Morphological and Molecular Identification of Dactylonectria macrodidyma as Causal Agent of a Severe Prunus lusitanica Dieback in Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. Morphological Observations
2.3. DNA Extraction, Amplification and Sequencing
2.4. Phylogenetic Analysis
2.5. Pathogenicity Test
3. Results
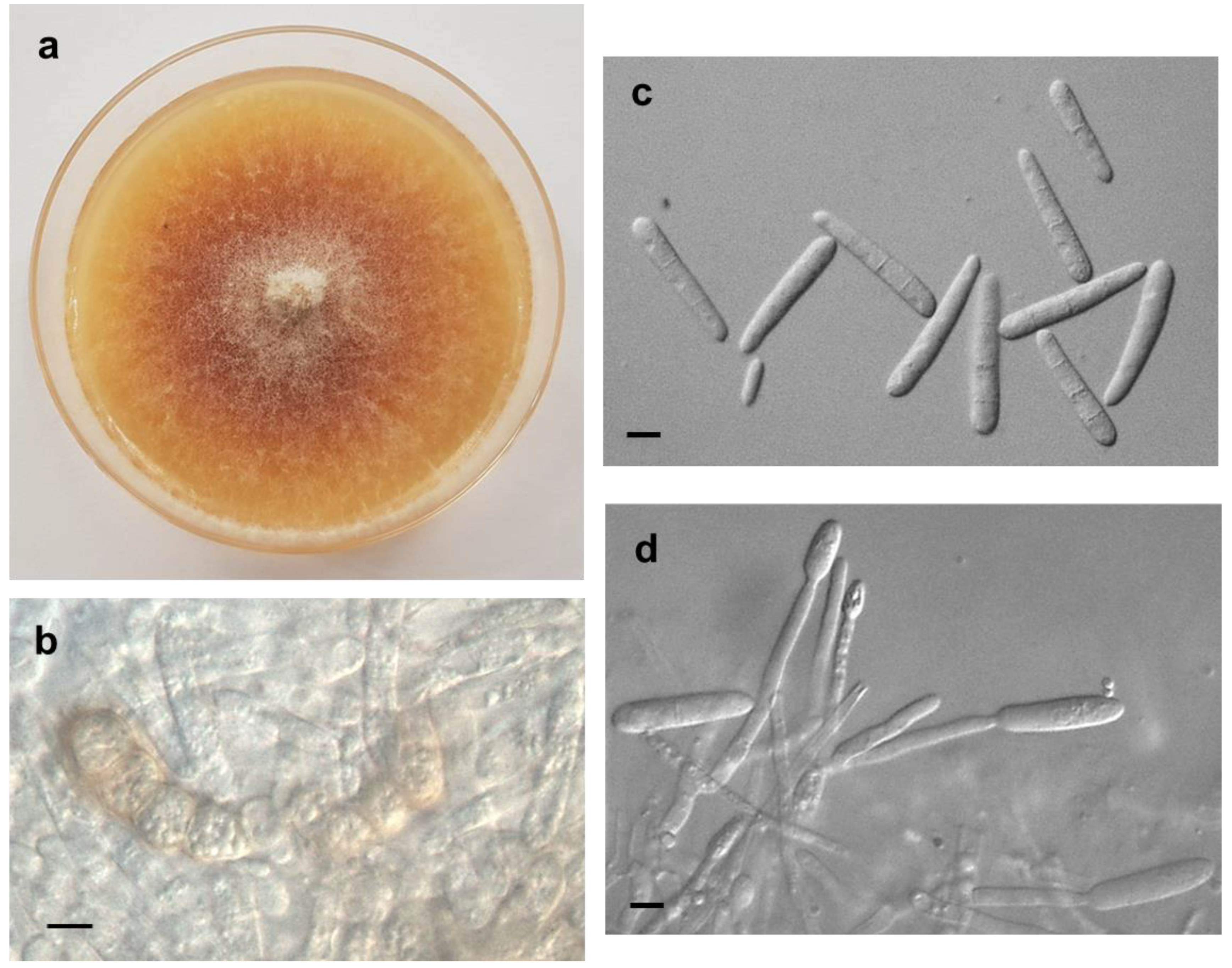
3.1. Morphological Observations
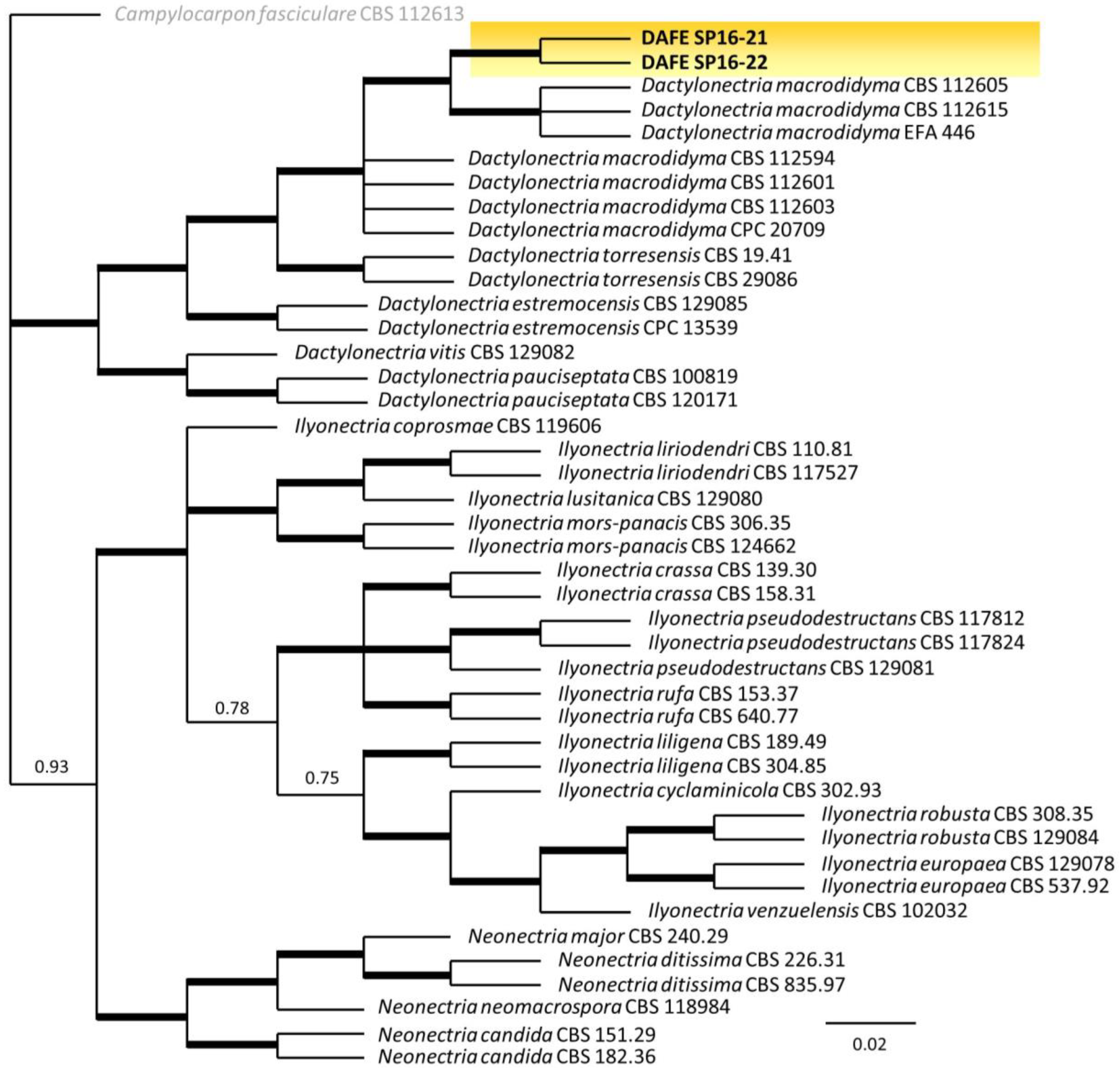
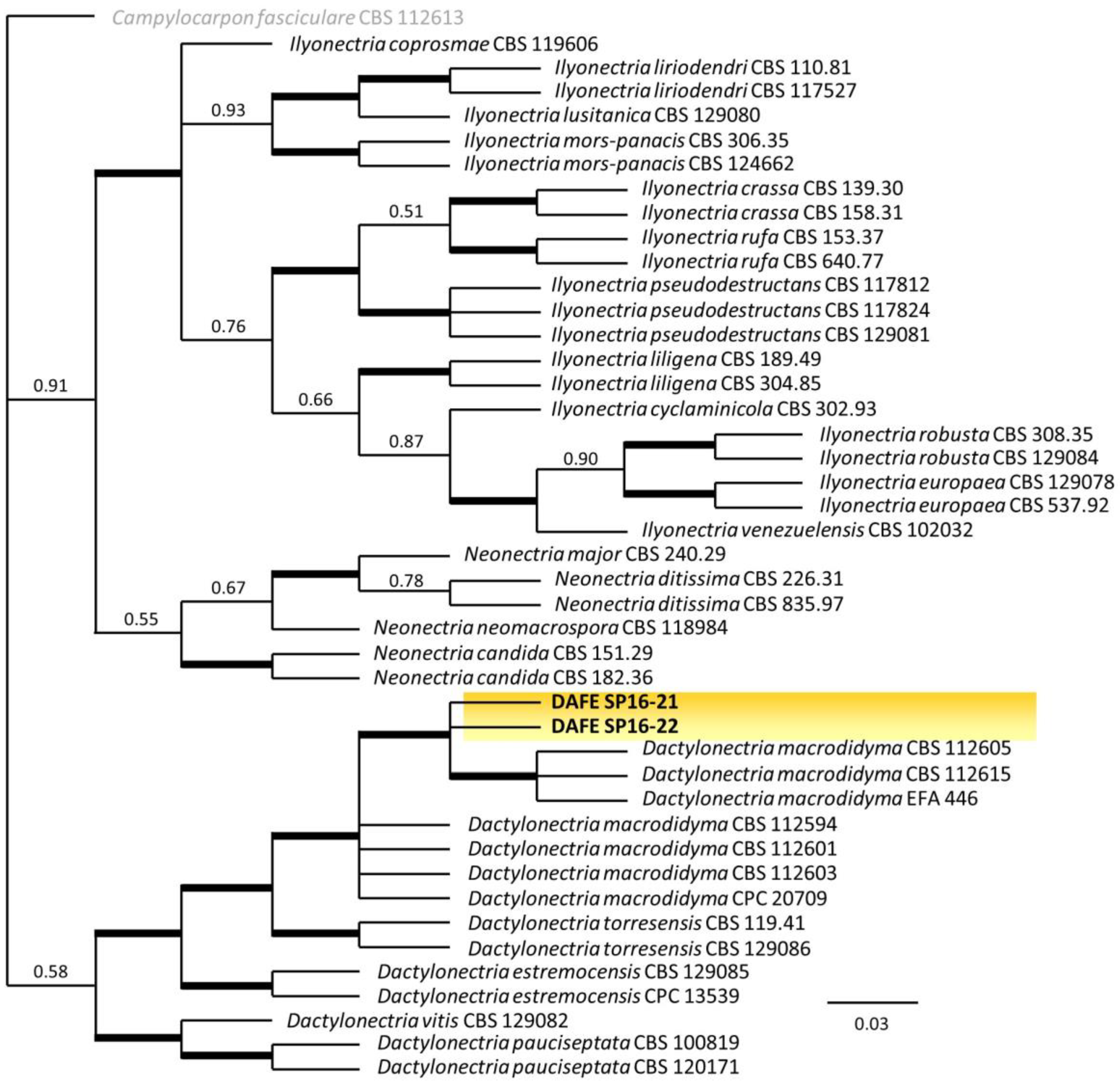
3.2. Phylogenetic Analysis
3.3. Pathogenicity Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rieuf, P. Parasites and saprophytes des plantes au Maroc. Les Cah. De La Rech. Agron. 1969, 27, 1–178. [Google Scholar]
- Bean, W.J. Trees and Shrubs Hardy in British Isles. Murray, J., Ed.; John Murray Pubs Ltd.: London, UK, 1981; Volume 1–4. [Google Scholar]
- Huxley, A.J.; Griffiths, M.; Levy, M. The New Royal Horticultural Society Dictionary of Gardening; Macmillan: London, UK, 1992; Volume 4. [Google Scholar]
- Hiemstra, J.A. Some general features of Verticillium wilts in trees. In A Compendium of Verticillium Wilts in Tree Species; Hiemstra, J.A., Harris, D.C., Eds.; Ponsen & Looijen: Wageningen, The Netherlands, 1998; pp. 5–11. [Google Scholar]
- Halstead, A.J.; Scrace, J.M. Pests and diseases of outdoor ornamentals, including hardy nursery stock. In Pest and Disease Management Handbook; Alford, D.V., Ed.; Blackwell Science: Oxford, UK, 2000; pp. 429–541. [Google Scholar]
- Cunnington, J.H.; Lawrie, A.C.; Pascoe, I.G. Genetic variation within Podosphaera tridactyla reveals a paraphyletic species complex with biological specialization towards specific Prunus subgenera. Mycol. Res. 2005, 109, 357–362. [Google Scholar] [CrossRef]
- Lázaro, B.B. Identification of the Main Plagues and Diseases that Affect the Azorean Plum—Prunus lusitanica L. ssp. azorica (Mouillef.) Franco. 2006, pp. 1–22. Available online: www.life-priolo.spea.pt/en/priolo-and-its-habitat/documents/ (accessed on 17 November 2022).
- Cave, G.L.; Randall-Schadel, B.; Redlin, S.C. Risk Analysis for Phytophthora Ramorum Werres, de Cock & Man In’t Veld, Causal Agent of Sudden Oak Death, Ramorum Leaf Blight, and Ramorum Dieback; USDA, APHIS, PPQ, CPHST, PERAL: Raleigh, NC, USA, 2008; p. 88. [Google Scholar]
- Joshi, V.; Jeffries, M.; Jesperson, G. Diseases diagnosed on commercial crops submitted to the British Columbia Ministry of Agriculture Plant Health Laboratory in 2011. Can. Plant Dis. Surv. 2012, 92, 7–17. [Google Scholar]
- Schulze, J.A.; Contreras, R.N. In Vivo chromosome doubling of Prunus lusitanica and preliminary morphological observations. HortScience 2017, 52, 332–337. [Google Scholar] [CrossRef]
- Agustí-Brisach, C.; Armengol, J. Black-foot disease of grapevine: An update on taxonomy, epidemiology and management strategies. Phytopathol. Mediterr. 2013, 52, 245–261. [Google Scholar]
- Tewoldemedhin, Y.T.; Mazzola, M.; Mostert, L.; McLeod, A. Cylindrocarpon species associated with apple tree roots in South Africa and their quantification using real-time PCR. Eur. J. Plant Pathol. 2011, 129, 637–651. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Haag, P.; Bowen, P.; O’Gorman, D.T. Grapevine trunk diseases in British Columbia: Incidence and characterization of the fungal pathogens associated with black foot disease of grapevine. Plant Dis. 2014, 98, 456–468. [Google Scholar] [CrossRef]
- Petit, E.; Barriault, E.; Baumgartner, K.; Wilcox, W.F.; Rolshausen, P.E. Cylindrocarpon species associated with black-foot of grapevine in Northeastern United States and Southeastern Canada. Am. J. Enol. Vitic. 2011, 62, 177–183. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Mayorquin, J.S.; Peacock, B.B.; Moreno, K.; Hajeri, S.; Yokomi, R.; Eskalen, A. Association of Neonectria macrodidyma with dry root rot of Citrus in California. J. Plant Pathol. Microbiol. 2016, 8, 391. [Google Scholar]
- Menkis, A.; Burokiene, D. Distribution and genetic diversity of the root-rot pathogen Neonectria macrodidyma in a forest nursery. For. Pathol. 2012, 42, 79–83. [Google Scholar] [CrossRef]
- Mora-Sala, B.; Cabral, A.; León, M.; Agustì-Brisach, C.; Armengol, J.; Abad-Campos, P. Survey, identification, and characterization of Cylindrocarpon-like asexual morphs in Spanish forest nurseries. Plant Dis. 2018, 102, 2083–2100. [Google Scholar] [CrossRef] [PubMed]
- Capote, N.; Del Río, M.Á.; Herencia, J.F.; Arroyo, F.T. Molecular and pathogenic characterization of Cylindrocarpon-like anamorphs causing root and basal rot of almonds. Plants 2022, 11, 984. [Google Scholar] [CrossRef]
- Aigoun-Mouhous, W.; Elena, G.; Cabral, A.; León, M.; Sabaou, N.; Armengol, J.; Chaouia, C.; Mahamedi, A.E.; Berraf-Tebbal, A. Characterization and pathogenicity of Cylindrocarpon-like asexual morphs associated with black foot disease in Algerian grapevine nurseries, with the description of Pleiocarpon algeriense sp. nov. Eur. J. Plant Pathol. 2019, 154, 887–901. [Google Scholar] [CrossRef]
- Wollenweber, H.W. Ramularia, Mycosphaerella, Nectria, Calonectria. Eine morphologisch pathologische studie zur abgrenzung von pilzgruppen mit cylindrischen und sichelförmigen konidienformen. Phytopathology 1913, 3, 198–242. [Google Scholar]
- Lombard, L.; Van der Merwe, N.A.; Groenewald, J.Z.; Crous, P.W. Lineages in Nectriaceae: Re-evaluating the generic status of Ilyonectria and allied genera. Phytopathol. Mediterr. 2014, 53, 515–532. [Google Scholar]
- Dhingra, O.D.; Sinclair, J.B. Basic Plant Pathology Methods, 2nd ed.; CRC Press: Boca Raton, FL, USA, 1995; p. 355. [Google Scholar]
- Nirenberg, H.I. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. In Mitteilungen Aus Der Biologischen Bundesanstalt Für Land- Und Forstwirtschaft (Berlin-Dahlem); Biologische Bundesanst. für Land- und Forstwirtschaft: Berlin, Germany, 1976; Volume 169, pp. 1–117. [Google Scholar]
- Halleen, F.; Schroers, H.F.; Groenewald, J.Z.; Crous, P.W. Novel species of Cylindrocarpon (Neonectria) and Campylocarpon gen. nov. associated with black foot disease of grapevines (Vitis spp.). Stud. Mycol. 2004, 50, 431–455. [Google Scholar]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour; Eyre Methuen: London, UK, 1978; p. 252. [Google Scholar]
- Pecchia, S.; Da Lio, D. Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. J. Microbiol. Methods 2018, 151, 118–128. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Groenewald, J.Z.; Risede, J.M.; Hywel-Jones, N.L. Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud. Mycol. 2004, 50, 415–429. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Cabral, A.; Groenewald, J.Z.; Rego, C.; Oliveira, H.; Crous, P.W. Cylindrocarpon root rot: Multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 2012, 11, 655–688. [Google Scholar] [CrossRef]
- Cabral, A.; Rego, C.; Nascimento, T.; Holiveira, H.; Groenewald, J.Z.; Crous, P.W. Multi-gene analysis and morphology reveal novel Ilyonectria species associated with black foot disease of grapevines. Fungal Biol. 2012, 116, 62–80. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.P.; Nouri, M.T.; Trouillas, F.P. Taxonomy and multi-locus phylogeny of cylindrocarpon-like species associated with diseased roots of grapevine and other fruit and nut crops in California. Fungal Syst. Evol. 2019, 4, 59–75. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11.0. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Chaverri, P.; Salgado, C.; Hirooka, Y.; Rossman, A.Y.; Samuels, G.J. Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud. Mycol. 2011, 68, 57–78. [Google Scholar] [CrossRef]
- Schroers, H.-J.; Zerjav, M.; Munda, A.; Halleen, F.; Crous, P.W. Cylindrocarpon pauciseptatum sp. nov., with notes on Cylindrocarpon species with wide, predominantly 3-septate macroconidia. Mycol. Res. 2008, 112, 82–92. [Google Scholar] [CrossRef]
- Dart, N.L.; Weeda, S.M. First report of Cylindrocarpon macrodidymum on Acer palmatum in Virginia. Plant Dis. 2011, 95, 1592. [Google Scholar] [CrossRef]
- Auger, J.; Pérez, I.; Esterio, M. Occurrence of root rot disease of cherimoya (Annona cherimola Mill.) caused by Dactylonectria macrodidyma in Chile. Plant Dis. 2015, 99, 1282. [Google Scholar] [CrossRef]
- Machín Barreiro, J.A. Identificación de Los Organismos Asociados a la Muerte de Plantas de Frutilla (Fragaria Ananassa Duch.) En El Departamento de Salto, Uruguay. Degree Thesis, Universidad de la República, Facultad de Agronomía, Montevideo, Uruguay, 2017; p. 41. Available online: https://www.colibri.udelar.edu.uy/jspui/bitstream/20.500.12008/18658/1/TTS_Mach%C3%ADnBarreiroJorgeAlex.pdf (accessed on 20 November 2022).
- Habibi, A.; Ghaderi, F. First record of Dactylonectria macrodidyma causing black root rot on strawberry. Mycologia Iranica 2020, 72, 241–246. [Google Scholar]
- Lombard, L.; Bezuidenhout, C.M.; Crous, P.W. Ilyonectria black foot rot associated with Proteaceae. Australas. Plant Pathol. 2013, 42, 337–349. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Peduto, F.; Gubler, W.D. First report of Ilyonectria macrodidyma causing root rot of olive trees (Olea europaea) in California. Plant Dis. 2012, 96, 1378. [Google Scholar] [CrossRef]
- El-ddin, S.N.; Haleem, R.A. First record of Dactylonectria macrodidyma: A cause of olive root rot in Iraq. For. Pathol. 2022, 52, e12729. [Google Scholar]
- Vitale, A.; Aiello, D.; Guarnaccia, V.; Perrone, G.; Stea, G.; Polizzi, G. First report of root rot caused by Ilyonectria (=Neonectria) macrodidyma on avocado (Persea americana) in Italy. J. Phytopathol. 2012, 160, 156–159. [Google Scholar] [CrossRef]
- Parkinson, L.E.; Shivas, R.G.; Dann, E.K. Pathogenicity of Nectriaceous fungi on avocado in Australia. Phytopathology 2017, 107, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.S.; Stenström, E.; Stenlid, J.; Finlay, R. Fungi in decayed roots of conifer seedlings from forest nurseries, afforested clearcuts and abandoned farmland. Plant Pathol. 2006, 55, 117–129. [Google Scholar] [CrossRef]
- van Der Merwe, R. Occurrence of Canker and Wood Rot Pathogens on Stone Fruit Propagation Material and Nursery Stone Fruit Trees. Master’s Thesis, Faculty of AgriSciences at the University of Stellenbosch, Stellenbosch, South Africa, 2019. Available online: https://scholar.sun.ac.za (accessed on 20 November 2022).
- Cabral, A.; Rego, C.; Crous, P.W.; Oliveira, H. Virulence and cross-infection potential of Ilyonectria spp. to grapevine. Phytopathol. Mediterr. 2012, 51, 340–354. [Google Scholar]
- Halleen, F.; Fourie, P.H.; Crous, P.W. A review of black-foot disease of grapevine. Phytopathol. Mediterr. 2006, 45, S55–S67. [Google Scholar]
- Ye, Q.; Zhang, W.; Jia, J.; Li, X.; Zhou, Y.; Han, C.; Wu, X.; Yan, J. Fungal pathogens associated with black foot of grapevine in China. Phytopathol. Mediterr. 2021, 60, 303–319. [Google Scholar] [CrossRef]
- Probst, C.M.; Ridgway, H.J.; Jaspers, M.V.; Jones, E.E. Pathogenicity of Ilyonectria liriodendri and Dactylonectria macrodidyma propagules in grapevines. Eur. J. Plant Pathol. 2019, 154, 405–421. [Google Scholar] [CrossRef]
- Probst, C.M.; Ridgway, H.J.; Jaspers, M.V.; Jones, E.E. Propagule and soil type affects the pathogenicity of Ilyonectria and Dactylonectria spp., the causal agents of black foot disease of grapevines. Vitis 2022, 61, 11–19. [Google Scholar]
- Wakelin, S.A.; Gomez-Gallego, M.; Jones, E.; Smaill, S.; Lear, G.; Lambie, S. Climate change induced drought impacts on plant diseases in New Zealand. Australas. Plant Pathol. 2018, 47, 101–114. [Google Scholar] [CrossRef]
- Marzialetti, P. Banca Dati Agrometeorologici Di Pistoia, 2018. Centro Sperimentale per Il Vivaismo. Available online: www.cespevi.it (accessed on 17 November 2022).
- Pendergrass, A.G.; Knutti, R.; Lehner, F.; Deser, C.; Sanderson, B.M. Precipitation variability increases in a warmer climate. Sci. Rep. 2017, 7, 17966. [Google Scholar] [CrossRef]
- World Meteorological Organization WMO Confirms 2016 as Hottest Year on Record, about 1.1 °C above Pre-Industrial Era. 2017. Available online: https://public.wmo.int/en/media/press-release/ (accessed on 26 November 2022).
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39. [Google Scholar] [CrossRef]
- Martínez-Diz, M.P.; Díaz-Losada, E.; Andrés-Sodupe, M.; Bujanda, R.; Maldonado-González, M.M.; Ojeda, S.; Yacoub, A.; Rey, P.; Gramaje, D. Field evaluation of biocontrol agents against black-foot and Petri diseases of grapevine. Pest Manag. Sci. 2021, 77, 697–708. [Google Scholar] [CrossRef]
- van Jaarsveld, W.J.; Halleen, F.; Bester, M.C.; Pierron, R.J.; Stempien, E.; Mostert, L. Investigation of Trichoderma species colonization of nursery grapevines for improved management of black foot disease. Pest Manag. Sci. 2021, 77, 397–405. [Google Scholar] [CrossRef]
- Halleen, F.; Crous, P.W.; Petrini, O. Fungi associated with healthy grapevine cuttings in nurseries, with special reference to pathogens involved in the decline of young vines. Australas. Plant Pathol. 2003, 32, 47–52. [Google Scholar] [CrossRef]
- Alaniz, S.; Abad-Campos, P.; García-Jiménez, J.; Armengol, J. Evaluation of fungicides to control Cylindrocarpon liriodendri and Cylindrocarpon macrodidymum in vitro, and their effect during the rooting phase in the grapevine propagation process. Crop Prot. 2011, 30, 489–494. [Google Scholar] [CrossRef]
- Dumroese, R.K.; James, R.L.; Wenny, D.L. Hot water and copper coatings in reused containers decrease inoculum of Fusarium and Cylindrocarpon and increase Douglas Fir seedling growth. Hortic. Sci. 2002, 37, 943–947. [Google Scholar] [CrossRef]



| Plant | Common Name | Disease | References |
|---|---|---|---|
| Acer palmatum | Japanese maple | Crown cankers and root rot | [39] |
| Annona cherimola | Cherimoya | Root-rot | [40] |
| Citrus | Trifoliate seedlings (Carrizo) | Dry Root Rot | [15] |
| Fragaria x ananassa | Strawberry | Black root rot | [41,42] |
| Ilex aquifolium | Holly | Root rot and dieback | [17] |
| Juniperus phoenicea | Phoenician juniper | Root rot and dieback | [17] |
| Leucospermum sp. | Pincushion | Black foot rot | [43] |
| Lonicera sp. | Honeysuckle | Root rot and dieback | [17] |
| Malus domestica | Apple | Apple Replant Disease, Root rot | [12,31] |
| Myrtus communis | Common myrtle | Root rot and dieback | [17] |
| Olea europea | Olive | Root rot | [44,45] |
| Persea americana | Avocado | Root rot, Black root rot | [46,47] |
| Picea abies | Norway spruce | Root dieback | [48] |
| Pinus halepensis | Aleppo pine | Root rot and dieback | [17] |
| Pinus sylvestris | Scots pine | Root-rot | [16,48] |
| Protea sp. | Sugarbush | Black foot rot | [43] |
| Prunus lusitanica | Portugal laurel | Crown rot | This study |
| Prunus dulcis | Almond | Root and basal rot | [18] |
| Prunus salicina | Plum | Crown infections | [49] |
| Pyracantha sp. | Firethorn | Root rot and dieback | [17] |
| Quercus faginea | Portuguese oak | Root rot and dieback | [17] |
| Quercus ilex | Holm oak | Root rot and dieback | [17] |
| Rosmarinus officinalis | Rosemary | Root rot and dieback | [17] |
| Vitis vinifera | Grape | Black foot | [13,50,51,52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecchia, S.; Caggiano, B.; Da Lio, D.; Resta, E. Morphological and Molecular Identification of Dactylonectria macrodidyma as Causal Agent of a Severe Prunus lusitanica Dieback in Italy. Horticulturae 2023, 9, 145. https://doi.org/10.3390/horticulturae9020145
Pecchia S, Caggiano B, Da Lio D, Resta E. Morphological and Molecular Identification of Dactylonectria macrodidyma as Causal Agent of a Severe Prunus lusitanica Dieback in Italy. Horticulturae. 2023; 9(2):145. https://doi.org/10.3390/horticulturae9020145
Chicago/Turabian StylePecchia, Susanna, Benedetta Caggiano, Daniele Da Lio, and Emilio Resta. 2023. "Morphological and Molecular Identification of Dactylonectria macrodidyma as Causal Agent of a Severe Prunus lusitanica Dieback in Italy" Horticulturae 9, no. 2: 145. https://doi.org/10.3390/horticulturae9020145
APA StylePecchia, S., Caggiano, B., Da Lio, D., & Resta, E. (2023). Morphological and Molecular Identification of Dactylonectria macrodidyma as Causal Agent of a Severe Prunus lusitanica Dieback in Italy. Horticulturae, 9(2), 145. https://doi.org/10.3390/horticulturae9020145






