Three-Dimensional Imaging in Agriculture: Challenges and Advancements in the Phenotyping of Japanese Quinces in Latvia
Abstract
:1. Introduction
1.1. Japanese Quinces
1.2. Three-Dimensional Technologies in Plant Phenotyping
2. Materials and Methods
- Jamovi (ver. 2.4) software: This was utilized for statistical computations [33].
- R language: In addition to Jamovi, it was also employed for statistical analyses [34].
- Python programming language: This was used for the development and implementation of algorithms.
- Libraries: Open3D (ver. 0.17) was pivotal in processing point cloud data, offering advanced functionalities for 3D modeling and object detection. NumPy (ver. 1.26) was used for data manipulation and computational tasks. PyQt5 (ver. 5.15.10) enabled the creation of intuitive graphical user interfaces.
2.1. Acquisition of Fruit Measurements Using the Manual Method
2.2. Three-Dimensional Data Acquisition and Postprocess
- The k-NN algorithm, renowned for its efficacy in pattern recognition [36], is specifically employed for color differentiation within the dataset. A fundamental prerequisite for the application of the k-NN algorithm is the initial training phase, wherein the algorithm is exposed to multiple color samples representative of the target object. This process is crucial for enabling the algorithm to discern and subsequently exclude background and irrelevant data points based on the defined color parameters of the object, in this case, Japanese quinces. For objects exhibiting a spectrum of colors, each distinctive hue is incorporated into the classification scheme to ensure comprehensive identification.
- In scenarios involving multiple fruits within a single point cloud, the need for discrete object detection becomes paramount. The “Imaginary Square” algorithm is ingeniously designed to address this challenge. Initiated at the point of maximum ‘y’ value within the point cloud, this algorithm progressively expands a conceptual square, encompassing an increasing number of data points. The expansion of the square continues as long as a sequential increase in data points is observed. The termination of square growth occurs upon reaching a plateau in point increment, suggesting the potential identification of an object. However, if objects are in close proximity, the algorithm may erroneously perceive them as a singular entity. To circumvent this, defining an upper limit on the object size becomes essential, beyond which further expansion of the square is deemed unnecessary.
- The third algorithm pivots on the concept of projecting the object onto a base plane. This is achieved by a reverse application of the k-NN algorithm, where the focus shifts from the object to the background. This reversal aids in the identification of outlier points delineating the base projection of the object. The inherent limitation of 3D imaging in capturing the area obscured by the fruit results in voids within the point cloud, which, in this context, represent the base projection of the fruit. The analysis commences in the region defined by the “Imaginary Square” algorithm, with each projection being scrutinized individually. This involves identifying the point with the maximum ‘y’ value on the projection perimeter and generating an ‘analysis point’ from which four vectors extend to the nearest perimeter points. This procedure, potentially iterated with slight adjustments to the analysis point, furnishes a detailed understanding of the projection’s perimeter, thereby inferring the spatial dimensions of the object situated above.
2.3. Experiment Conditions and Specifications
3. Results
3.1. Characterization of Japanese Quince Fruit Parameters Using Manual Measurement Techniques
3.2. Characterization of Japanese Quince Fruits Utilizing a 3D Imaging-Based Methodology
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hellín, P.; Vila, R.; Jordán, M.J.; Laencina, J.; Rumpunen, K.; Ros, J.M. Charasteristics and Composition of Chaenomeles Fruit Juice. In Japanese Quince Potential Fruit Crop for Northen Europe; Department of Crop Science, Swedish University of Agricultural Sciences: Uppsala, Sweden, 2003; Volume 184, pp. 127–140. [Google Scholar]
- Krasnova, I.; Ruisa, S.; Segliņa, D. Investigations of the Biochemical Composition of Chaenomeles japonica Fruits. Cheminė Technol. 2007, 4, 16–20. [Google Scholar]
- Górnaś, P.; Siger, A.; Juhņeviča, K.; Lācis, G.; Šnē, E.; Segliņa, D. Cold-Pressed Japanese Quince (Chaenomeles japonica (Thunb.) Lindl. Ex Spach) Seed Oil as a Rich Source of α-Tocopherol, Carotenoids and Phenolics: A Comparison of the Composition and Antioxidant Activity with Nine Other Plant Oils. Eur. J. Lipid Sci. Technol. 2014, 116, 563–570. [Google Scholar] [CrossRef]
- Zvikas, V.; Urbanaviciute, I.; Bernotiene, R.; Kulakauskiene, D.; Morkunaite, U.; Balion, Z.; Majiene, D.; Liaudanskas, M.; Viskelis, P.; Jekabsone, A.; et al. Investigation of Phenolic Composition and Anticancer Properties of Ethanolic Extracts of Japanese Quince Leaves. Foods 2020, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Ruisa, S. Studies on Japanese Quince (Chaenomeles japonica) in Latvia; Department of Horticulture Plant Breeding, SLU Balsgard: Uppsala, Sweden, 1996; pp. 204–207. [Google Scholar]
- Rumpunen, K. Chaenomeles: Potential New Fruit Crop for Northern Europe. In Trends in New Crops and New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, VA, USA, 2002; pp. 385–392. [Google Scholar]
- Ruisa, S.; Lacis, G. Characterization of Chaenomeles japonica Genetic Resources. Hortic. Veg. Grow. 2001, 20, 50–60. [Google Scholar]
- Seglina, D.; Krasnova, I.; Heidemane, G.; Ruisa, S. Influence of Drying Technology on the Quality of Dried Candied Chaenomeles japonica during Storage. Latv. J. Agron. 2009, 12, 113–118. [Google Scholar]
- Rumpunen, K.; Kviklys, D.; Kauppinen, S.; Ruisa, S.; Tigerstedt, P.M. Breeding Strategies for the Fruit Crop Japanese Quince (Chaenomeles japonica); Department of Horticulture Plant Breeding, SLU Balsgard: Uppsala, Sweden, 2003; pp. 59–80. [Google Scholar]
- Rumpunen, K.; Kviklys, D. Combining Ability and Patterns of Inheritance for Plant and Fruit Traits in Japanese Quince (Chaenomeles japonica). Euphytica 2003, 132, 139–149. [Google Scholar] [CrossRef]
- Weber, C. Cultivars in the Genus Chaenomeles. Bull. Pop. Inf. Arnold Arbor. Harv. Univ. 1963, 3, 17–75. [Google Scholar]
- Ruisa, S.; Rubauskis, E. Evaluation of the Selected Genotypes of Chaenomeles japonica. In Proceedings of the International Scientific Conference “Environmentally Friendly Fruit Growing”, Tartu, Estonia, 7–9 September 2005; Volume 223, pp. 69–75. [Google Scholar]
- Kaufmane, E.; Ruisa, S. Breeding of New Cultivars of the Fruit Crop Japanese Quince (Chaenomeles japonica) in Latvia. Acta Horticult. 2020, 1281, 51–58. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Gardner, K.M.; Money, D.; Sawler, J.; Bloom, J.S.; Moffett, P.; Chao, C.T.; Schwaninger, H.; Fazio, G.; Zhong, G.; et al. Genome to Phenome Mapping in Apple Using Historical Data. Plant Genome 2016, 9, plantgenome2015.11.0113. [Google Scholar] [CrossRef]
- Paulus, S.; Schumann, H.; Kuhlmann, H.; Léon, J. High-Precision Laser Scanning System for Capturing 3D Plant Architecture and Analysing Growth of Cereal Plants. Biosyst. Eng. 2014, 121, 1–11. [Google Scholar] [CrossRef]
- Paulus, S. Measuring Crops in 3D: Using Geometry for Plant Phenotyping. Plant Methods 2019, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Omasa, K.; Hosoi, F.; Konishi, A. 3D Lidar Imaging for Detecting and Understanding Plant Responses and Canopy Structure. J. Exp. Bot. 2006, 58, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Paulus, S.; Dupuis, J.; Riedel, S.; Kuhlmann, H. Automated Analysis of Barley Organs Using 3D Laser Scanning: An Approach for High Throughput Phenotyping. Sensors 2014, 14, 12670–12686. [Google Scholar] [CrossRef] [PubMed]
- Dornbusch, T.; Lorrain, S.; Kuznetsov, D.; Fortier, A.; Liechti, R.; Xenarios, I.; Fankhauser, C. Measuring the Diurnal Pattern of Leaf Hyponasty and Growth in Arabidopsis—A Novel Phenotyping Approach Using Laser Scanning. Funct. Plant Biol. 2012, 39, 860. [Google Scholar] [CrossRef] [PubMed]
- Fasoula, D.A.; Ioannides, I.M.; Omirou, M. Phenotyping and Plant Breeding: Overcoming the Barriers. Front. Plant Sci. 2020, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.; Boudon, F.; Godin, C. 3D Plant Phenotyping: All You Need Is Labelled Point Cloud Data. In Computer Vision—ECCV 2020 Workshops; Bartoli, A., Fusiello, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12540, pp. 244–260. ISBN 978-3-030-65413-9. [Google Scholar]
- Boogaard, F.P.; Van Henten, E.J.; Kootstra, G. Improved Point-Cloud Segmentation for Plant Phenotyping through Class-Dependent Sampling of Training Data to Battle Class Imbalance. Front. Plant Sci. 2022, 13, 838190. [Google Scholar] [CrossRef] [PubMed]
- Harandi, N.; Vandenberghe, B.; Vankerschaver, J.; Depuydt, S.; Van Messem, A. How to Make Sense of 3D Representations for Plant Phenotyping: A Compendium of Processing and Analysis Techniques. Plant Methods 2023, 19, 60. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, F.; Pang, S.; Zhao, X.; Chen, L.; Liu, J.; Xue, B.; Xu, G.; Li, L.; Jing, H.; et al. Crop 3D—A LiDAR Based Platform for 3D High-Throughput Crop Phenotyping. Sci. China Life Sci. 2018, 61, 328–339. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-Field High Throughput Phenotyping and Cotton Plant Growth Analysis Using LiDAR. Front. Plant Sci. 2018, 9, 16. [Google Scholar] [CrossRef]
- Zhu, R.; Sun, K.; Yan, Z.; Yan, X.; Yu, J.; Shi, J.; Hu, Z.; Jiang, H.; Xin, D.; Zhang, Z.; et al. Analysing the Phenotype Development of Soybean Plants Using Low-Cost 3D Reconstruction. Sci. Rep. 2020, 10, 7055. [Google Scholar] [CrossRef]
- Ju, C.; Zhao, Y.; Wu, F.; Li, R.; Yang, T.; Zhong, X.; Yan, F.; Liu, T.; Sun, C. Three-Dimensional Reconstruction of Maize Plants and Extraction of Phenotypic Parameters Based on Laser Point Cloud Data. Rev. Square 2021. [Google Scholar] [CrossRef]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. A Novel Mesh Processing Based Technique for 3D Plant Analysis. BMC Plant Biol. 2012, 12, 63. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Yang, H.; Zhang, C.; Tang, Y. A Hierarchical Growth Method for Extracting 3D Phenotypic Trait of Apple Tree Branch in Edge Computing. Wirel. Netw. 2023. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Jin, X.; Bian, L.; Ge, Y. High-Throughput Phenotyping of Plant Leaf Morphological, Physiological, and Biochemical Traits on Multiple Scales Using Optical Sensing. Crop J. 2023, 11, 1303–1318. [Google Scholar] [CrossRef]
- Gu, F.; Song, Z.; Zhao, Z. Single-Shot Structured Light Sensor for 3D Dense and Dynamic Reconstruction. Sensors 2020, 20, 1094. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Slaughter, D.; Max, N.; Maloof, J.; Sinha, N. Structured Light-Based 3D Reconstruction System for Plants. Sensors 2015, 15, 18587–18612. [Google Scholar] [CrossRef] [PubMed]
- Haleem, A.; Javaid, M.; Singh, R.P.; Rab, S.; Suman, R.; Kumar, L.; Khan, I.H. Exploring the Potential of 3D Scanning in Industry 4.0: An Overview. Int. J. Cogn. Comput. Eng. 2022, 3, 161–171. [Google Scholar] [CrossRef]
- The Jamovi Project. Jamovi (Version 2.4) [Computer Software]; The Jamovi Project: San Diego, CA, USA, 2023. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Zivid Zivid Studio. Available online: https://www.zivid.com/zivid-studio-software (accessed on 14 November 2023).
- Uddin, S.; Haque, I.; Lu, H.; Moni, M.A.; Gide, E. Comparative Performance Analysis of K-Nearest Neighbour (KNN) Algorithm and Its Different Variants for Disease Prediction. Sci. Rep. 2022, 12, 6256. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, X.; Au, W.; Kang, H.; Chen, C. Intelligent Robots for Fruit Harvesting: Recent Developments and Future Challenges. Precis. Agric. 2022, 23, 1856–1907. [Google Scholar] [CrossRef]
- Li, B.; Lecourt, J.; Bishop, G. Advances in Non-Destructive Early Assessment of Fruit Ripeness towards Defining Optimal Time of Harvest and Yield Prediction—A Review. Plants 2018, 7, 3. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, Z.; Li, D.; Liu, X. Phenotypic Techniques and Applications in Fruit Trees: A Review. Plant Methods 2020, 16, 107. [Google Scholar] [CrossRef] [PubMed]




| Projector brightness | 0.25x to 1.8x; 1x = 400 lumens |
| The field of view angle | 0 (Directly above) |
| Resolution | 1920 × 1200 (2.3 Mpixel), Native 3D Color |
| Point cloud output | 3D (XYZ) + Color (RGB) + SNR |
| Exposure time (minimum per pattern projection) | 6.5 ms |
| Focus distance | 1000 mm |
| Optimal working distance | 700 to 1500 mm |
| Camera distance from objects | 1000 mm |
| Field of view | 702 × 432 |
| Spatial resolution | 0.37 mm and 3.71 × 10−4 mm per distance (z) |
| Capture time | 200 ms |
| Point precision in Euclidian distance | 110 µm |
| Local Planarity Precision in Euclidian distance | 190 µm |
| Genotype | Fruit Length (mm) | Fruit Width (mm) | |
|---|---|---|---|
| N | Ada | 30 | 30 |
| Alfa | 30 | 30 | |
| Darius | 30 | 30 | |
| Rasa | 30 | 30 | |
| Rondo | 30 | 30 | |
| SR1-1 | 30 | 30 | |
| SR1-2 | 30 | 30 | |
| SR1-3 | 30 | 30 | |
| SR1-4 | 30 | 30 | |
| SR1-5 | 30 | 30 | |
| SR1-6 | 30 | 30 | |
| Median | Ada | 49 | 47 |
| Alfa | 40 | 44 | |
| Darius | 37 | 42 | |
| Rasa | 44 | 52 | |
| Rondo | 46 | 47 | |
| SR1-1 | 45 | 47 | |
| SR1-2 | 40 | 47 | |
| SR1-3 | 43 | 50 | |
| SR1-4 | 37 | 40 | |
| SR1-5 | 43 | 45 | |
| SR1-6 | 46 | 47 | |
| Standard deviation | Ada | 4 | 3 |
| Alfa | 3 | 2 | |
| Darius | 4 | 4 | |
| Rasa | 4 | 4 | |
| Rondo | 6 | 4 | |
| SR1-1 | 6 | 3 | |
| SR1-2 | 3 | 3 | |
| SR1-3 | 4 | 4 | |
| SR1-4 | 4 | 3 | |
| SR1-5 | 4 | 3 | |
| SR1-6 | 4 | 3 |
| Genotype | Average Fruit Weight (g) | Maximum Fruit Weight (g) | Characteristics of the Fruit |
|---|---|---|---|
| SR1-1 | 53 | 130 | Round and slightly flattened with a smooth surface, exhibiting mild ribbing and a notably deep inflorescence. |
| SR1-2 | 45 | 78 | Bright yellow and homogeneous in appearance, these are barrel-shaped with significant puncture and rust characteristics. |
| SR1-3 | 64 | 122 | Dark yellow, round, and slightly flattened; characterized by prominent red dots and brown dotted rust; they also display a ribbed texture. |
| SR1-4 | 34 | 71 | Predominantly bright yellow, round, and barrel-shaped, with some assuming a pear-shaped (pyriform) form. They have a very smooth surface and are mostly free from puncture. |
| SR1-5 | 42 | 110 | Yellow, round, and barrel-shaped with a smooth texture; slight ribbing at the tip, aesthetically pleasing, and with a few red dots; near the inflorescence, there is slight brown rust. |
| SR1-6 | 56 | 105 | Smooth, attractive, and yellow, varying from round or oval, to bottle-shaped. Some exhibit pronounced red dots and slight russeting in the form of small brown dots or stripes. |
| Rasa | 48 | 75 | Yellow and rounded, exhibiting mild ribbing. In some years, they assume a pear-shaped (pyriform) appearance. |
| Darius | 34 | 45 | Oblong and yellow, characterized by a smooth and homogeneous surface. |
| Rondo | 52 | 67 | Yellow and oblong, featuring a deep flower bed and generally homogeneous in appearance. |
| Ada | 55 | 78 | Dark yellow with a pink wreath, oblong, and maintaining a homogeneous texture. |
| Alfa | 53 | 67 | Yellow with pronounced rust spots, rounded, slightly ribbed, and featuring a deep flower bed. |
| Genotype | Fruit Length (mm) | Fruit Width (mm) | |
|---|---|---|---|
| N | Ada | 19 | 19 |
| Alfa | 19 | 19 | |
| Darius | 15 | 15 | |
| Rasa | 23 | 23 | |
| Rondo | 20 | 20 | |
| SR1-1 | 16 | 16 | |
| SR1-2 | 29 | 29 | |
| SR1-3 | 25 | 25 | |
| SR1-4 | 29 | 29 | |
| SR1-5 | 17 | 17 | |
| SR1-6 | 29 | 29 | |
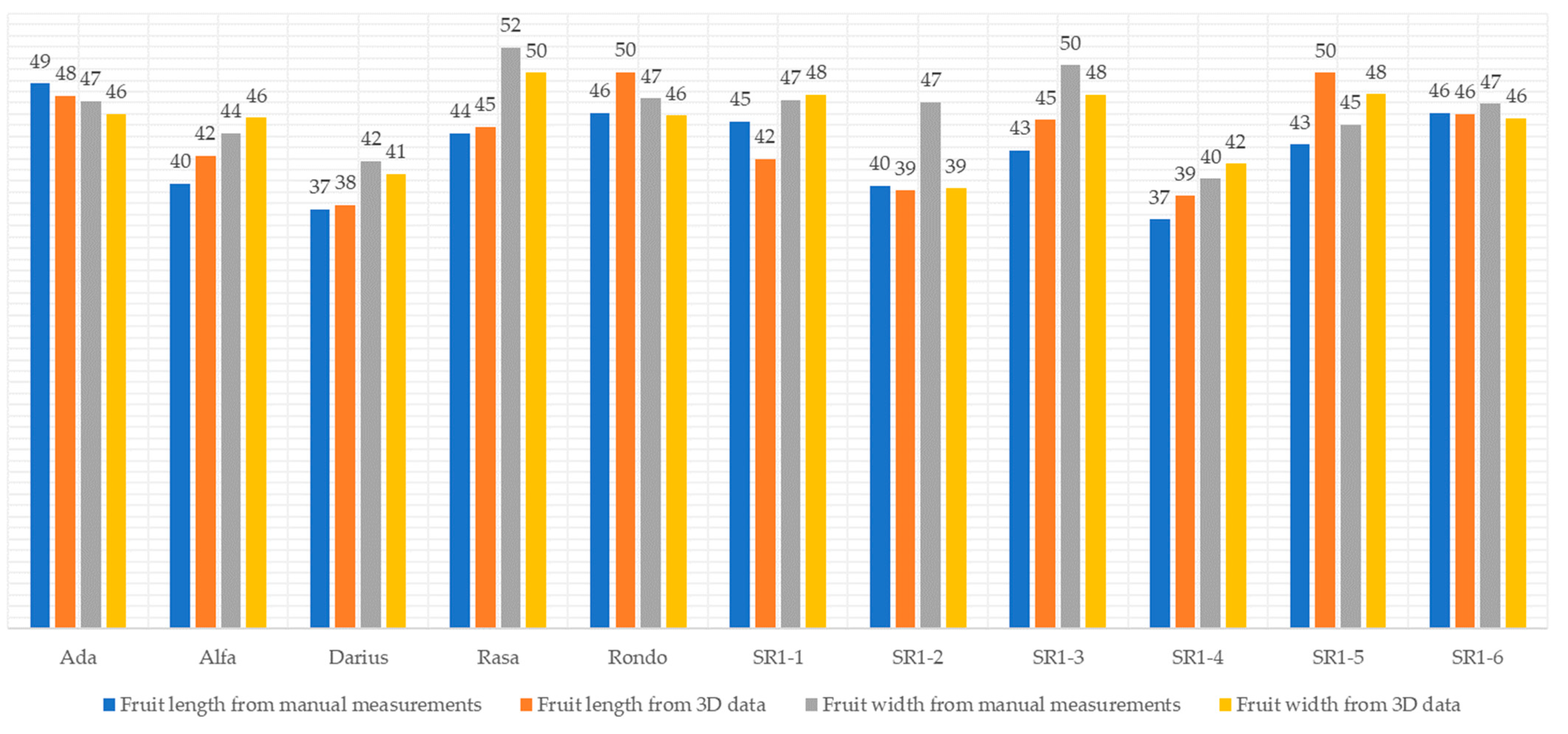
| Median | Ada | 48 | 46 |
| Alfa | 42 | 46 | |
| Darius | 38 | 41 | |
| Rasa | 45 | 50 | |
| Rondo | 50 | 46 | |
| SR1-1 | 42 | 48 | |
| SR1-2 | 39 | 39 | |
| SR1-3 | 45 | 48 | |
| SR1-4 | 39 | 42 | |
| SR1-5 | 50 | 48 | |
| SR1-6 | 46 | 46 | |
| Standard deviation | Ada | 3 | 4 |
| Alfa | 3 | 4 | |
| Darius | 2 | 4 | |
| Rasa | 5 | 5 | |
| Rondo | 10 | 6 | |
| SR1-1 | 2 | 6 | |
| SR1-2 | 3 | 7 | |
| SR1-3 | 3 | 4 | |
| SR1-4 | 4 | 4 | |
| SR1-5 | 3 | 6 | |
| SR1-6 | 5 | 3 |
| Genotype | Fruit Length (mm) | Fruit Width (mm) | |
|---|---|---|---|
| Median | Ada | −2 | 3 |
| Alfa | −3 | 0 | |
| Darius | −2 | 2 | |
| Rasa | 0 | −1 | |
| Rondo | −7 | 2 | |
| SR1-1 | 3 | −2 | |
| SR1-2 | 2 | 6 | |
| SR1-3 | −3 | 2 | |
| SR1-4 | −2 | −3 | |
| SR1-5 | −5 | −5 | |
| SR1-6 | 0 | 1 | |
| Standard deviation | Ada | 4 | 3 |
| Alfa | 5 | 4 | |
| Darius | 3 | 6 | |
| Rasa | 5 | 4 | |
| Rondo | 13 | 6 | |
| SR1-1 | 5 | 5 | |
| SR1-2 | 4 | 6 | |
| SR1-3 | 4 | 3 | |
| SR1-4 | 3 | 4 | |
| SR1-5 | 4 | 5 | |
| SR1-6 | 5 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaufmane, E.; Edelmers, E.; Sudars, K.; Namatēvs, I.; Nikulins, A.; Strautiņa, S.; Kalniņa, I.; Peter, A. Three-Dimensional Imaging in Agriculture: Challenges and Advancements in the Phenotyping of Japanese Quinces in Latvia. Horticulturae 2023, 9, 1347. https://doi.org/10.3390/horticulturae9121347
Kaufmane E, Edelmers E, Sudars K, Namatēvs I, Nikulins A, Strautiņa S, Kalniņa I, Peter A. Three-Dimensional Imaging in Agriculture: Challenges and Advancements in the Phenotyping of Japanese Quinces in Latvia. Horticulturae. 2023; 9(12):1347. https://doi.org/10.3390/horticulturae9121347
Chicago/Turabian StyleKaufmane, Edīte, Edgars Edelmers, Kaspars Sudars, Ivars Namatēvs, Arturs Nikulins, Sarmīte Strautiņa, Ieva Kalniņa, and Astile Peter. 2023. "Three-Dimensional Imaging in Agriculture: Challenges and Advancements in the Phenotyping of Japanese Quinces in Latvia" Horticulturae 9, no. 12: 1347. https://doi.org/10.3390/horticulturae9121347
APA StyleKaufmane, E., Edelmers, E., Sudars, K., Namatēvs, I., Nikulins, A., Strautiņa, S., Kalniņa, I., & Peter, A. (2023). Three-Dimensional Imaging in Agriculture: Challenges and Advancements in the Phenotyping of Japanese Quinces in Latvia. Horticulturae, 9(12), 1347. https://doi.org/10.3390/horticulturae9121347











