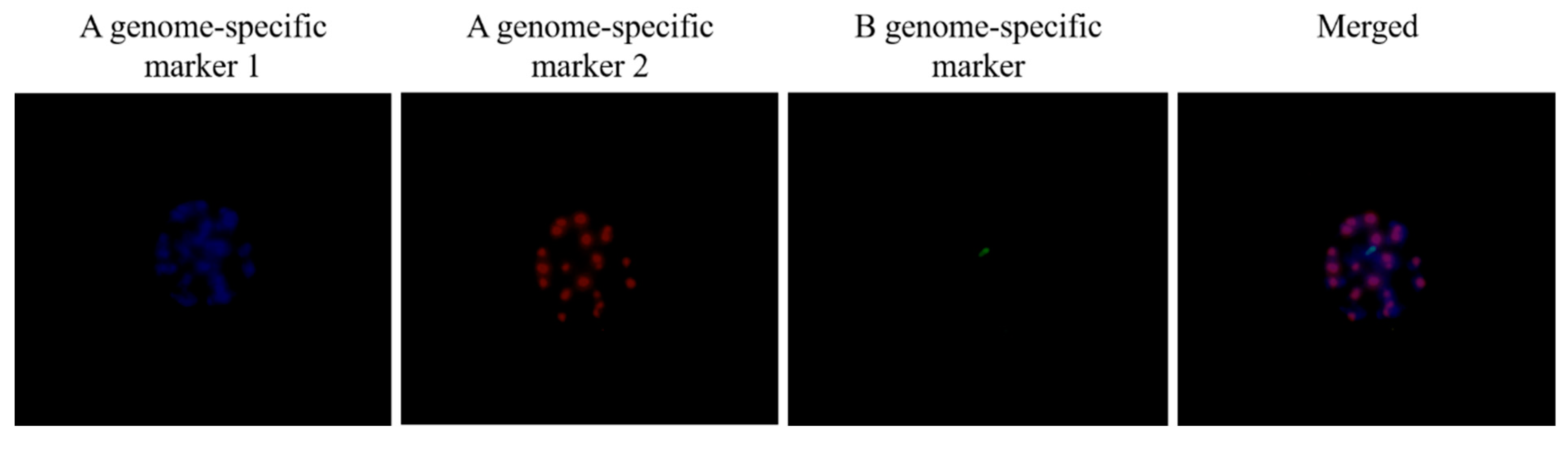
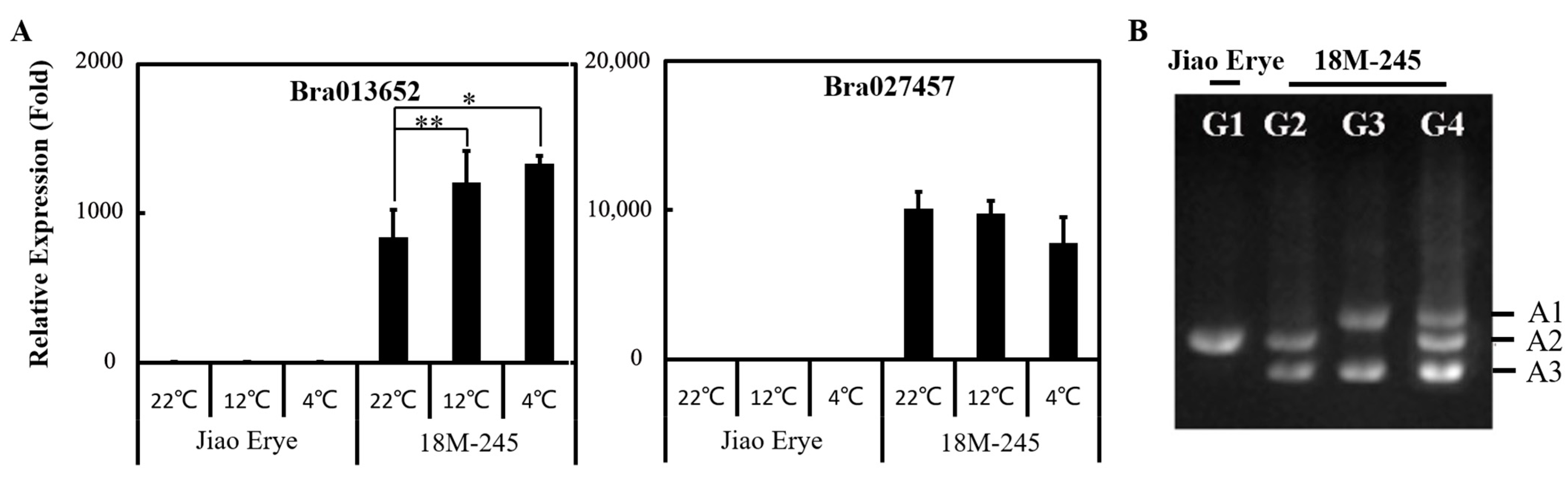
3.3. Identification of a Monosomic Alien Chromosome Addition Line in 18M-245
3.4. LDOX Transferred from the Brassica B Genome Is Highly Expressed in 18M-245
3.5. BjuB014115 Is Necessary for the Purple Color Trait in 18M-245
3.6. BjuB014115 Is Regulated by MYB and bHLH Transcription Factors from the Brassica A Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001, 127, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, M.K.; Burbulis, I.E.; Winkel-Shirley, B. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol. Biol. 1999, 40, 45–54. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Zhou, H.; Lin-Wang, K.; Wang, F.; Espley, R.V.; Ren, F.; Zhao, J.; Ogutu, C.; He, H.; Jiang, Q.; Allan, A.C.; et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019, 221, 1919–1934. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. Cell Mol. Biol. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- Feyissa, D.N.; Løvdal, T.; Olsen, K.M.; Slimestad, R.; Lillo, C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 2009, 230, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. Cell Mol. Biol. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Hayashi, K.; Matsumoto, S.; Tsukazaki, H.; Kondo, T.; Kubo, N.; Hirai, M. Mapping of a novel locus regulating anthocyanin pigmentation in Brassica rapa. Breed. Sci. 2010, 60, 76–80. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, D.; Yu, S.; Liu, J.; Wang, D.; Zhang, F.; Yu, Y.; Zhao, X.; Lu, G.; Su, T. Mapping the BrPur gene for purple leaf color on linkage group A03 of Brassica rapa. Euphytica 2014, 199, 293–302. [Google Scholar] [CrossRef]
- He, Q.; Wu, J.; Xue, Y.; Zhao, W.; Li, R.; Zhang, L. The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hortic. Res. 2020, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Wu, J.; Zheng, S.; Cheng, F.; Liu, B.; Liang, J.; Cui, Y.; Wang, X. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). Mol. Breed. 2015, 35, 113. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, K.; Wu, J.; Guo, N.; Liang, J.; Wang, X.; Cheng, F. QTL-Seq and sequence assembly rapidly mapped the gene BrMYBL2.1 for the purple trait in Brassica rapa. Sci. Rep. 2020, 10, 2328. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zhang, S.; Zhang, S.; Fei, L. Research on creation of Purple Chinese Cabbage germplasm. Acta Hortic. Sin. 2006, 33, 1032. [Google Scholar]
- Zhang, M.K.; Zhang, L.G.; Gong, Z.H.; Hui, M.X. Screening RAPD markers linked to purple trait of Chinese Cabbage and its chromosome location. Acta Bot. Boreali-Occident. Sin. 2008, 7, 82–88. [Google Scholar]
- Lee, H.; Oh, I.-N.; Kim, J.; Jung, D.; Cuong, N.P.; Kim, Y.; Lee, J.; Kwon, O.; Park, S.U.; Lim, Y.; et al. Phenolic compound profiles and their seasonal variations in new red-phenotype head-forming Chinese cabbages. LWT 2018, 90, 433–439. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Choi, S.R.; Chhapekar, S.S.; Kim, M.S.; Singh, S.; Yi, S.Y.; Oh, S.H.; Kim, H.; Lee, C.Y.; Oh, M.H.; et al. Red Chinese cabbage transcriptome analysis reveals structural genes and multiple transcription factors regulating reddish purple color. Int. J. Mol. Sci. 2020, 21, 2901. [Google Scholar] [CrossRef]
- Xie, L.; Li, F.; Zhang, S.; Zhang, H.; Qian, W.; Li, P.; Zhang, S.; Sun, R. Mining for candidate genes in an introgression line by using RNA sequencing: The anthocyanin overaccumulation phenotype in Brassica. Front. Plant Sci. 2016, 7, 1245. [Google Scholar] [CrossRef]
- Zhou, R.N.; Shi, R.; Jiang, S.M.; Yin, W.B.; Wang, H.H.; Chen, Y.H.; Hu, J.; Wang, R.R.; Zhang, X.Q.; Hu, Z.M. Rapid EST isolation from chromosome 1R of rye. BMC Plant Biol. 2008, 8, 28. [Google Scholar] [CrossRef]
- Jiang, S.M.; Yin, W.B.; Hu, J.; Shi, R.; Zhou, R.N.; Chen, Y.H.; Zhou, G.H.; Wang, R.R.; Song, L.Y.; Hu, Z.M. Isolation of expressed sequences from a specific chromosome of Thinopyrum intermedium infected by BYDV. Genome 2009, 52, 68–76. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef]
- Lei, L.; Li, Y.; Wang, Q.; Xu, J.; Chen, Y.; Yang, H.; Ren, D. Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 2014, 203, 1146–1160. [Google Scholar] [CrossRef]
- Guzzo, F.; Campagnari, E.; Levi, M. A new FISH protocol with increased sensitivity for physical mapping with short probes in plants. J. Exp. Bot. 2000, 51, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, H.; Zhang, F.; Yangjun, Y.U.; Zhao, X.; Shuancang, Y.U.; Wang, W.; Tongbing, S.U.; Guixiang, L.U. Identification and RNA-seq of new Purple Chinese Cabbage 15NG28 progenies. Acta Agric. Boreali-Sin. 2017, 6, 14–24. [Google Scholar]
- Yang, T.; Ma, H.; Zhang, J.; Wu, T.; Song, T.; Tian, J.; Yao, Y. Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit. Plant J. 2019, 100, 572–590. [Google Scholar] [CrossRef]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Liu, C.; Jun, J.H.; Dixon, R.A. MYB5 and MYB14 Play Pivotal Roles in Seed Coat Polymer Biosynthesis in Medicago truncatula. Plant Physiol. 2014, 165, 1424–1439. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Fan, K.; Li, Z.; Jia, Q.; Lin, W.; Zhang, Y. Phytochrome-interacting factor 4 (PIF4) negatively regulates anthocyanin accumulation by inhibiting PAP1 transcription in Arabidopsis seedlings. Plant Sci. 2021, 303, 110788. [Google Scholar] [CrossRef]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Fang, H.; Wang, Y.; Zhang, J.; Zhang, Z.; Wang, N.; Chen, X. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 72. [Google Scholar] [CrossRef]
- Lelivelt, C.L.C.; Lange, W.; Dolstra, O. Intergeneric crosses for the transfer of resistance to the beet cyst nematode from Raphanus sativus to Brassica napus. Euphytica 1993, 68, 111–120. [Google Scholar] [CrossRef]
- Zhang, S.; Li, P.; Qian, W.; Zhang, S.; Li, F.; Zhang, H.; Wang, X.; Sun, R. Mapping and expression profiling reveal an inserted fragment from purple mustard involved anthocyanin accumulation in Chinese cabbage. Euphytica 2016, 212, 83–95. [Google Scholar] [CrossRef]
- Gould, K.S. Nature’s swiss army knife: The diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 2004, 314–320. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, L.J.; Gao, H.N.; Wang, X.F.; Li, Z.W.; Li, Y.Y. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 2022, 189, 2044–2060. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. Cell Mol. Biol. 2008, 53, 814–827. [Google Scholar] [CrossRef]



| Class | Compound | Content (ng/gDW) | Fold Change (18M-245 vs. Jiao Erye) | p Value | |
|---|---|---|---|---|---|
| Jiao Erye | 18M-245 | ||||
| Cyanidin | Cyanidin-3-O-glucoside | 31.88 | 26.39 | 0.83 | 0.702 |
| Cyanidin | Cyanidin-3,5,3’-O-triglucoside | 0.01 | 17.81 | 1191.97 | 0.005 |
| Cyanidin | Cyanidin-3-O-(6-O-p-coumaroyl)-glucoside | 0.00 | 5.95 | 0.148 | |
| Cyanidin | Cyanidin-3-O-sophoroside | 0.00 | 3.29 | 0.003 | |
| Cyanidin | Cyanidin-3,5-O-diglucoside | 0.00 | 2.47 | 0.001 | |
| Cyanidin | Cyanidin-3-(6′′-caffeylsophoroside)-5-glucoside | 0.00 | 1.19 | 0.002 | |
| Cyanidin | Cyanidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 0.00 | 0.99 | 0.019 | |
| Cyanidin | Cyanidin-3-(6-O-p-caffeoyl)-glucoside | 0.70 | 0.81 | 0.817 | |
| Cyanidin | Cyanidin-3-O-sambubioside-5-O-glucoside | 0.00 | 0.03 | 0.022 | |
| Cyanidin | Cyanidin-3-O-xyloside | 0.00 | 0.02 | 0.048 | |
| Cyanidin | Cyanidin-3-O-(6′′-ferulylsophoroside)-5-glucoside | 0.01 | 0.01 | 1.16 | 0.922 |
| Cyanidin | Cyanidin-3-O-sambubioside | 0.00 | 0.01 | 0.012 | |
| Pelargonidin | Pelargonidin-3-sophoroside-5-glucoside | 0.00 | 0.05 | 0.009 | |
| Pelargonidin | Pelargonidin-3-O-galactoside | 0.29 | 0.04 | 0.14 | 0.167 |
| Pelargonidin | Pelargonidin-3-O-glucoside | 0.00 | 0.03 | 0.007 | |
| Pelargonidin | Pelargonidin-3-O-sophoroside | 0.00 | 0.03 | 0.010 | |
| Pelargonidin | Pelargonidin-3-O-sophoroside-5-O-(malonyl)-glucoside | 0.00 | 0.02 | 0.040 | |
| Pelargonidin | Pelargonidin-3-O-(6-O-p-coumaroyl)-glucoside | 0.00 | 0.01 | 0.324 | |
| Pelargonidin | Pelargonidin-3-O-rutinoside | 0.01 | 0.00 | 0.15 | 0.165 |
| Peonidin | Peonidin-3,5-O-diglucoside | 0.01 | 0.06 | 4.44 | 0.018 |
| Peonidin | Peonidin-3-O-glucoside | 0.00 | 0.05 | 0.013 | |
| Peonidin | Peonidin-3-sophoroside-5-glucoside | 0.00 | 0.05 | 0.023 | |
| Peonidin | Peonidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 0.00 | 0.02 | 0.023 | |
| Peonidin | Peonidin-3-O-(6-O-p-coumaroyl)-glucoside | 0.00 | 0.01 | 0.015 | |
| Peonidin | Peonidin-3-(caffeoyl-glucosyl-glucoside)-5-glucoside | 0.00 | 0.00 | 0.044 | |
| Delphinidin | Delphinidin-3-O-sophoroside | 0.66 | 0.54 | 0.82 | 0.832 |
| Delphinidin | Delphinidin-3,5-O-diglucoside | 0.08 | 0.12 | 1.41 | 0.649 |
| Delphinidin | Delphinidin-3-O-(6-O-malonyl-beta-D-glucoside) | 0.00 | 0.04 | 8.78 | 0.088 |
| Delphinidin | Delphinidin-3-O-sambubioside | 0.00 | 0.01 | 0.016 | |
| Petunidin | Petunidin-3-O-(6-O-malonyl-beta-D-glucoside) | 0.03 | 0.04 | 1.29 | 0.763 |
| Petunidin | Petunidin-3-O-glucoside | 0.00 | 0.01 | 0.218 | |
| Petunidin | Petunidin-3-O-sambubioside | 0.00 | 0.00 | 0.00 | |
| Malvidin | Malvidin-3-O-(6-O-malonyl-beta-D-glucoside) | 0.00 | 0.05 | Inf | 0.012 |
| Malvidin | Malvidin | 0.00 | 0.00 | 0.23 | 0.525 |
| flavonoid | Kaempferol-3-O-rutinoside | 0.00 | 18.12 | 0.020 | |
| flavonoid | Quercetin-3-O-glucoside | 5.63 | 2.23 | 0.40 | 0.539 |
| flavonoid | Naringenin-7-O-glucoside | 0.21 | 0.13 | 0.65 | 0.760 |
| Gene ID | Gene Name | Green Reads Count | Purple Reads Count | log2Ratio (Green/Purple) | p-Value | FDR |
|---|---|---|---|---|---|---|
| Bra027796 | BrEGL3 | 90 | 176 | −1.06819 | 0.457398 | 1 |
| Bra037887 | BrTTG8 | 3 | 4408 | −10.5618 | 6.57 × 10−7 | 0.00207 |
| Bra009770 | BtTTG1 | 528 | 1069 | −1.11916 | 0.427705 | 1 |
| Bra007957 | BrMYBL2-1 | 4 | 463 | −6.911 | 0.000222 | 0.038438 |
| Bra016164 | BrMYBL2-2 | 45 | 2875 | −6.0951 | 0.000392 | 0.055515 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, X.; Zhang, D.; Zhao, H.; Su, T.; Zhao, X.; Wang, W.; Li, P.; Yu, Y.; Wang, J.; Yu, S.; et al. Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage. Horticulturae 2023, 9, 146. https://doi.org/10.3390/horticulturae9020146
Xin X, Zhang D, Zhao H, Su T, Zhao X, Wang W, Li P, Yu Y, Wang J, Yu S, et al. Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage. Horticulturae. 2023; 9(2):146. https://doi.org/10.3390/horticulturae9020146
Chicago/Turabian StyleXin, Xiaoyun, Deshuang Zhang, Hong Zhao, Tongbing Su, Xiuyun Zhao, Weihong Wang, Peirong Li, Yangjun Yu, Jiao Wang, Shuancang Yu, and et al. 2023. "Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage" Horticulturae 9, no. 2: 146. https://doi.org/10.3390/horticulturae9020146
APA StyleXin, X., Zhang, D., Zhao, H., Su, T., Zhao, X., Wang, W., Li, P., Yu, Y., Wang, J., Yu, S., & Zhang, F. (2023). Identification of a Monosomic Alien Chromosome Addition Line Responsible for the Purple Color Trait in Heading Chinese Cabbage. Horticulturae, 9(2), 146. https://doi.org/10.3390/horticulturae9020146





