Exploring the Potential Biocontrol Isolates of Trichoderma asperellum for Management of Collar Rot Disease in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Isolation of the Fungus
2.2. Morphological Observations
2.3. Molecular Identification and Phylogenetic Analysis
2.4. In Vitro Antagonistic Assay
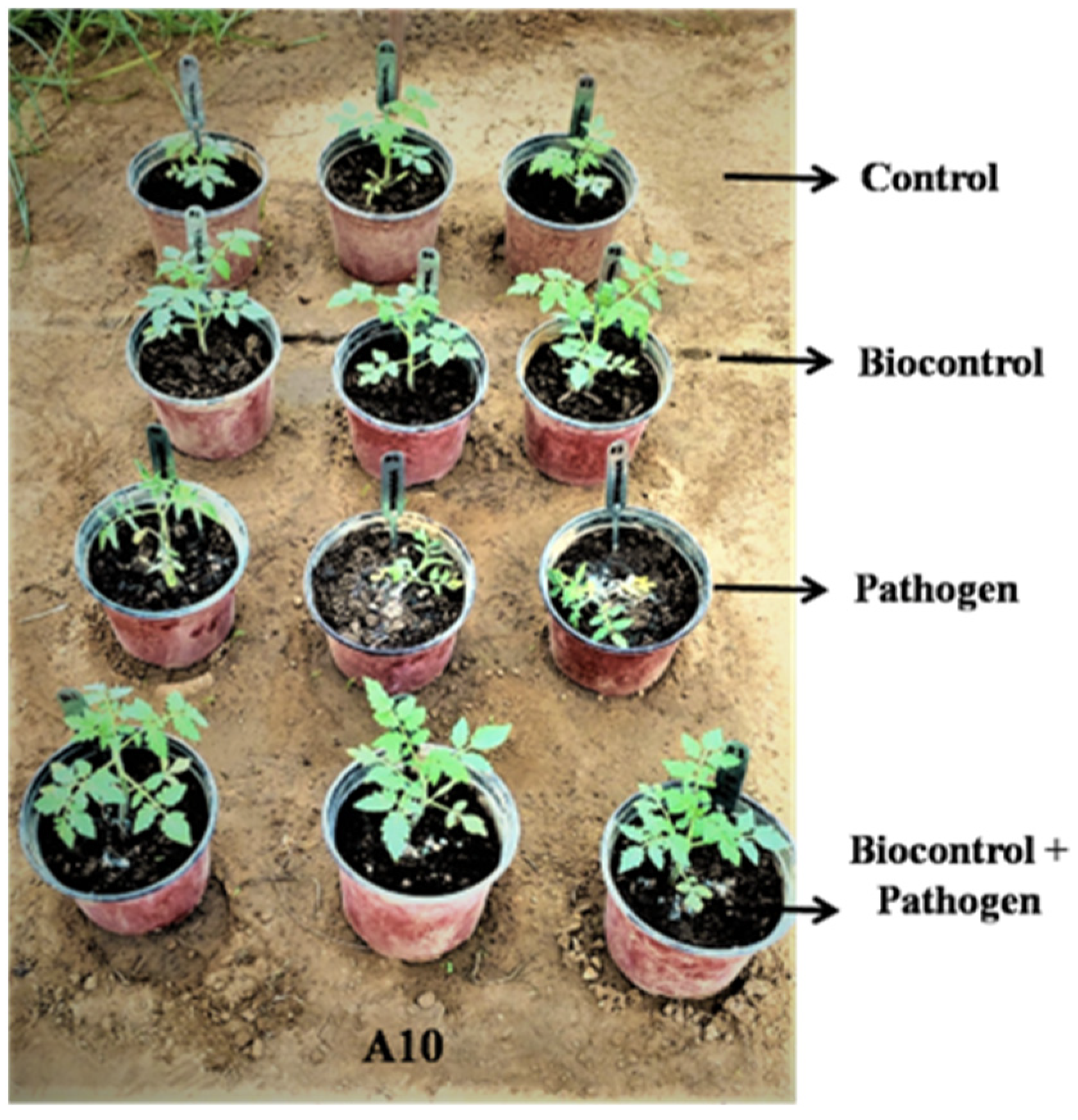
2.5. In Planta Bio-Control Assay
2.6. Biochemical Analysis
2.6.1. Cellulase Assay
2.6.2. β-1,3 Glucanase Assay
2.6.3. β-1,4 Glucanase Assay
2.6.4. Chitinase Assay
Preparation of Colloidal Chitin
Enzyme Assay
2.6.5. Protease Assay
2.7. Secondary Metabolites Profiling
2.7.1. Extraction and Separation of Antifungal Metabolites
2.7.2. GC-MS Analysis
2.8. Thin Layer Chromatography (TLC) Assay
2.9. Data Analysis
3. Results
3.1. Morphological Identification of T. asperellum Isolates
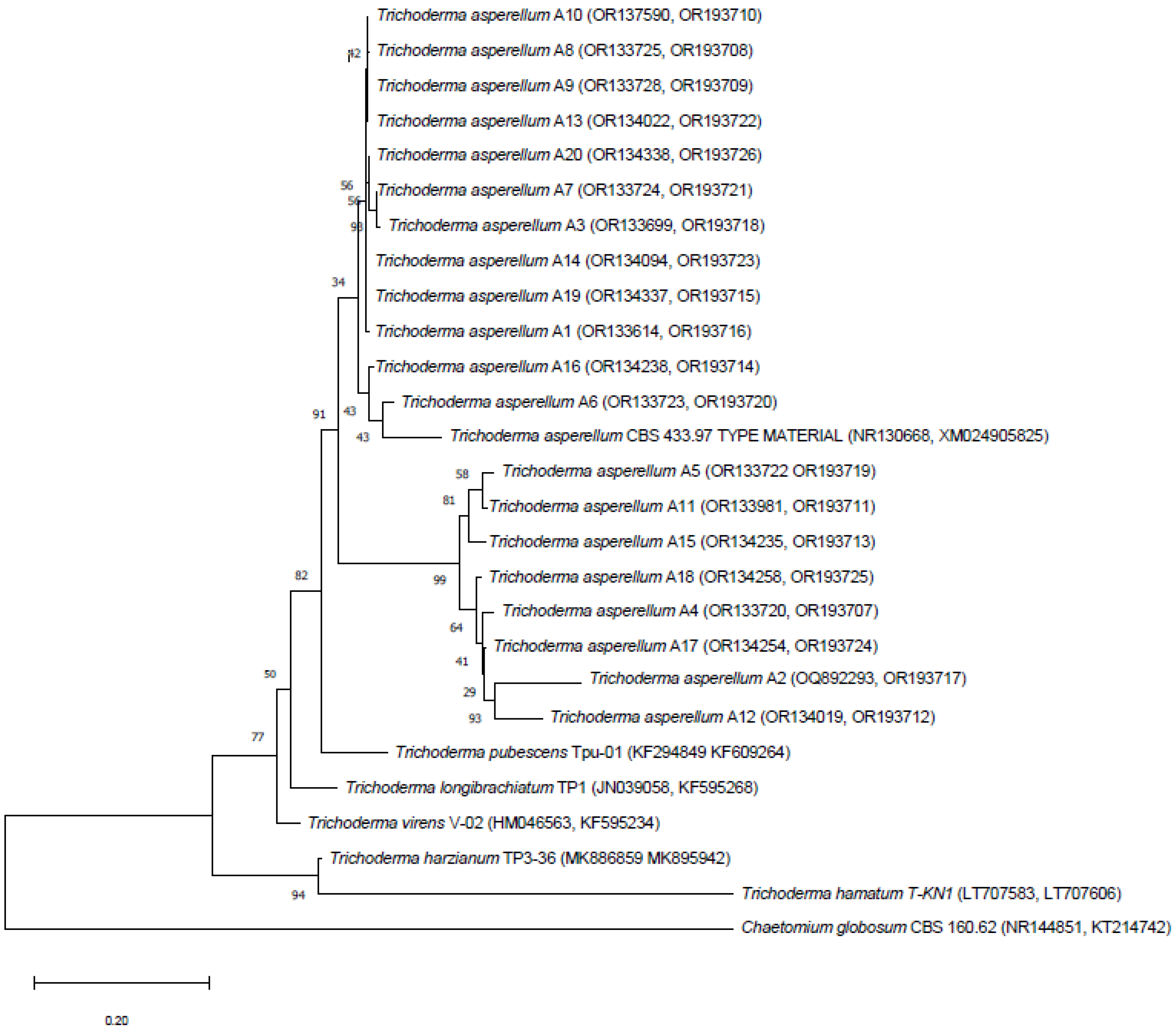
3.2. Molecular Identification and Phylogenetic Analysis
3.3. In Vitro Antagonistic Assay
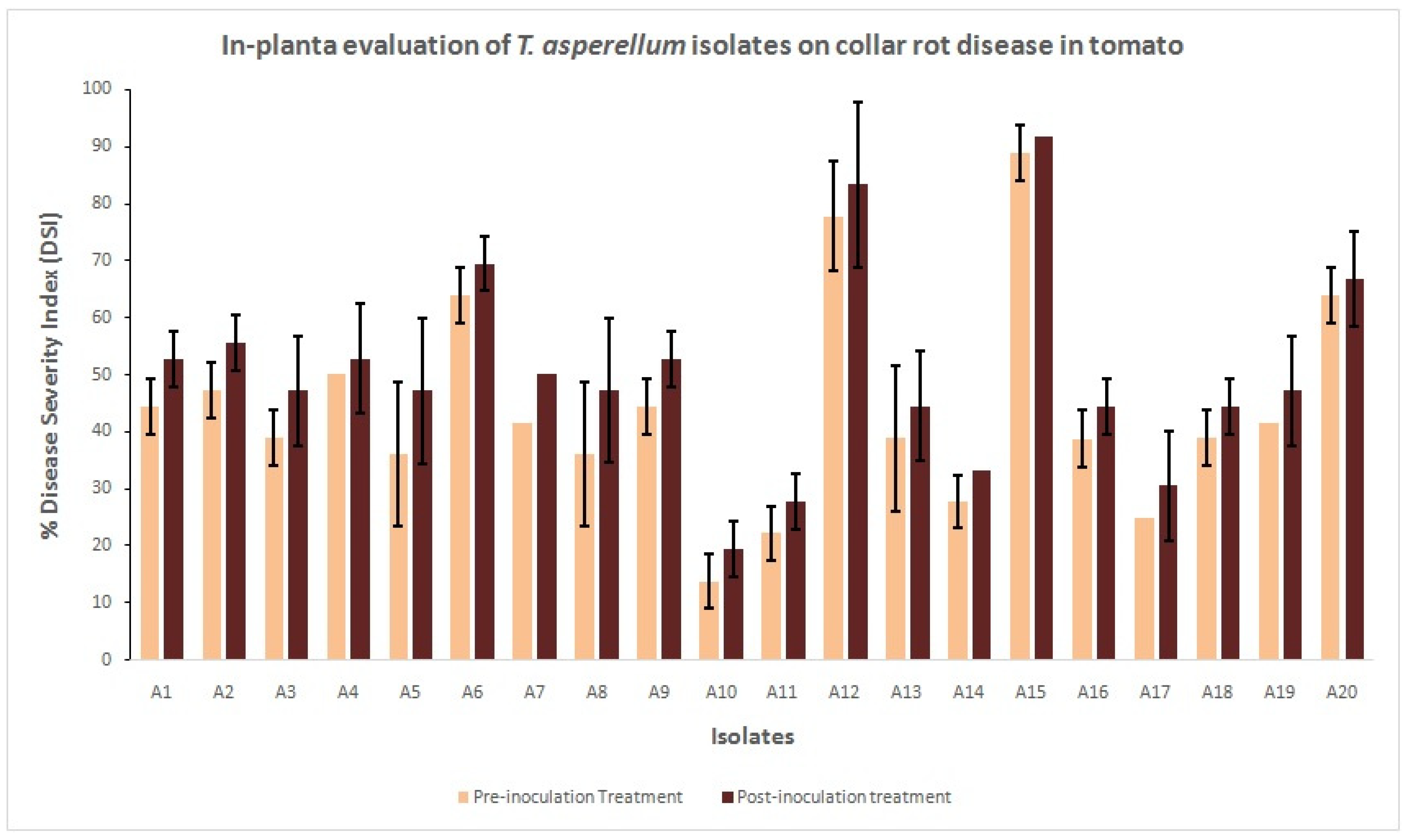
3.4. In Planta Bio-Control Assay
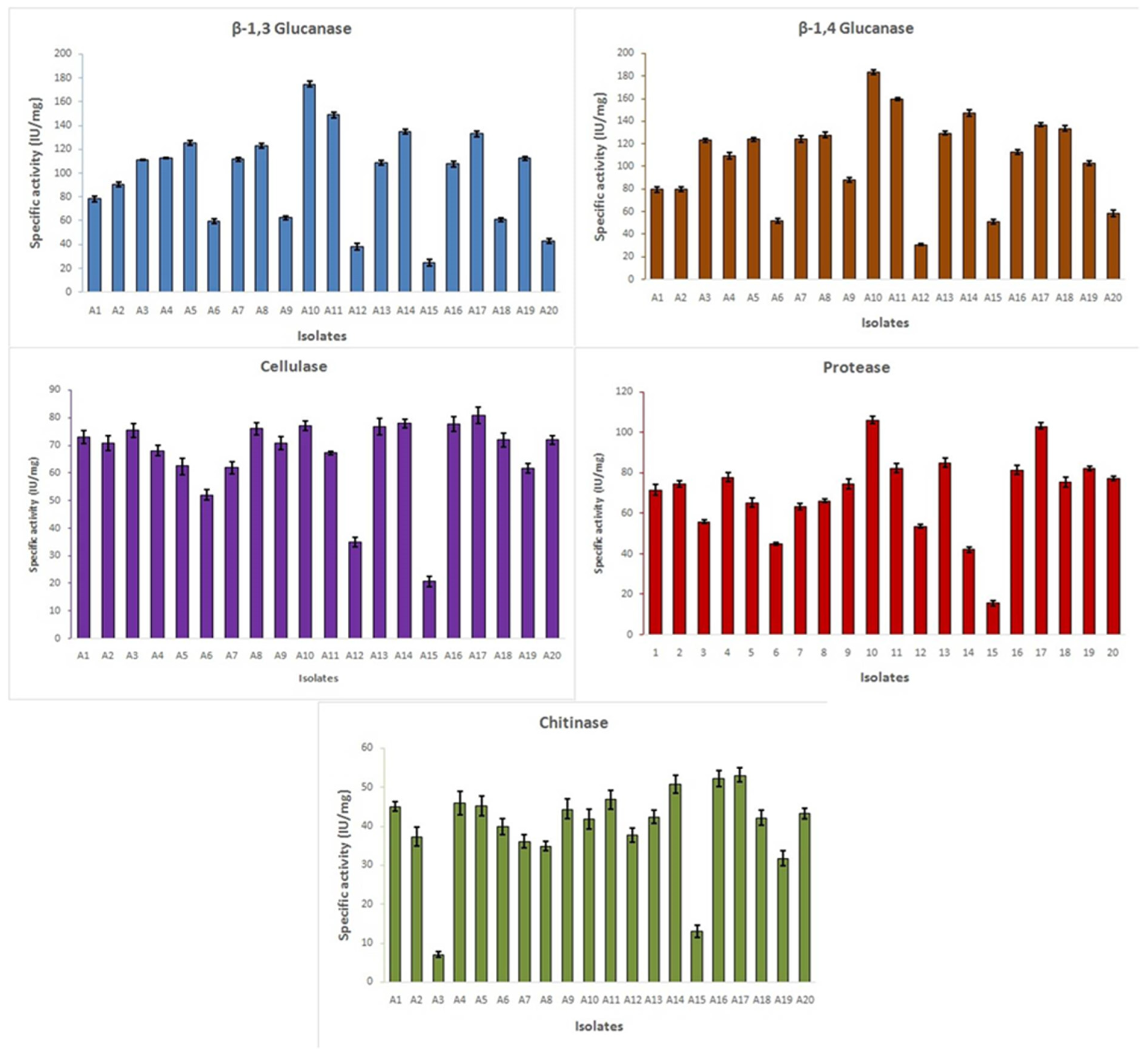
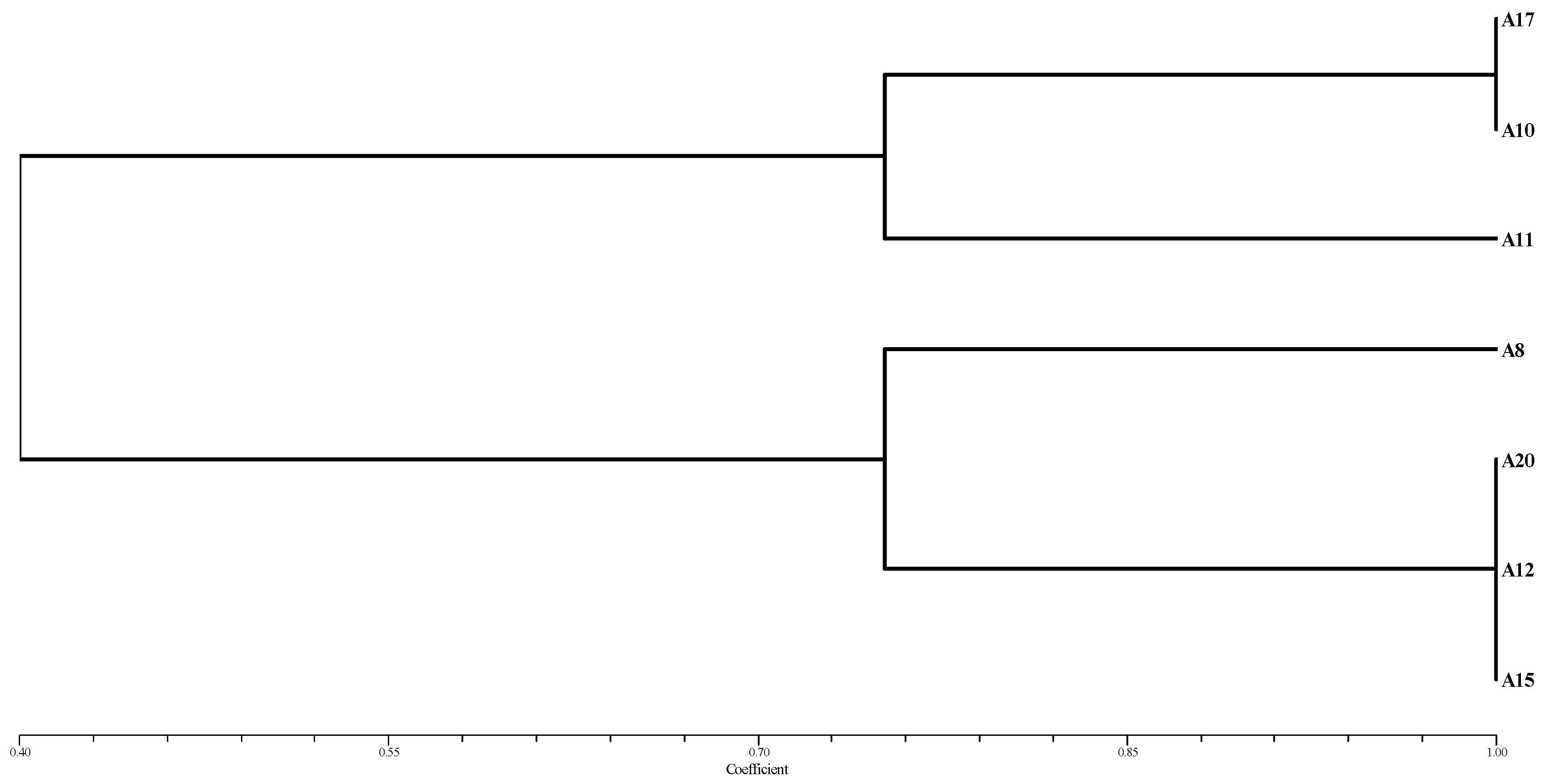
3.5. Enzyme Assay
3.6. Comparative Analysis of Volatile Organic Compounds of T. asperellum Isolates
3.7. TLC Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nalim, F.A.; Starr, J.L.; Woodard, K.E.; Segner, S.; Keller, N.P. Mycelial compatibility groups in Texas peanut field populations of Sclerotium rolfsii. Phytopathology 1995, 85, 1507–1512. [Google Scholar] [CrossRef]
- Aycock, R. Stem Rot and Other Diseases Caused by Sclerotium Rolfsii or the Status of Rolf’s Fungus after 70 Years; North Carolina Agricultural Experiment Station Technical Bulletin No.175; North Carolina Agricultural Experiment Station: Salisbury, NC, USA, 1966; pp. 1–202. [Google Scholar]
- Cilliers, A.; Herselman, L.; Pretorius, Z. Genetic variability within and among mycelial compatibility groups of Sclerotium rolfsii in South Africa. Phytopathology 2000, 90, 1026–1031. [Google Scholar] [CrossRef]
- Harlton, C.E.; Uvesque, C.A.; Punja, Z.K. Genetic diversity in Sclerotium (Athelia) rolfsii and related species. Phytopathology. 1995, 85, 1269–1281. [Google Scholar] [CrossRef]
- Bera, S.K.; Kasundra, S.V.; Kamdar, J.H.; Ajay, B.C.; Lal, C.; Thirumalasmy, P.P. Variable response of interspecific breeding lines of groundnut to Sclerotium rolfsii infection under field and laboratory conditions. Electron. J. Plant Breed. 2014, 5, 22–29. [Google Scholar]
- Bosamia, T.C.; Dodia, S.M.; Mishra, G.P.; Ahmad, S.; Joshi, B.; Thirumalaisamy, P.P.; Kumar, N.; Rathnakumar, A.L.; Sangh, C.; Kumar, A.; et al. Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea L.) using comparative RNA-Seq analysis of resistant and susceptible genotypes. PLoS ONE 2020, 15, e0236823. [Google Scholar] [CrossRef] [PubMed]
- Mahato, A.; Biswas, M.K.; Patra, S. Prevalence of collar rot of tomato caused by Sclerotium rolfsii (Sacc.) under the red and lateritic zone of West Bengal, India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3231–3236. [Google Scholar] [CrossRef]
- Punja, Z.K. The biology, ecology and control of Sclerotium rolfsii. Annu. Rev. Phytopathol. 1985, 23, 97–127. [Google Scholar] [CrossRef]
- Guzmán-Guzmán, P.; Kumar, A.; de Los Santos-Villalobos, S.; Parra-Cota, F.I.; Orozco-Mosqueda, M.D.C.; Fadiji, A.E.; Hyder, S.; Babalola, O.O.; Santoyo, G. Trichoderma species: Our best fungal allies in the biocontrol of plant diseases—A review. Plants 2023, 12, 432. [Google Scholar] [CrossRef] [PubMed]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The secrets of a Multitalented Biocontrol Agent. Plants 2020, 7, 962. [Google Scholar] [CrossRef]
- Verma, M.; Brar, S.K.; Tyagi, R.D.; Surampalli, R.Y.; Valéro, J.R. Antagonistic Fungi, Trichoderma Spp.: Panoply of Biological Control. Biochem. Eng. J. 2007, 37, 1–20. [Google Scholar] [CrossRef]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 2023, 21, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Hjeljord, L.; Tronsmo, A.; Harman, G.E.; Kubicek, C.P. Trichoderma and Gliocladium in biological control: An overview. In Trichoderma and Gliocladium, Enzymes, Biological Control and Commercial Applications; Kubicek, C.P., Harman, G.E., Eds.; Taylor and Francis: London, UK, 1998; pp. 131–151. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Pentilla, M.E. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In Trichoderma and Gliocladium. Vol.2. Enzymes, Biological Control and Commercial Applications; Harman, G.E., Kubicek, C.P., Eds.; Taylor and Francis Ltd.: London, UK, 1998; pp. 49–71. [Google Scholar]
- Howell, C.R. The role of antibiosis in bio-control. In Trichoderma and Gliocladium; Harman, G.E., Kubicek, C.P., Eds.; Taylor and Francis: London, UK, 1998; Volume 2, pp. 173–184. [Google Scholar]
- Howell, C.R. Mechanisms employed by Trichoderma species in the biological control of plant diseases: The history and evolution of current concepts. Plant Dis. 2003, 87, 4–10. [Google Scholar] [CrossRef]
- Paparizas, G.C. Trichoderma and Gliocladium: Biology, ecology and potential for bio-control. Annu. Rev. Phytopathol. 1985, 23, 23–54. [Google Scholar] [CrossRef]
- Narasimha Murthy, K.; Nirmala Devi, D.; Srinivas, C. Efficacy of Trichoderma asperellum against Ralstonia solanacearum under greenhouse conditions. Ann. Plant Sci. 2013, 2, 342–350. [Google Scholar]
- Wu, Q.; Sun, R.; Ni, M.; Yu, J.; Li, Y.; Yu, C.; Dou, K.; Ren, J.; Chen, J. Identification of a novel fungus, Trichoderma asperellum GDFS1009, and comprehensive evaluation of its bio-control efficacy. PLoS ONE 2017, 12, e0179957. [Google Scholar] [CrossRef]
- Moussa, Z.; Alanazi, Y.F.; Khateb, A.M.; Eldadamony, N.M.; Ismail, M.M.; Saber, W.I.; Darwish, D.B.E. Domiciliation of Trichoderma asperellum Suppresses Globiosporangium ultimum and Promotes Pea Growth, Ultrastructure, and Metabolic Features. Microorganisms 2023, 11, 198. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Ichikawa, H.; Naznin, H.A.; Kogure, A.; Hyakumachi, M. Systemic resistance induced in Arabidopsis thaliana by Trichoderma asperellum SKT-1, a microbial pesticide of seed borne diseases of rice. Pest Manag. Sci. 2011, 68, 60–66. [Google Scholar] [CrossRef]
- Shoresh, M.; Yedidia, I.; Chet, I. Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 2005, 95, 76–84. [Google Scholar] [CrossRef]
- Herrera, W.; Valbuena, O.; Pavone-Maniscalco, D. Formulation of Trichoderma asperellum TV190 for biological control of Rhizoctonia solani on corn seedlings. Egypt. J. Biol. Pest Control. 2020, 30, 44. [Google Scholar] [CrossRef]
- Brotman, Y.; Briff, E.; Viterbo, A.; Chet, I. Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 2008, 147, 779–789. [Google Scholar] [CrossRef]
- Guo, R.; Ji, S.; Wang, Z.; Zhang, H.; Wang, Y.; Liu, Z. Trichoderma asperellum xylanases promote growth and induce resistance in poplar. Microbiol. Res. 2021, 248, 126767. [Google Scholar] [CrossRef]
- Srinivasa, N.; Sriram, S.; Singh, C.; Shivashankar, K.S. Secondary metabolites approach to study the bio-efficacy of Trichoderma asperellum isolates in India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1105–1123. [Google Scholar] [CrossRef][Green Version]
- Degani, O.; Khatib, S.; Becher, P.; Gordani, A.; Harris, R. Trichoderma asperellum secreted 6-Pentyl-α-Pyrone to control Magnaporthiopsis maydis, the maize late wilt disease agent. Biology 2021, 10, 897. [Google Scholar] [CrossRef] [PubMed]
- Geraldine, A.M.; Lopes, F.A.C.; Carvalho, D.D.C.; Barbosa, E.T.; Rodrigues, A.R.; Brandão, R.S.; Ulhoa, C.J.; Junior, M.L. Cell wall-degrading enzymes and parasitism of sclerotia are key factors on field bio-control of white mold by Trichoderma spp. Biol. Control. 2013, 67, 308–316. [Google Scholar] [CrossRef]
- Qualhato, T.F.; Lopes, F.A.C.; Steindorff, A.S.; Brandao, R.S.; Jesuino, R.S.A.; Ulhoa, C.J. Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: Evaluation of antagonism and hydrolytic enzyme production. Biotechnol. Lett. 2013, 35, 1461–1468. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Yu, C.; Dou, K.; Wang, M.; Li, Y.; Chen, J. Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced bio-control of Fusarium oxysporum f. sp. cucumerinum. Biol. Control. 2016, 94, 37–46. [Google Scholar] [CrossRef]
- Silva, B.D.S.; Ulhoa, C.J.; Batista, K.A.; Yamashita, F.; Fernandes, K.F. Potential fungal inhibition by immobilized hydrolytic enzymes from Trichoderma asperellum. J. Agric. Food Chem. 2011, 59, 8148–8154. [Google Scholar] [CrossRef]
- Snehal, D.F. Isolation and identification of fungi from contaminated soil to build biological resource as biocontrol activity. Macromol. Ind. J. 2017, 12, 105. [Google Scholar]
- Cullings, K.W. Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Mol. Ecol. 1992, 1, 233–240. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Thokala, P.; Narayanasamy, P.; Kamil, D.; Choudhary, S.P. Polyphasic taxonomy of Indian Trichoderma species. Phytotaxa 2021, 502, 1–27. [Google Scholar] [CrossRef]
- Garcia, E.F. Screening of fungal antagonist to control Sclerotium cepivorum. In New Approaches in Biological Control of Soil Borne Diseases; Jensen, D.F., Ed.; IOBCM/PRS Bull.: Copenhagen, Denmark, 1991; pp. 79–81. [Google Scholar]
- Morton, D.T.; Stroube, N.H. Antagonistic and stimulatory effect of microorganism upon Sclerotium rolfsii. Phytopathology 1955, 45, 419–420. [Google Scholar]
- Dennis, C.; Webster, J. Antagonistic properties of species groups of Trichoderma. II. Production of volatile antibiotics. Trans. Brit. Mycol. Soc. 1971, 57, 363–369. [Google Scholar] [CrossRef]
- Quimio, A.J.; Cumagun, C.J. Workbook on Tropical fungi: Collection, Isolation and Identification; The Mycological Society of the Philippines, Inc.: Laguna, Philippines, 2001. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Nelson, N.A. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 1944, 153, 375. [Google Scholar] [CrossRef]
- Thrane, C.; Jensen, D.F.; Tronsmo, A. Substrate colonization, strain competition, enzyme production in vitro, and bio-control of Pythium ultimum by Trichoderma spp. isolates P1 and T3. Eur. J. Plant Pathol. 2000, 106, 215–225. [Google Scholar] [CrossRef]
- Vessey, J.C.; Pegg, G.F. Autolysis and chitinase production in cultures of Verticillium albo-atrum. Trans. Br. Mycol. Soc. 1973, 60, 133–143. [Google Scholar] [CrossRef]
- Sahai, A.S.; Manocha, M.S. Chitinases of fungi and plants: Their involvement in morphogenesis and host-parasite interaction. FEMS Microbiol. Rev. 1993, 11, 317–338. [Google Scholar] [CrossRef]
- Chaithra, M.; Prameeladevi, T.; Prasad, L.; Kundu, A.; Bhagyasree, S.N.; Subramanian, S.; Kamil, D. Metabolomic diversity of local strains of Beauveria bassiana (Balsamo) Vuillemin and their efficacy against the cassava mite, Tetranychustruncatus Ehara (Acari: Tetranychidae). PLoS ONE 2022, 17, e0277124. [Google Scholar] [CrossRef]
- Pandian, R.T.P.; Raja, M.; Kumar, A.; Sharma, P. Morphological and molecular characterization of Trichoderma asperellum strain Ta13. Indian Phytopathol. 2016, 69, 297–303. [Google Scholar]
- Sriram, S.; Savitha, M.J.; Rohini, H.S.; Jalali, S.K. The most widely used fungal antagonist for plant disease management in India, Trichoderma viride is Trichoderma asperellum as confirmed by oligonucleotide barcode and morphological characters. Curr. Sci. 2013, 2023, 1332–1340. [Google Scholar]
- Chaithra, M.; Prameeladevi, T.; Bhagyasree, S.N.; Prasad, L.; Subramanian, S.; Kamil, D. Multilocus sequence analysis for population diversity of indigenous entomopathogenic fungus Beauveria bassiana and its bio-efficacy against the cassava mite, Tetranychus truncatus Ehara (Acari: Tetranychidae). Front. Microbiol. 2022, 13, 1007017. [Google Scholar] [CrossRef]
- Poosapati, S.; Ravulapalli, P.D.; Tippirishetty, N.; Vishwanathaswamy, D.K.; Chunduri, S. Selection of high temperature and salinity tolerant Trichoderma isolates with antagonistic activity against Sclerotium rolfsii. SpringerPlus 2014, 3, 641. [Google Scholar] [CrossRef]
- Manjunath, M.; Singh, A.; Tripathi, A.N.; Prasanna, R.; Rai, A.B.; Singh, B. Bioprospecting the fungicides compatible Trichoderma asperellum isolate effective against multiple plant pathogens in vitro. J. Environ. Biol. 2017, 38, 553. [Google Scholar] [CrossRef]
- Akash, A.U.; Ramya, V.; Uma Devi, G.; Pushpavalli, S.N.C.V.L.; Triveni, S. Antagonist activities of native rhizosphere micro-flora against groundnut stem rot pathogen, Sclerotium rolfsii Sacc. Egypt. J. Biol. Pest Control. 2022, 32, 133. [Google Scholar] [CrossRef]
- Guzman-Valle, P.; Bravo-Luna, L.; Montes-Belmont, R.; Guigon-Lopez, C.; Sepulveda-Jimenez, G. Induction of resistance to Sclerotium rolfsii in different varieties of onion by inoculation with Trichoderma asperellum. Eur. J. Plant Pathol. 2014, 138, 223–229. [Google Scholar] [CrossRef]
- El-Komy, M.H.; Al-Qahtani, R.M.; Ibrahim, Y.E.; Almasrahi, A.A.; Al-Saleh, M.A. Soil application of Trichoderma asperellum strains significantly improves Fusarium root and stem rot disease management and promotes growth in cucumbers in semi-arid regions. Eur. J. Plant Pathol. 2022, 162, 637–653. [Google Scholar] [CrossRef]
- Ayyandurai, M.; Akila, R.; Manonmani, K.; Harish, S.; Mini, M.L.; Vellaikumar, S. Deciphering the mechanism of Trichoderma spp. consortia possessing volatile organic compounds and antifungal metabolites in the suppression of Sclerotium rolfsii in groundnut. Physiol. Mol. Plant Pathol. 2023, 125, 102005. [Google Scholar] [CrossRef]
- Harman, G.E. Overview of mechanisms and uses of Trichoderma spp. Phytopathology 2006, 96, 190–194. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, J.; Yang, K.; Wang, Y.; Tian, Y.; Liang, Z. Transcriptomic and metabonomic insights into the bio-control mechanism of Trichoderma asperellum M45a against watermelon Fusarium wilt. PLoS ONE 2022, 17, e0272702. [Google Scholar] [CrossRef]
- Degani, O.; Rabinovitz, O.; Becher, P.; Gordani, A.; Chen, A. Trichoderma longibrachiatum and Trichoderma asperellum confer growth promotion and protection against late wilt disease in the field. J. Fungi. 2021, 7, 444. [Google Scholar] [CrossRef]
- El-Komy, M.H.; Saleh, A.A.; Ibrahim, Y.E.; Hamad, Y.K.; Molan, Y.Y. Trichoderma asperellum strains confer tomato protection and induce its defense-related genes against the Fusarium wilt pathogen. Tropical Plant Pathol. 2016, 41, 277–287. [Google Scholar] [CrossRef]
- Karuppiah, V.; He, A.; Lu, Z.; Wang, X.; Li, Y.; Chen, J. Trichoderma asperellum GDFS1009-mediated maize resistance against Fusarium graminearum stalk rot and mycotoxin degradation. Biol. Control. 2022, 174, 105026. [Google Scholar] [CrossRef]
- Rivera-Mendez, W.; Obregon, M.; Moran-Diez, M.E.; Hermosa, R.; Monte, E. Trichoderma asperellum bio-control activity and induction of systemic defenses against Sclerotium cepivorum in onion plants under tropical climate conditions. Biol. Control. 2020, 141, 104145. [Google Scholar] [CrossRef]
- Inbar, J.; Chet, I. The role of recognition in the induction of specific chitinases during mycoparasitism by Trichoderma harzianum. Microbiology 1995, 141, 2823–2829. [Google Scholar] [CrossRef]
- El-Komy, M.H.; Saleh, A.A.; Eranthodi, A.; Molan, Y.Y. Characterization of novel Trichoderma asperellum isolates to select effective bio-control agents against tomato Fusarium wilt. Plant Pathol. J. 2015, 31, 50. [Google Scholar] [CrossRef] [PubMed]
- Samuels, G.J.; Lieckfeldt, E.L.K.E.; Nirenberg, H.I. Trichoderma asperellum, a new species with warted conidia, and redescription of T. viride. Sydowia 1999, 51, 71–88. [Google Scholar]
- Kangjam, V.; Pongener, N.; Singh, R.; Banik, S.; Daiho, L.; Ao, N.T. In vitro Screening for Potential Antagonistic Activity of Trichoderma Isolates against Sclerotium rolfsii Causing Collar Rot of French Bean. Environ. Ecol. 2023, 41, 623–632. [Google Scholar]
- Srinivasa, N.; Kamil, D.; Singh, C.; Singode, A.; Gupta, D. Molecular and Biochemical Characterization of Potential Isolates of Trichoderma Species Effective against Soil-Borne Pathogens. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3132–3149. [Google Scholar] [CrossRef]
- Bhardwaj, N.R.; Kumar, J. Characterization of volatile secondary metabolites from Trichoderma asperellum. J. Appl. Nat. Sci. 2017, 9, 954–959. [Google Scholar] [CrossRef]
- Scudeletti, D.; Crusciol, C.A.C.; Bossolani, J.W.; Moretti, L.G.; Momesso, L.; Servaz Tubana, B.; De Castro, S.G.Q.; De Oliveira, E.F.; Hungria, M. Trichoderma asperellum inoculation as a tool for attenuating drought stress in sugarcane. Front. Plant Sci. 2021, 12, 645542. [Google Scholar] [CrossRef] [PubMed]
- Degani, O.; Gordani, A. New Antifungal Compound, 6-Pentyl-α-Pyrone, against the Maize Late Wilt Pathogen, Magnaporthiopsis maydis. Agronomy 2022, 12, 2339. [Google Scholar] [CrossRef]
- Man, L.I.U.; Qichen, N.I.U.; Shuxia, Y.I.N.; Ziyue, W.A.N.G. Inhibitory activity of 6-pentyl-2H-pyran-2-one against the pathogenesis fungi of dollar spot and its control efficacy. J. Pestic. Sci. 2023, 25, 104–116. [Google Scholar] [CrossRef]
- Hao, J.; Wuyun, D.; Xi, X.; Dong, B.; Wang, D.; Quan, W.; Zhang, Z.; Zhou, H. Application of 6-Pentyl-α-Pyrone in the Nutrient Solution Used in Tomato Soilless Cultivation to Inhibit Fusarium oxysporum HF-26 Growth and Development. Agronomy 2023, 13, 1210. [Google Scholar] [CrossRef]
- Aggarwal, R.; Tewari, A.K.; Srivastava, K.D.; Singh, D.V. Role of antibiosis in the biological control of spot blotch (Cochliobolus sativus) of wheat by Chaetomium globosum. Mycopathologia 2004, 157, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Saputra, H.; Puspota, F.; Tjandrawati, T. Production of an antibacterial compound against the plant pathogen Erwinia carotovora subsp. carotovora by the bio-control strain Gliocladium sp. TN C73. J. Agric. Tech. 2013, 9, 1157–1165. [Google Scholar]
- Adnan, L.A.; Sathishkumar, P.; Yusoff, A.R.M.; Hadibarata, T. Metabolites characterisation of laccase mediated Reactive Black 5 biodegradation by fast growing ascomycete fungus Trichoderma atroviride F03. Int. Biodeterior. Biodegrad. 2015, 104, 274–282. [Google Scholar] [CrossRef]
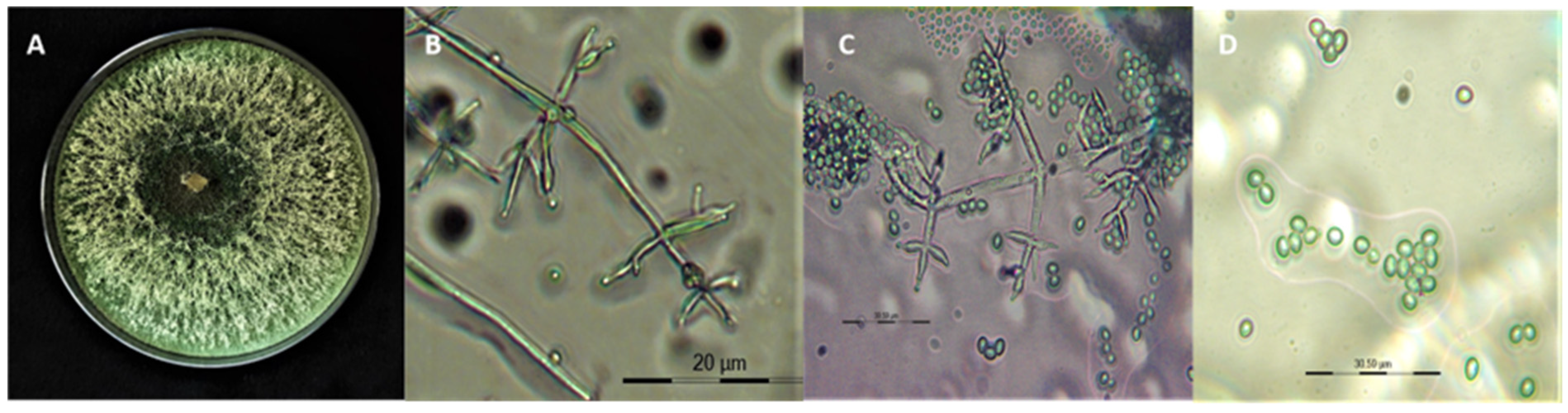



| Enzymes | Substrates | Supplier and Product Number | Standards (1 mg/mL Stock) | OD (in nm) |
|---|---|---|---|---|
| Cellulase | Cellulose (0.5%) | Sigma-Aldrich—435236 | Glucose | 530 |
| β-1,3 glucanase | Laminarin (3.2 mg/mL distilled water) | Sigma-Aldrich—L9634 | Glucose | 530 |
| β-1,4 glucanase | Carboxy methyl cellulose (1%) | Himedia—GRM329 | Glucose | 575 |
| Chitinase | Colloidal chitin (0.5%) | Himedia—GRM1356 | NAG | 420 |
| Protease | Casein (1%) | Sigma-Aldrich—C9801 | Tyrosine | 280 |
| Isolates Code | Collection Source and Location | GPS Location | ITS a | β-Tubulin a | Colony Characteristics | |
|---|---|---|---|---|---|---|
| Latitude | Longitude | |||||
| A1 | Soil, IARI | 28°38′ N | 77°10′ E | OR133614 | OR193716 | Abundant mycelium with dark green spores |
| A2 | Soil, IARI | 28°38′ N | 77°10′ E | OQ892293 | OR193717 | Abundant mycelium with yellowish-green spores |
| A3 | Rhizosphere, Guntur | 16°18′ N | 80°27′ E | OR133699 | OR193718 | Less aerial mycelium with dark green centered spores |
| A4 | Groundnut field, TN | 11°39′ N | 78°12′ E | OR133720 | OR193707 | Abundant mycelium with yellowish-green spores |
| A5 | Vegetable field, TN | 11°3′ N | 77°17′ E | OR133722 | OR193719 | Dense dark green spores with less aerial mycelium form rings |
| A6 | Soil, IIHR, KA | 13°7′ N | 72°29′ E | OR133723 | OR193720 | Dense dark green spores with cottony mycelium |
| A7 | Soil, Akola | 20°42′ N | 76°59′ E | OR133724 | OR193721 | Cottony mycelium with light green spores |
| A8 | Soil, Tirupathi | 13°37′ N | 79°25′ E | OR133725 | OR193708 | Dark green spores forming rings with cottony mycelium |
| A9 | Soil, Navsari | 20°57′ N | 72°55′ E | OR133728 | OR193709 | Dense light to dark greenish spores with aerial mycelium |
| A10 | Soil, Navsari | 20°57′ N | 72°55′ E | OR137590 | OR193710 | Fast-growing, dense dark green spores with aerial mycelium |
| A11 | Soil, Jaipur | 26°55′ N | 75°49′ E | OR133981 | OR193711 | Fast-growing, abundant cottony mycelium forms spores at later |
| A12 | Veg. field, IARI | 28°38′ N | 77°10′ E | OR134019 | OR193712 | Dark green-centered spores with aerial mycelium |
| A13 | Chilli field, IARI | 28°38′ N | 77°10′ E | OR134022 | OR193722 | Aerial mycelium with abundant yellowish to light green spores |
| A14 | Soil, Lucknow | 26°50′ N | 80°55′ E | OR134094 | OR193723 | Abundant aerial mycelium with dense dark green spores |
| A15 | Soil, Varanasi | 25°19′ N | 82°58′ E | OR134235 | OR193713 | Less mycelium, slow growing with very less spores |
| A16 | Soil, Barracpore | 22°45′ N | 88°22′ E | OR134238 | OR193714 | Dense light green spores with aerial mycelium |
| A17 | Soil, IIHR | 13°7′ N | 72°29′ E | OR134254 | OR193724 | Fast-growing, dense aerial mycelium forms spores at later |
| A18 | Soil, Navsari | 20°57′ N | 72°55′ E | OR134258 | OR193725 | Dense light green spores abundant throughout the plate |
| A19 | Soil, Navsari | 20°57′ N | 72°55′ E | OR134337 | OR193715 | Dense yellowish to light green spores with less mycelium |
| A20 | Soil, NBPGR | 28°38′ N | 77°10′ E | OR134338 | OR193726 | Dense dark green spores with less aerial mycelium |
| Isolates | Percent Inhibition of A. rolfsii Growth | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dual Culture Assay | Volatile Assay | |||||||||
| R1 | R2 | R3 | Average | SD | R1 | R2 | R3 | Average | SD | |
| A1 | 64.58 | 64.22 | 64.15 | 64.32 f | 0.23 | 40.61 | 41.50 | 41.85 | 41.32 k | 0.64 |
| A2 | 64.62 | 64.50 | 63.66 | 64.26 f | 0.52 | 45.20 | 44.00 | 44.50 | 44.57 j | 0.60 |
| A3 | 66.69 | 66.20 | 65.90 | 66.26 e | 0.40 | 48.15 | 46.20 | 46.50 | 46.95 i | 1.05 |
| A4 | 68.62 | 67.50 | 68.22 | 68.11 d | 0.57 | 50.16 | 48.26 | 48.80 | 49.07 h | 0.98 |
| A5 | 68.89 | 67.82 | 66.95 | 67.89 d | 0.97 | 50.88 | 49.60 | 49.22 | 49.90 h | 0.87 |
| A6 | 50.61 | 51.20 | 52.33 | 51.38 h | 0.87 | 28.06 | 26.30 | 26.75 | 27.04 m | 0.91 |
| A7 | 67.10 | 67.85 | 68.45 | 67.80 d | 0.68 | 51.66 | 50.20 | 49.24 | 50.37 gh | 1.22 |
| A8 | 69.23 | 68.50 | 68.00 | 68.58 d | 0.62 | 53.24 | 51.76 | 51.90 | 52.30 f | 0.82 |
| A9 | 62.31 | 60.50 | 61.45 | 61.42 g | 0.91 | 46.32 | 46.90 | 46.15 | 46.46 i | 0.39 |
| A10 | 95.23 | 94.60 | 94.15 | 94.66 a | 0.54 | 72.10 | 70.15 | 70.60 | 70.95 a | 1.02 |
| A11 | 82.64 | 80.69 | 84.59 | 82.64 b | 1.95 | 65.72 | 63.16 | 62.38 | 63.75 c | 1.75 |
| A12 | 30.58 | 31.22 | 31.00 | 30.93 i | 0.33 | 22.64 | 22.55 | 21.80 | 22.33 n | 0.46 |
| A13 | 70.54 | 71.66 | 71.50 | 71.23 c | 0.61 | 60.50 | 61.25 | 61.80 | 61.18 d | 0.65 |
| A14 | 70.12 | 71.50 | 70.88 | 70.83 c | 0.69 | 60.12 | 59.00 | 59.25 | 59.46 e | 0.59 |
| A15 | 22.82 | 18.92 | 20.87 | 20.87 j | 1.95 | 11.20 | 12.30 | 11.15 | 11.55 op | 0.65 |
| A16 | 68.23 | 67.77 | 68.00 | 68.00 d | 0.23 | 59.88 | 58.20 | 56.30 | 58.13 e | 1.79 |
| A17 | 80.46 | 82.10 | 81.00 | 81.19 b | 0.84 | 69.21 | 67.24 | 68.35 | 68.27 b | 0.99 |
| A18 | 65.91 | 65.25 | 64.20 | 65.12 ef | 0.86 | 52.65 | 51.60 | 51.25 | 51.83 fg | 0.73 |
| A19 | 68.70 | 68.45 | 68.00 | 68.38 d | 0.35 | 50.12 | 49.45 | 50.00 | 49.86 h | 0.36 |
| A20 | 52.35 | 51.00 | 53.70 | 52.35 h | 1.35 | 31.22 | 29.45 | 28.60 | 29.76 l | 1.34 |
| CD @ 5% | 1.497 | 1.603 | ||||||||
| SEm ± | 0.823 | 0.943 | ||||||||
| CV (%) | 1.411 | 2.034 | ||||||||
| Isolates | % Disease Severity Index (DSI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-Inoculation Treatment | Post-Inoculation Treatment | |||||||||
| R1 | R2 | R3 | Average | SD | R1 | R2 | R3 | Average | SD | |
| A1 | 50 | 41.67 | 41.67 | 44.45 | 4.81 | 50 | 50 | 58.33 | 52.78 | 4.81 |
| A2 | 50 | 50 | 41.67 | 47.22 | 4.81 | 50 | 58.33 | 58.33 | 55.55 | 4.81 |
| A3 | 41.67 | 41.67 | 33.33 | 38.89 | 4.82 | 58.33 | 41.67 | 41.67 | 47.22 | 9.62 |
| A4 | 50 | 50 | 50 | 50.00 | 0.00 | 58.33 | 58.33 | 41.67 | 52.78 | 9.62 |
| A5 | 50 | 25 | 33.3 | 36.11 | 12.73 | 58.33 | 50 | 33.3 | 47.22 | 12.75 |
| A6 | 66.67 | 66.67 | 58.33 | 63.89 | 4.82 | 66.67 | 66.67 | 75 | 69.45 | 4.81 |
| A7 | 41.67 | 41.67 | 41.67 | 41.67 | 0.00 | 50 | 50 | 50 | 50.00 | 0.00 |
| A8 | 50 | 25 | 33.3 | 36.11 | 12.73 | 58.33 | 50 | 33.37 | 47.22 | 12.71 |
| A9 | 50 | 41.67 | 41.67 | 44.48 | 4.81 | 50 | 58.33 | 50 | 52.78 | 4.81 |
| A10 | 16.6 | 8.3 | 16.6 | 13.83 | 4.79 | 16.6 | 16.6 | 25 | 19.40 | 4.85 |
| A11 | 25 | 25 | 16.6 | 22.20 | 4.85 | 25 | 25 | 33.33 | 27.78 | 4.81 |
| A12 | 66.67 | 83.33 | 83.33 | 77.77 | 9.62 | 66.67 | 91.67 | 91.67 | 83.33 | 14.43 |
| A13 | 50 | 41.67 | 25 | 38.89 | 12.73 | 33.33 | 50 | 50 | 44.44 | 9.62 |
| A14 | 25 | 25 | 33 | 27.67 | 4.62 | 33.33 | 33.33 | 33.33 | 33.33 | 0.00 |
| A15 | 91.67 | 91.67 | 83.33 | 88.89 | 4.82 | 91.67 | 91.67 | 91.67 | 91.67 | 0.00 |
| A16 | 41.67 | 41.67 | 33 | 38.78 | 5.01 | 41.67 | 41.67 | 50 | 44.45 | 4.81 |
| A17 | 25 | 25 | 25 | 25.00 | 0.00 | 25 | 25 | 41.67 | 30.56 | 9.62 |
| A18 | 41.67 | 41.67 | 33.33 | 38.89 | 4.82 | 41.67 | 41.67 | 50 | 44.45 | 4.81 |
| A19 | 41.67 | 41.67 | 41.67 | 41.67 | 0.00 | 41.67 | 41.67 | 58.33 | 47.22 | 9.62 |
| A20 | 66.67 | 66.67 | 58.33 | 63.89 | 4.82 | 66.67 | 75 | 58.33 | 66.67 | 8.34 |
| Isolates | Specific Activity in IU/mg | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β-1,3 Glucanase | β-1,4 Glucanase | Cellulase | Protease | Chitinase | ||||||
| Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | |
| A1 | 78.25 i | 2.00 | 79.60 k | 2.30 | 73.00 cde | 2.30 | 71.53 f | 2.80 | 45.07 bcd | 1.28 |
| A2 | 90.25 h | 2.00 | 79.93 k | 2.40 | 70.94 ef | 2.70 | 74.53 ef | 1.60 | 37.33 fg | 2.32 |
| A3 | 111.00 ef | 0.25 | 123.13 g | 1.80 | 75.44 bcd | 2.50 | 55.81 h | 0.80 | 7.02 j | 0.77 |
| A4 | 112.75 e | 0.25 | 109.56 h | 2.90 | 68.09 fg | 1.80 | 77.83 d | 2.40 | 45.85 bc | 2.96 |
| A5 | 125.50 d | 2.00 | 124.18 fg | 1.70 | 62.47 h | 3.00 | 65.33 g | 2.40 | 45.17 bcd | 2.60 |
| A6 | 59.50 j | 2.00 | 52.10 m | 2.20 | 52.04 i | 1.90 | 44.85 i | 0.80 | 39.88 ef | 2.13 |
| A7 | 111.58 ef | 1.50 | 124.28 fg | 2.70 | 61.92 h | 2.30 | 63.38 g | 1.70 | 36.07 g | 1.62 |
| A8 | 122.92 d | 1.80 | 127.68 ef | 2.50 | 76.04 bc | 2.10 | 66.08 g | 0.90 | 34.85 gh | 1.24 |
| A9 | 62.40 j | 1.70 | 88.25 j | 2.10 | 70.87 efg | 2.40 | 74.53 ef | 2.50 | 44.37 bcd | 2.52 |
| A10 | 174.68 a | 2.30 | 183.48 a | 2.40 | 77.09 b | 1.80 | 106.06 a | 1.90 | 41.87 de | 2.54 |
| A11 | 149.08 b | 2.40 | 159.60 b | 1.30 | 67.26 g | 0.70 | 82.33 bc | 2.40 | 46.82 b | 2.39 |
| A12 | 37.93 l | 2.80 | 30.73 n | 0.90 | 34.99 j | 1.60 | 53.53 h | 0.90 | 37.65 fg | 1.76 |
| A13 | 108.96 fg | 1.80 | 129.58 e | 1.40 | 76.75 b | 2.90 | 85.13 b | 2.40 | 42.40 de | 1.77 |
| A14 | 134.68 c | 2.10 | 147.25 c | 2.60 | 77.94 ab | 1.50 | 42.03 i | 1.20 | 50.81 a | 2.32 |
| A15 | 24.79 m | 2.90 | 51.28 m | 1.90 | 20.84 k | 1.90 | 15.53 j | 1.30 | 13.07 i | 1.52 |
| A16 | 107.64 g | 2.50 | 112.95 h | 1.80 | 77.82 ab | 2.60 | 81.35 c | 2.20 | 52.17 a | 1.95 |
| A17 | 133.18 c | 2.40 | 136.85 d | 1.60 | 80.92 a | 2.90 | 103.08 a | 1.57 | 53.09 a | 1.85 |
| A18 | 60.91 j | 1.60 | 133.43 d | 2.40 | 72.04 de | 2.50 | 75.43 de | 2.41 | 42.12 de | 1.88 |
| A19 | 112.36 e | 1.90 | 102.83 i | 1.80 | 61.75 h | 1.80 | 82.05 c | 1.23 | 31.77 h | 1.98 |
| A20 | 43.10 k | 1.80 | 58.38 l | 2.90 | 72.02 de | 1.60 | 77.15 de | 1.26 | 43.22 cd | 1.33 |
| CD @ 5% | 3.318 | 3.543 | 3.651 | 3.052 | 3.315 | |||||
| SEm ± | 4.044 | 4.609 | 4.896 | 3.421 | 4.035 | |||||
| CV (%) | 2.050 | 1.992 | 3.327 | 2.647 | 5.082 | |||||
| Compounds | Functional Group | Molecular Formula | Isolates | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A10 | A17 | A12 | A15 | A8 | A11 | A20 | ||||||||||
| * RA % | RT ** | * RA % | RT ** | * RA % | RT ** | *RA % | RT ** | * RA % | RT ** | * RA % | RT ** | * RA % | RT ** | |||
| Acetic acid | Acid | CH3COOH | 3.99 ± 1.12 | 5.68 | 2.35 ± 0.42 | 4.83 | 0.53 ± 0.03 | 6.88 | 1.47 ± 0.39 | 12.53 | 0.15 ± 0.02 | 9.43 | 1.28 ± 0.62 | 16.52 | 0.40 ± 0.02 | 8.24 |
| n-Propyl acetate | Ester | C5H10O2 | 0.21 ± 0.06 | 5.82 | - | - | - | - | 0.40 ± 0.02 | 13.05 | - | - | - | - | - | - |
| 2,3-Butanediol | Alcohol | C4H10O2 | 4.99 ± 1.07 | 8.27 | - | - | 6.57 ± 1.32 | - | - | 23.41 ± 4.56 | 64.5 | 7.24 ± 1.64 | 42.67 | - | - | |
| 2-Furan carboxaldehyde | Aldehyde | C5H4O2 | 0.12 ± 0.03 | 10.55 | - | - | 0.97 ± 0.05 | - | - | 0.90 ± 0.02 | 14.2 | 1.88 ± 0.51 | 24.52 | - | - | |
| 5-Methyl furfural | Aldehyde | C6H6O2 | 0.12 ± 0.01 | 10.55 | 0.07 ± 0.02 | 12.65 | 0.97 ± 0.06 | - | - | - | - | 0.94 ± 0.11 | 18.97 | - | - | |
| Dodecane | Hydrocarbon | C12H26 | 0.04 ± 0.01 | 18.92 | - | - | - | - | - | - | - | - | - | - | - | - |
| 1-Tetradecene | Hydrocarbon | C14H28 | 0.12 ± 0.04 | 27.33 | - | - | - | - | - | - | - | - | - | - | 0.18 ± 0.01 | 10.32 |
| Tetradecane | Hydrocarbon | C14H30 | 0.27 ± 0.13 | 27.65 | 0.68 ± 0.14 | 26.45 | 1.41 ± 0.42 | 12.35 | - | - | 0.31 ± 0.01 | 18.9 | 0.89 ± 0.07 | 10.58 | 0.44 ± 0.02 | 11.47 |
| α- Pyrone | Ketone | C5H4O2 | 2.19 ± 0.82 | 33.39 | 0.11 ± 0.03 | 12.88 | 0.12 ± 0.01 | 18.43 | - | - | 0.04 ± 0.01 | 12.7 | 1.43 ± 0.22 | 20.34 | 5.14 ± 1.94 | 22.25 |
| δ-2,4-Dienolactone | Lactone | C7H12 | 8.43 ± 1.53 | 33.85 | 0.06 ± 0.01 | 8.69 | 1.23 ± 0.47 | 15.30 | - | - | 0.21 ± 0.05 | 8.4 | 2.76 ± 0.13 | 12.76 | 0.13 ± 0.04 | 7.86 |
| 2H-Pyran-2-one | Ketone | C5H4O2 | 17.39 ± 3.45 | 33.48 | 1.24 ± 0.32 | 13.80 | 5.23 ± 1.94 | 19.21 | - | - | 2.49 ± 0.86 | 15.7 | 9.87 ± 2.41 | 18.65 | 8.89 ± 2.32 | 20.34 |
| (E)-6-Pent-1-enylpyran-2-one | Ketone | C21H26O2 | 0.43 ± 0.05 | 34.55 | 2.18 ± 0.48 | 15.35 | 0.1 ± 0.02 | 17.33 | - | - | 5.88 ± 1.23 | 13.8 | 0.59 ± 0.04 | 18.43 | 2.26 ± 0.91 | 13.66 |
| 1-Hexadecene | Hydrocarbon | C16H32 | 0.12 ± 0.02 | 35.49 | - | - | - | - | 0.09 ± 0.01 | 15.92 | 0.49 ± 0.03 | 27.3 | 1.09 ± 0.32 | 18.60 | 1.11 ± 0.07 | 15.97 |
| Hexadecane | Hydrocarbon | C16H32 | 0.33 ± 0.08 | 35.74 | 3.83 ± 0.71 | 19.55 | 2.07 ± 0.72 | 14.73 | 0.08 ± 0.01 | 15.95 | 0.99 ± 0.12 | 27.6 | 1.91 ± 0.62 | 19.71 | - | - |
| Cyclohexadecane | Hydrocarbon | C16H32 | 0.25 ± 0.02 | 49.10 | 1.09 ± 0.32 | 12.75 | - | - | 0.11 ± 0.03 | 15.91 | - | - | - | - | - | - |
| 1-Eicosene | Ester | C20H40 | 0.25 ± 0.04 | 49.10 | 5.63 ± 1.79 | 5.68 | 0.86 ± 0.06 | 28.45 | 0.19 ± 0.04 | 5.43 | - | - | 1.03 ± 0.04 | 29.56 | 1.18 ± 0.11 | 24.70 |
| 1-Docosanol | Alcohol | C22H46O | 0.25 ± 0.01 | 49.10 | - | - | - | - | - | - | 0.27 ± 0.07 | 11.3 | - | - | 0.46 ± 0.06 | 26.85 |
| Eicosane | Hydrocarbon | C20H42 | 0.25 ± 0.02 | 49.28 | - | - | - | - | 0.14 ± 0.04 | 8.35 | 1.40 ± 0.09 | 8.25 | 2.10 ± 0.90 | 28.98 | - | - |
| Hexadecanoic acid | Acid | C16H32O2 | 4.12 ± 1.23 | 50.29 | 9.42 ± 2.96 | 13.67 | - | - | - | - | - | - | - | - | - | - |
| 9,12-Octa decadienoic acid | Acid | C18H32O2 | 2.83 ± 0.90 | 55.19 | 2.59 ± 0.41 | 24.56 | 0.37 ± 0.08 | 8.43 | 0.04 ± 0.01 | 18.90 | - | - | - | - | - | - |
| Ethyl linoleate | Ester | C20H36O2 | 0.35 ± 0.12 | 54.41 | - | - | 0.14 ± 0.04 | 12.56 | 0.04 ± 0.01 | 13.50 | - | - | - | - | - | - |
| Tetracosane | Hydrocarbon | C24H50 | 0.11 ± 0.02 | 60.65 | 1.06 ± 0.02 | 13.80 | 0.26 ± 0.03 | 13.45 | - | - | 0.33 ± 0.04 | 6.76 | 1.37 ± 0.46 | 13.08 | 0.58 ± 0.04 | 27.59 |
| Harziandione | Ketone | C20H30O2 | 0.24 ± 0.01 | 64.23 | - | - | - | - | - | - | - | - | 6.70 ± 1.94 | 63.77 | 4.49 ± 2.07 | 58.88 |
| Hexacosane | Hydrocarbon | C26H54 | 0.05 ± 0.01 | 65.71 | 0.57 ± 0.12 | 11.65 | 0.14 ± 0.01 | 19.58 | 0.03 ± 0.01 | 2.56 | 0.15 ± 0.03 | 12.34 | 0.33 ± 0.04 | 19.42 | 0.25 ± 0.03 | 15.20 |
| Heptadecane | Hydrocarbon | C17H36 | - | - | 0.32 ± 0.08 | 13.09 | - | - | - | - | - | - | 0.56 ± 0.05 | 18.91 | 0.12 ± 0.02 | 10.24 |
| 1-Octadecene | Hydrocarbon | C18H36 | - | - | 0.83 ± 0.27 | 12.60 | 0.41 ± 0.02 | 13.08 | 0.09 ± 0.02 | 14.29 | 0.44 ± 0.06 | 14.0 | - | - | 0.46 ± 0.07 | 8.65 |
| Octadecane | Hydrocarbon | C18H36 | - | - | 2.23 ± 1.01 | 12.60 | 0.62 ± 0.03 | 15.48 | 0.20 ± 0.03 | 9.20 | 1.91 ± 0.23 | 13.2 | 2.16 ± 0.91 | 16.77 | 1.07 ± 0.38 | 18.55 |
| Docosene | Hydrocarbon | C22H44 | - | - | 4.07 ± 1.53 | 32.55 | 0.24 ± 0.03 | 11.35 | 0.06 ± 0.01 | 12.11 | - | - | 1.86 ± 0.11 | 15.69 | 0.98 ± 0.23 | 13.34 |
| Octadecanoic acid | Acid | C18H36O2 | 0.95 ± 0.23 | 55.70 | 3.87 ± 1.25 | 23.90 | 0.75 ± 0.11 | 11.87 | 0.13 ± 0.02 | 7.88 | - | - | 0.07 ± 0.01 | 7.24 | - | - |
| Tetracosahexaene | Hydrocarbon | C24H38 | - | - | 0.72 ± 0.09 | 24.76 | 0.15 ± 0.01 | 18.68 | - | - | - | - | - | - | 1.44 ± 0.71 | 14.21 |
| Dimethyl Sulfoxide | Organosulfur | C2H6OS | - | - | - | - | 0.79 ± 0.06 | 33.70 | - | - | - | - | - | - | 0.30 ± 0.06 | 24.45 |
| Chemical groups | Content (%) | |||||||||||||||
| Acids | 11.89 | 18.23 | 1.65 | 1.64 | 0.15 | 1.35 | 0.40 | |||||||||
| Esters | 0.81 | 5.63 | 1.0 | 0.63 | - | 1.03 | 1.18 | |||||||||
| Alcohols | 5.24 | - | 6.57 | - | 23.68 | 7.24 | 0.46 | |||||||||
| Aldehydes | 0.24 | 0.07 | 1.94 | - | 0.90 | 2.82 | - | |||||||||
| Hydrocarbons | 1.54 | 15.4 | 5.3 | 0.80 | 6.02 | 12.27 | 6.63 | |||||||||
| Ketones | 20.25 | 3.53 | 5.45 | - | 8.41 | 18.59 | 20.78 | |||||||||
| Lactones | 8.43 | 0.06 | 1.23 | - | 0.21 | 2.76 | 0.13 | |||||||||
| Organosulfur | - | - | 0.79 | - | - | - | 0.30 | |||||||||
| Total | 48.40 | 42.92 | 23.93 | 3.07 | 39.37 | 46.06 | 29.88 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanmugaraj, C.; Kamil, D.; Kundu, A.; Singh, P.K.; Das, A.; Hussain, Z.; Gogoi, R.; Shashank, P.R.; Gangaraj, R.; Chaithra, M. Exploring the Potential Biocontrol Isolates of Trichoderma asperellum for Management of Collar Rot Disease in Tomato. Horticulturae 2023, 9, 1116. https://doi.org/10.3390/horticulturae9101116
Shanmugaraj C, Kamil D, Kundu A, Singh PK, Das A, Hussain Z, Gogoi R, Shashank PR, Gangaraj R, Chaithra M. Exploring the Potential Biocontrol Isolates of Trichoderma asperellum for Management of Collar Rot Disease in Tomato. Horticulturae. 2023; 9(10):1116. https://doi.org/10.3390/horticulturae9101116
Chicago/Turabian StyleShanmugaraj, C., Deeba Kamil, Aditi Kundu, Praveen Kumar Singh, Amrita Das, Zakir Hussain, Robin Gogoi, P. R. Shashank, R. Gangaraj, and M. Chaithra. 2023. "Exploring the Potential Biocontrol Isolates of Trichoderma asperellum for Management of Collar Rot Disease in Tomato" Horticulturae 9, no. 10: 1116. https://doi.org/10.3390/horticulturae9101116
APA StyleShanmugaraj, C., Kamil, D., Kundu, A., Singh, P. K., Das, A., Hussain, Z., Gogoi, R., Shashank, P. R., Gangaraj, R., & Chaithra, M. (2023). Exploring the Potential Biocontrol Isolates of Trichoderma asperellum for Management of Collar Rot Disease in Tomato. Horticulturae, 9(10), 1116. https://doi.org/10.3390/horticulturae9101116







