Biocontrol Potential of Trichoderma asperellum CMT10 against Strawberry Root Rot Disease
Abstract
1. Introduction
2. Materials and Methods
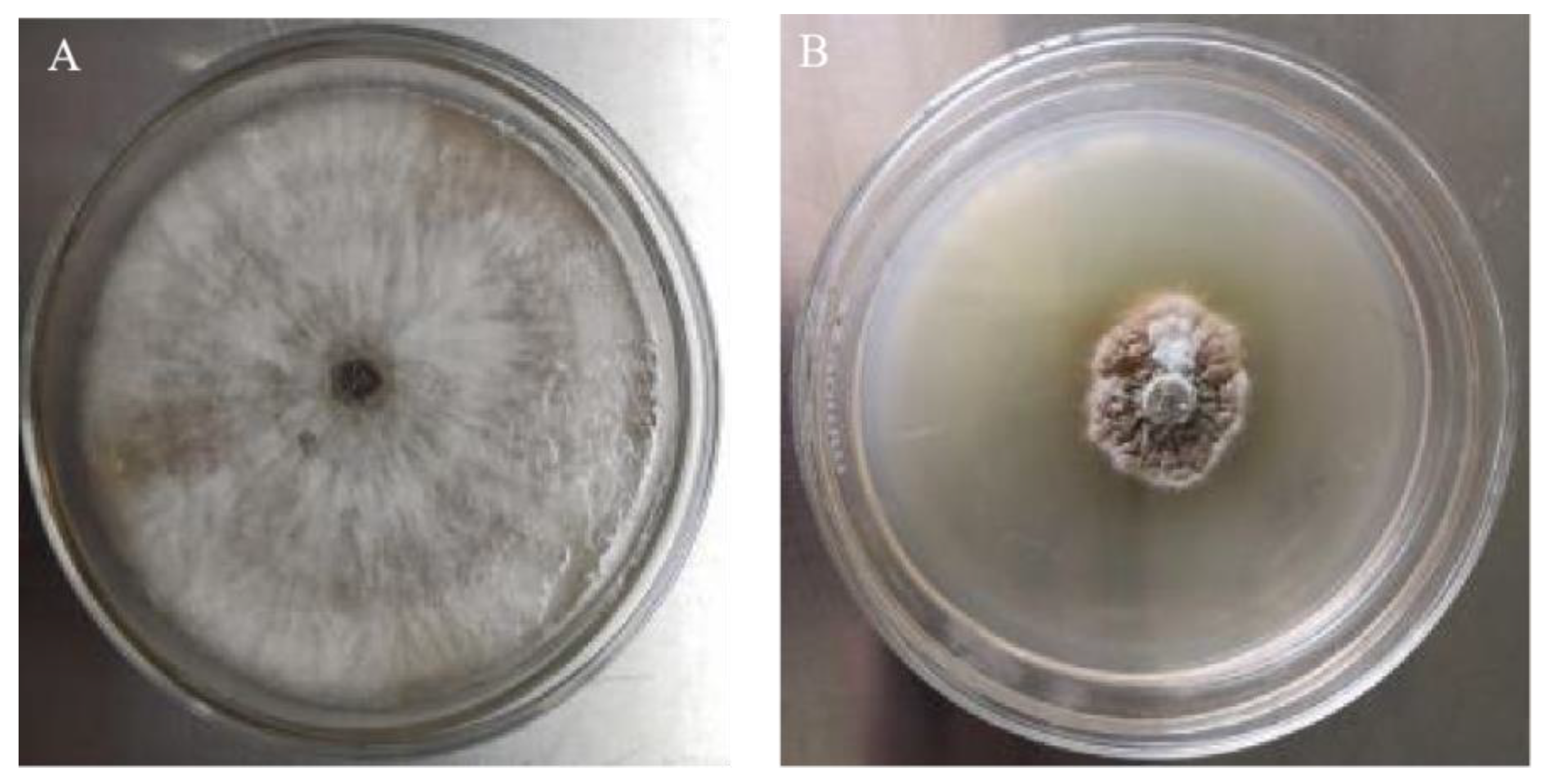
2.1. Plant Pathogen and Plant Materials
2.2. Isolation and Screening of Trichoderma Strains
2.3. Morphological and Molecular Identification of Trichoderma CMT10
2.4. In Vitro Biocontrol of Trichoderma CMT10 against N. clavispora
2.4.1. Inhibitory Effects of Volatile Compounds
2.4.2. Inhibitory Effects of Soluble Compounds
2.4.3. Hyperparasitism of Trichoderma CMT10
2.5. Biochemical Properties of Trichoderma CMT10
2.6. Control Effects of Trichoderma CMT10 on Strawberry Root Rot
2.7. Growth-Promoting Effects of Trichoderma CMT10 on Strawberry Seedlings
2.8. Data Statistics and Analysis
3. Results
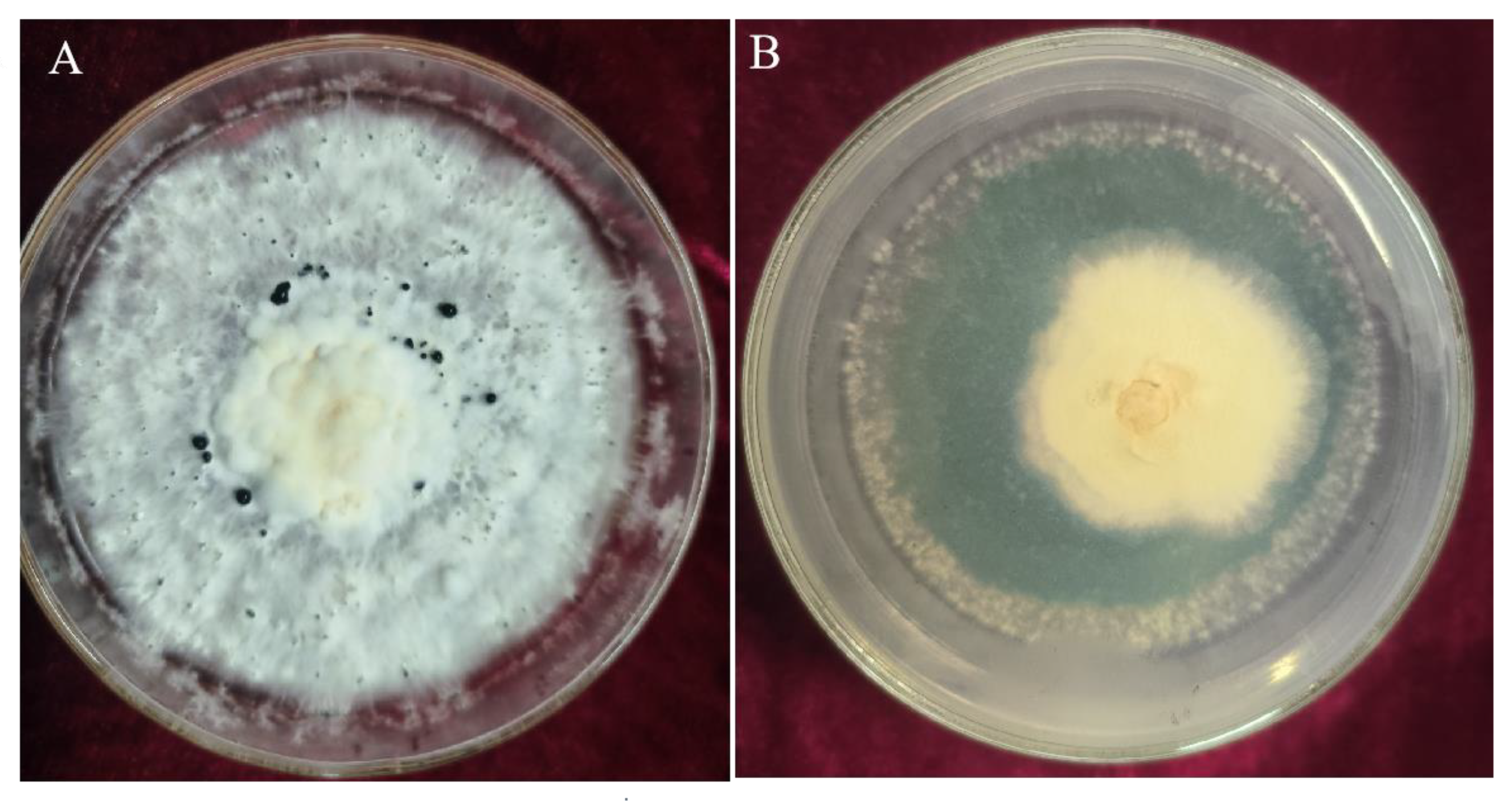
3.1. Screening of Trichoderma Strains with Inhibitory Effects on N. clavispora
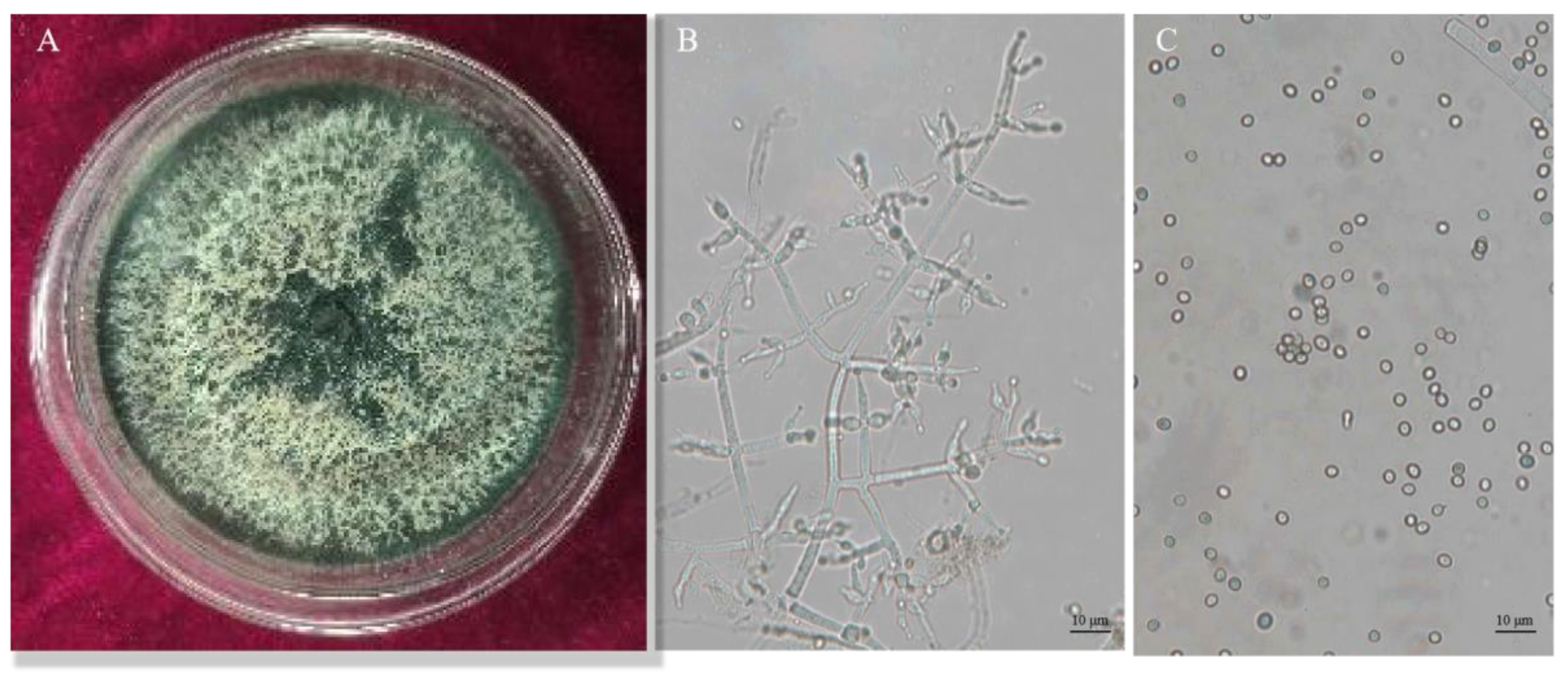
3.2. Identification of Trichoderma CMT10
3.3. In Vitro Biocontrol of Trichoderma CMT10 against N. clavispora
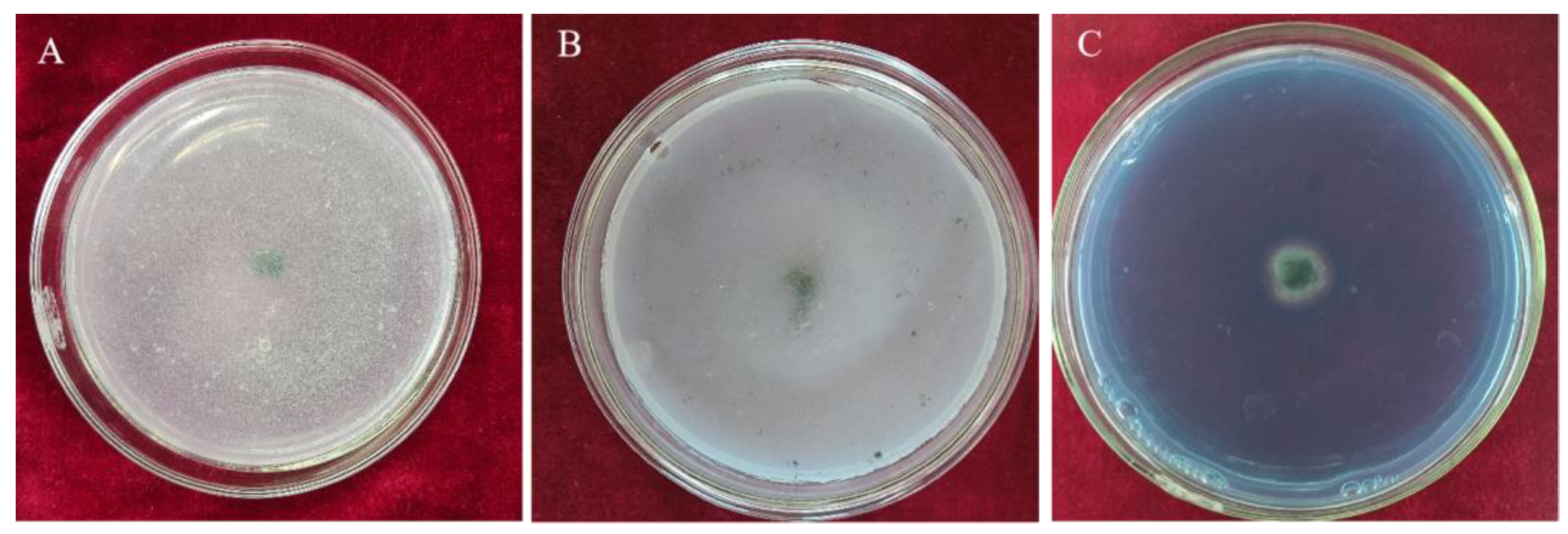
3.3.1. Inhibition Rates of Volatile Metabolites from Trichoderma CMT10 on N. clavispora
3.3.2. Inhibition Rates of Soluble Metabolites from Trichoderma CMT10 on N. clavispora
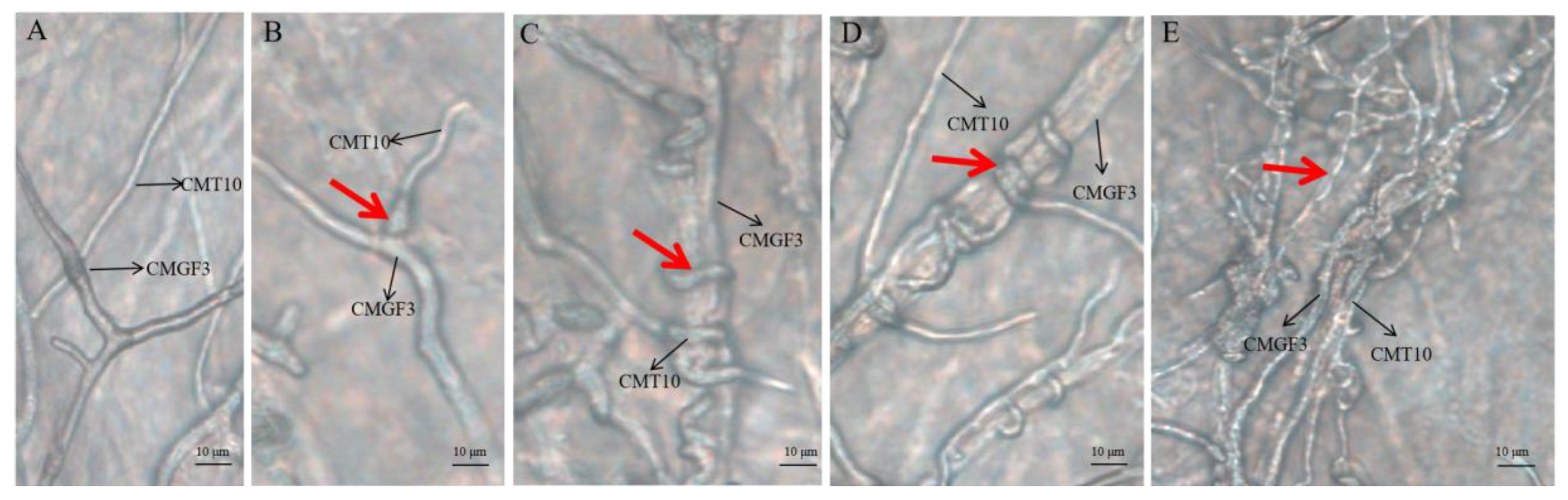
3.3.3. Hyperparasitism of Trichoderma CMT10 on N. clavispora
3.4. Determination of Biochemical Properties of T. asperellum CMT10
3.5. Biocontrol Efficiency of T. asperellum CMT10 against Strawberry Root Rot
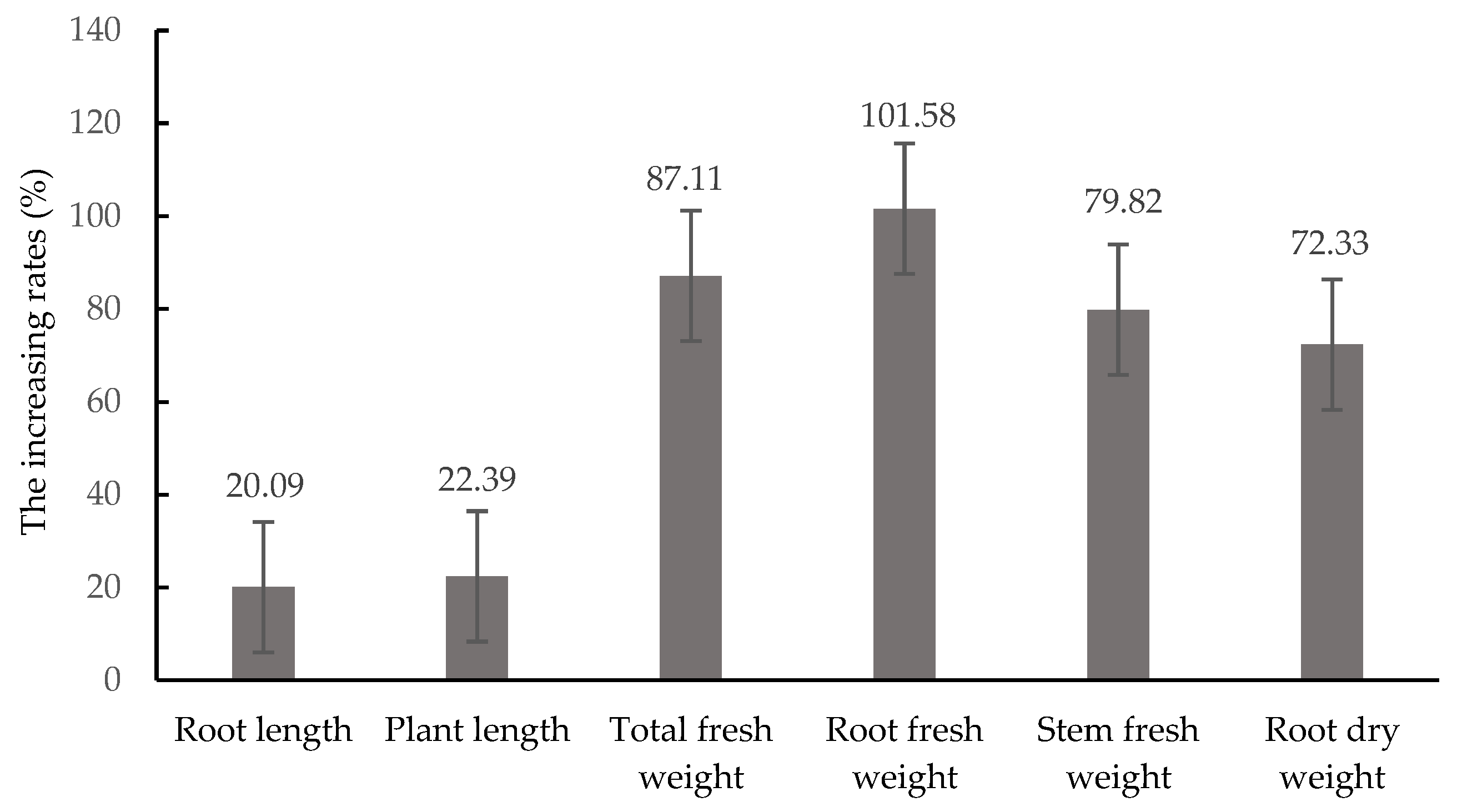
3.6. Growth-Promoting Efficiency of T. asperellum CMT10 on Strawberry Seedlings
4. Discussion
4.1. Significance of Exploring Biocontrol Resources for Strawberry Root Rot
4.2. Biocontrol Mechanism of T. asperellum CMT10
4.3. Practical Application of T. asperellum CMT10
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Basu, A.; Nguyen, A.; Betts, N.M.; Lyons, T.J. Strawberry as a functional food: An evidence-based review. Crit. Rev. Food Sci. Nutr. 2014, 54, 790–806. [Google Scholar] [CrossRef]
- Ji, Y.; Li, X.; Gao, Q.H.; Geng, C.; Duan, K. Colletotrichum species pathogenic to strawberry: Discovery history, global diversity, prevalence in China, and the host range of top two species. Phytopathol. Res. 2022, 4, 42. [Google Scholar] [CrossRef]
- Wang, M.Y.; Du, Y.Q.; Cai, W.W.; Wang, Z.H.; Zhu, J.Q. Effect of complex antagonistic bacteria on controlling strawberry root rot. J. Agric. Sci. Technol. 2020, 22, 100–110. [Google Scholar]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Iqbal, M.; Jamshaid, M.; Zahid, M.A.; Andreasson, E.; Vetukuri, R.A.; Stenberg, J.A. Biological control of strawberry crown rot, root rot and grey mould by the beneficial fungus Aureobasidium pullulans. Bio-Control 2021, 66, 535–545. [Google Scholar] [CrossRef]
- Baggio, J.S.; Cordova, L.G.; Peres, N.A. Sources of inoculum and survival of 22 Macrophomina phaseolina in Florida strawberry fields. Plant Dis. 2019, 103, 2417–2424. [Google Scholar] [CrossRef]
- Feliziani, E.; Romanazzi, G. Postharvest decay of strawberry fruit: Etiology, epidemiology, and disease management. J. Berry Res. 2016, 6, 47–63. [Google Scholar] [CrossRef]
- Hong, S.; Kim, T.Y.; Won, S.J.; Moon, J.H.; Ajuna, H.B.; Kim, K.; Ahn, Y.S. Control of fungal diseases and fruit yield improvement of strawberry using Bacillus velezensis CE100. Microorganisms 2022, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Yang, H.M.; Jin, J.; Yan, H.H.; Wang, J.P.; Zhang, R.Q. Synergistic activity of antagonistic Trichoderma spp. and Rhizoctonia solani increases disease severity on strawberry petioles. Eur. J. Plant Pathol. 2022, 164, 375–389. [Google Scholar] [CrossRef]
- Abdel-lateif, K.S. Trichoderma as biological control weapon against soil borne plant pathogens. Afr. J. Biotechnol. 2017, 16, 2299–2306. [Google Scholar]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar]
- Ferreira, F.V.; Musumeci, M.A. Trichoderma as biological control agent: Scope and prospects to improve efficacy. World J. Microbiol. Biotechnol. 2021, 37, 90. [Google Scholar] [CrossRef]
- Zhang, H.; Du, G.D.; Song, Y.N.; Lu, X.F.; Ying, N. Screening, identification and the effect validation of Trichoderma against the root rot of strawberry. J. Shenyang Agric. Univ. 2015, 46, 654–660. [Google Scholar]
- Mercado, J.A.; Barceló, M.; Pliego, C.; Rey, M.; Caballero, J.L.; Muñoz-Blanco, J.; Ruano-Rosa, D.; López-Herrera, C.; de Los Santos, B.; Romero-Muñoz, F.; et al. Expression of the β-1,3-glucanase gene bgn13.1 from Trichoderma harzianum in strawberry increases tolerance to crown rot diseases but interferes with plant growth. Transgenic Res. 2015, 24, 979–989. [Google Scholar] [CrossRef]
- Rees, H.J.; Drakulic, J.; Cromey, M.G.; Bailey, A.M.; Foster, G.D. Endophytic Trichoderma spp. can protect strawberry and privet plants from infection by the fungus Armillaria mellea. PLoS ONE 2022, 17, e0271622. [Google Scholar] [CrossRef]
- Mirzaeipour, Z.; Bazgir, E.; Zafari, D.; Darvishnia, M. Selection and biocontrol efficiency of Trichoderma isolates against Rhizoctonia root rot and their growth promotion effects on strawberry plants. J. Plant Pathol. 2023, 105, 1563–1579. [Google Scholar] [CrossRef]
- Debbi, A.; Boureghda, H.; Monte, E.; Hermosa, R. Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in Algeria and a biocontrol strategy with indigenous Trichoderma spp. Front. Microbiol. 2018, 9, 282. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, M.F.; Arnão, E.; Warner, A.J.; Subedi, A.; Rocha, L.F.; Srour, A.; Bond, J.P.; Fakhoury, A.M. Trichoderma isolates inhibit Fusarium virguliforme growth, reduce root rot, and induce defense-related genes on soybean seedlings. Plant Dis. 2020, 104, 1949–1959. [Google Scholar] [CrossRef] [PubMed]
- Shaigan, S.; Seraji, A.; Moghaddam, S.A. Identification and investigation on antagonistic effect of Trichoderma spp. on tea seedlings white foot and root rot (Sclerotium rolfsii Sacc.) in vitro condition. Pak. J. Biol. Sci. 2008, 11, 2346–2350. [Google Scholar] [CrossRef] [PubMed]
- Yang, H. Classification and Identification of Trichoderma; China Land Press: Beijing, China, 2009; pp. 14–20. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- O’donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Damodaran, T.; Rajan, S.; Muthukumar, M.; Ram, G.; Yadav, K.; Kumar, S.; Ahmad, I.; Nidhi, K.; Mishra, V.K.; Vinay, K.; et al. Biological management of banana Fusarium wilt caused by Fusarium oxysporum f. sp. cubense tropical race 4 using antagonistic fungal isolate CSR-T-3 (Trichoderma reesei). Front. Microbiol. 2020, 11, 595845. [Google Scholar] [CrossRef] [PubMed]
- Kexiang, G.; Xiaoguang, L.; Yonghong, L.; Tianbo, Z.; Shuliang, W. Potential of Trichoderma harzianum and T. atroviride to Control Botryosphaeria berengeriana f. sp. piricola, the cause of apple ring rot. J. Phytopathol. 2002, 150, 271–276. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Shin, S.H.; Lim, Y.; Lee, S.E.; Yang, N.W.; Rhee, J.H. CAS agar diffusion assay for the measurement of siderophores in biological fluids. J. Microbiol. Methods 2001, 44, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Liaqat, F.; Eltem, R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 2016, 6, 120. [Google Scholar] [CrossRef]
- Yao, C.X.; Li, X.J.; Li, Q.; Xing, G.Z.; Fang, W.Y.; Li, C.H.; Zhang, Y.Y.; Yao, C.X.; Xu, M.; Li, F.F.; et al. Screening and identification of antagonistic bacteria against tobacco Fusarium root rot and evaluation of their effects on growth promoting and disease control. Chin. J. Biol. Control 2021, 37, 1066–1072. [Google Scholar]
- Brick, J.M.; Bostock, R.M.; Silversone, S.E. Rapid in situ assay for indole acetic acid production by bacteria immobilized on nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef]
- Vestberg, M.; Kukkonen, S.; Saari, K.; Parikka, P.; Huttunen, J.; Tainio, L.; Devos, N.; Weekers, F.; Kevers, C.; Thonart, P.; et al. Microbial inoculation for improving the growth and health of micropropagated strawberry. Appl. Soil Ecol. 2004, 27, 243–258. [Google Scholar] [CrossRef]
- Zhang, M.; Kong, Z.; Fu, H.; Shu, X.; Xue, Q.; Lai, H.; Guo, Q. Rhizosphere microbial ecological characteristics of strawberry root rot. Front. Microbiol. 2023, 14, 1286740. [Google Scholar] [CrossRef] [PubMed]
- Elad, Y.; Chet, I.; Henis, Y. Biological control of Rhizoctonia solani in strawberry fields by Trichoderma harzianum. Plant Soil 1981, 60, 245–254. [Google Scholar] [CrossRef]
- Hernández-Muñiz, P.; Borrero, C.; Ordóñez-Martín, J.; Pastrana, A.M.; Avilés, M. Optimization of the use of industrial wastes in Anaerobic soil disinfestation for the control of Fusarium wilt in strawberry. Plants 2023, 12, 3185. [Google Scholar] [CrossRef] [PubMed]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef]
- Yao, X.; Guo, H.; Zhang, K.; Zhao, M.; Ruan, J.; Chen, J. Trichoderma and its role in biological control of plant fungal and nematode disease. Front. Microbiol. 2023, 14, 1160551. [Google Scholar] [CrossRef]
- Mohiddin, F.A.; Padder, S.A.; Bhat, A.H.; Ahanger, M.A.; Shikari, A.B.; Wani, S.H.; Bhat, F.A.; Nabi, S.U.; Hamid, A.; Bhat, N.A.; et al. Phylogeny and optimization of Trichoderma harzianum for Chitinase production: Evaluation of their antifungal behaviour against the prominent soil borne phyto-pathogens of temperate India. Microorganisms 2021, 9, 1962. [Google Scholar] [CrossRef] [PubMed]
- Risoli, S.; Cotrozzi, L.; Sarrocco, S.; Nuzzaci, M.; Pellegrini, E.; Vitti, A. Trichoderma-induced resistance to Botrytis cinerea in Solanum species: A meta-analysis. Plants 2022, 11, 180. [Google Scholar] [CrossRef]
- Kottb, M.; Gigolashvili, T.; Großkinsky, D.K.; Piechulla, B. Trichoderma volatiles effecting Arabidopsis: From inhibition to protection against phytopathogenic fungi. Front. Microbiol. 2015, 6, 995. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.A.A.; Najeeb, S.; Hussain, S.; Xie, B.; Li, Y. Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic fungi. Microorganisms 2020, 29, 817. [Google Scholar] [CrossRef]
- Naglot, A.; Goswami, S.; Rahman, I.; Shrimali, D.D.; Yadav, K.K.; Gupta, V.K.; Veer, V. Antagonistic potential of native Trichoderma viride strain against potent tea fungal pathogens in north east India. Plant Pathol. J. 2015, 31, 278–289. [Google Scholar] [CrossRef]
- Manganiello, G.; Sacco, A.; Ercolano, M.R.; Vinale, F.; Lanzuise, S.; Pascale, A.; Napolitano, M.; Lombardi, N.; Lorito, M.; Woo, S.L. Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic acid. Front. Microbiol. 2018, 9, 1966. [Google Scholar] [CrossRef]
- Shaw, S.; Le Cocq, K.; Paszkiewicz, K.; Moore, K.; Winsbury, R.; de Torres Zabala, M.; Studholme, D.J.; Salmon, D.; Thornton, C.R.; Grant, M.R. Transcriptional reprogramming underpins enhanced plant growth promotion by the biocontrol fungus Trichoderma hamatum GD12 during antagonistic interactions with Sclerotinia sclerotiorum in soil. Mol. Plant Pathol. 2016, 17, 1425–1441. [Google Scholar] [CrossRef]
- Hewedy, O.A.; Abdel Lateif, K.S.; Seleiman, M.F.; Shami, A.; Albarakaty, F.M.; El-Meihy, R.M. Phylogenetic diversity of Trichoderma strains and their antagonistic potential against soil-borne pathogens under stress conditions. Biology 2020, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Larran, S.; Santamarina Siurana, M.P.; Roselló Caselles, J.; Simón, M.R.; Perelló, A. In vitro antagonistic activity of Trichoderma harzianum against Fusarium sudanense causing seedling blight and seed rot on wheat. ACS Omega 2020, 5, 23276–23283. [Google Scholar] [CrossRef] [PubMed]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The secrets of a multitalented biocontrol agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Jogaiah, S.; Abdelrahman, M.; Tran, L.P.; Ito, S.I. Different mechanisms of Trichoderma virens-mediated resistance in tomato against Fusarium wilt involve the jasmonic and salicylic acid pathways. Mol. Plant Pathol. 2018, 19, 870–882. [Google Scholar] [CrossRef]




| Genea | Primer | Primer Sequence (5′-3′) | PCR Conditions | Reference |
|---|---|---|---|---|
| ITS | ITS1 ITS4 | TCCGTAGGTGAACCTGCGG | 94 °C for 5 min (94 °C for 30 s, 55 °C for 30 s and 72 °C for 40 s) × 35 cycles, 72 °C for 7 min | [21] |
| TCCTCCGCTTATTGATATGC | ||||
| tef-1α | TEF-F | TGGGCCATCAACTGAGAAAGA | 94 °C for 5 min (94 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min) × 35 cycles, 72 °C for 7 min | [22] |
| TEF-R | TCTCCCTACACTTCAACTGCACA |
| Code | Culture Accession Number(s) | Original Name | Accession no. ITS | Accession no. tef-1α |
|---|---|---|---|---|
| 1 | CEN1463 | T. asperellum | MK714888 | MK696646 |
| 2 | T34 | T. asperellum | LC123614 | EU077228 |
| 3 | ZJSX5002 | T. asperellum | JQ040324 | JQ040480 |
| 4 | KUFA0403 | T. asperellum | OM169354 | OP132635 |
| 5 | RM-28 | T. asperellum | MK092975 | MK095221 |
| 6 | TR5 | T. longibrachiatum | KC859426 | KC572116 |
| 7 | Tr5 | T. harzianum | OP938774 | OP948262 |
| 8 | DUCC001 | T. citrinoviride | JF700484 | JF700485 |
| 9 | S206 | T. caerulescens | JN715590 | JN715624 |
| 10 | TW20050 | T. gamsii | KU523894 | KU523895 |
| 11 | YMF1.02659 | T. kunmingense | KJ742800 | KJ742802 |
| 12 | CBS 121219 | T. yunnanense | GU198302 | GU198243 |
| Treatments | Colony Diameter (cm) | Inhibition Rate (%) |
|---|---|---|
| CMT10 | 2.93 ± 0.153 | 65.49 a |
| CMT4 | 4.13 ± 0.058 | 51.37 b |
| CMGF3 | 8.50 ± 0.000 | - |
| Treatments | Disease Index | Control Efficiency (%) |
|---|---|---|
| CMGF3 | 84.00 ± 0.04 a | - |
| CMT10 | 0.00 ± 0.00 c | - |
| CMGF3 + CMT10 | 31.00 ± 0.61 b | 63.09 ± 0.07 a |
| Control | 0.00 ± 0.00 c | - |
| Treatments | Plant Height (cm) | Root Length (cm) | Total Fresh Weight (g) | Root Fresh Weight (g) | Stem Fresh Weight (g) | Root Dry Weight (g) |
|---|---|---|---|---|---|---|
| CMT10 | 12.57 ± 1.35 a | 23.75 ± 2.18 a | 13.55 ± 3.53 a | 7.18 ± 3.37 a | 6.37 ± 2.08 a | 2.66 ± 1.00 a |
| Control | 10.53 ± 1.41 b | 19.67 ± 2.70 b | 7.61 ± 1.66 b | 3.87 ± 1.59 b | 3.74 ± 0.61 b | 1.56 ± 0.50 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Yang, R.; Wang, Z.; Ma, Y.; Ren, W.; Wei, D.; Ye, W. Biocontrol Potential of Trichoderma asperellum CMT10 against Strawberry Root Rot Disease. Horticulturae 2024, 10, 246. https://doi.org/10.3390/horticulturae10030246
Liu P, Yang R, Wang Z, Ma Y, Ren W, Wei D, Ye W. Biocontrol Potential of Trichoderma asperellum CMT10 against Strawberry Root Rot Disease. Horticulturae. 2024; 10(3):246. https://doi.org/10.3390/horticulturae10030246
Chicago/Turabian StyleLiu, Ping, Ruixian Yang, Zuhua Wang, Yinhao Ma, Weiguang Ren, Daowei Wei, and Wenyu Ye. 2024. "Biocontrol Potential of Trichoderma asperellum CMT10 against Strawberry Root Rot Disease" Horticulturae 10, no. 3: 246. https://doi.org/10.3390/horticulturae10030246
APA StyleLiu, P., Yang, R., Wang, Z., Ma, Y., Ren, W., Wei, D., & Ye, W. (2024). Biocontrol Potential of Trichoderma asperellum CMT10 against Strawberry Root Rot Disease. Horticulturae, 10(3), 246. https://doi.org/10.3390/horticulturae10030246







