A Multicultivar Approach for Grape Bunch Weight Estimation Using Image Analysis
Abstract
:1. Introduction
- Code 202: bunch length;
- Code 203: bunch width;
- Code 204: bunch compactness;
- Code 208: bunch shape;
- Code 220: berry length;
- Code 221: berry width;
- Code 222: berry size uniformity;
2. Materials and Methods
2.1. Site Characterization and Trial Description
2.2. Data Set
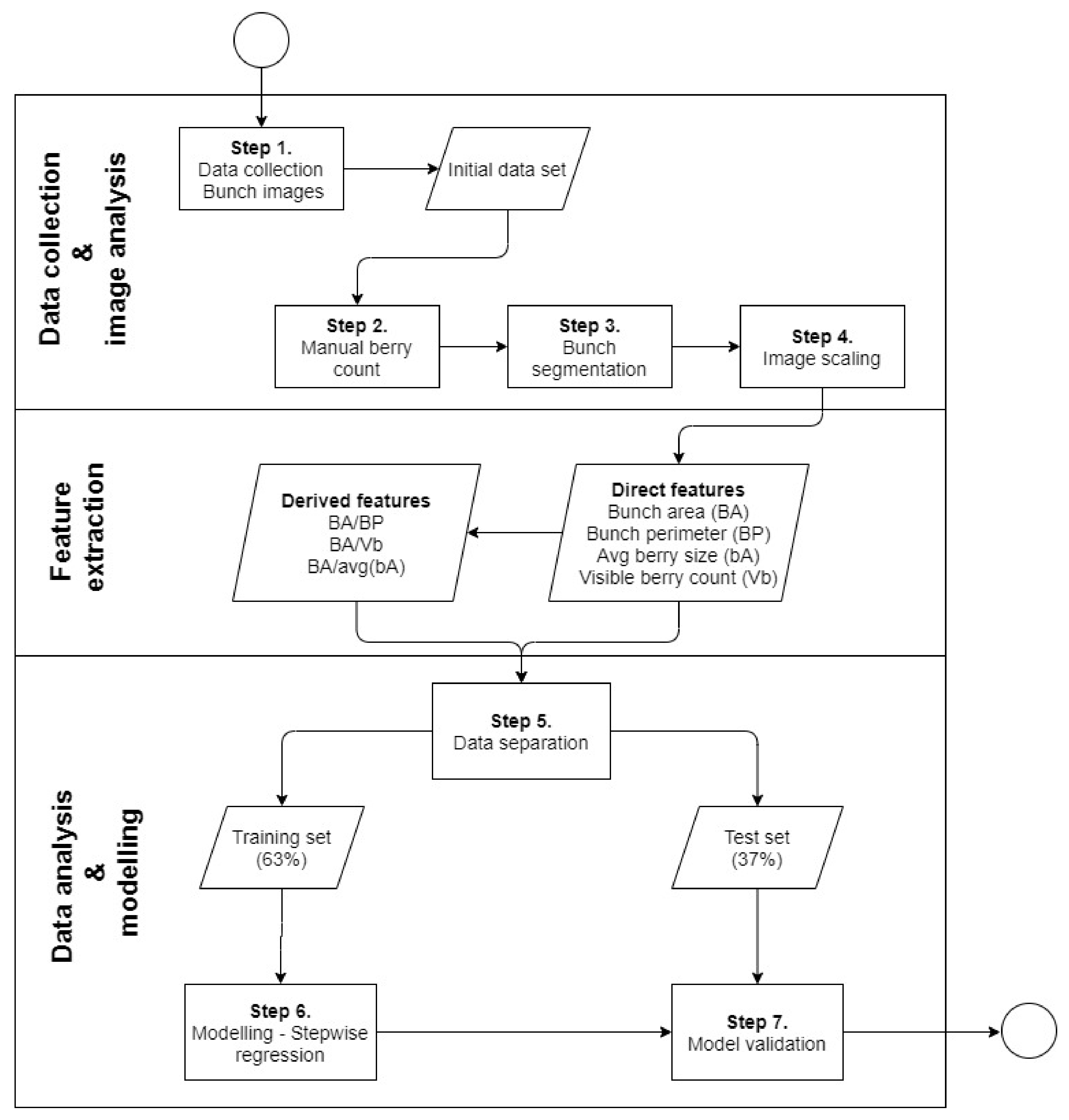
2.3. Data Collection and Image Analysis
2.4. Data Analysis
- ratio between bunch area and visible berries (BA_Vb), which represents the average area occupied by each visible berry;
- ratio between bunch area and bunch perimeter (BA_BP), which represents the relationship between the area that berries occupy and their perimeter, and;
- ratio between bunch area and the average bA of each variety (BA_avg(bA)), which represents the average number of berries per unit of bunch area.
3. Results
3.1. Characterization of Bunch Features
3.2. Relationships between Image Features and Bunch Weight
3.3. Multiple Regression Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nuske, S.; Achar, S.; Bates, T.; Narasimhan, S.; Singh, S. Yield Estimation in Vineyards by Visual Grape Detection. In Proceedings of the 2011 IEEE/RSJ International Conference on Intelligent Robots and Systems, San Francisco, CA, USA, 25–30 September 2011; pp. 2352–2358. [Google Scholar]
- Dunn, G.M.; Martin, S.R. Yield prediction from digital image analysis: A technique with potential for vineyard assessments prior to harvest. Aust. J. Grape Wine Res. 2008, 10, 196–198. [Google Scholar] [CrossRef]
- Bramley, R.; Ouzman, J.; Boss, P. Variation in vine vigour, grape yield and vineyard soils and topography as indicators of variation in the chemical composition of grapes, wine and wine sensory attributes. Aust. J. Grape Wine Res. 2011, 17, 217–229. [Google Scholar] [CrossRef]
- Victorino, G.; Braga, R.; Lopes, C.M. The effect of topography on the spatial variability of grapevine vegetative and repro-ductive components. In Proceedings of the I Congresso Luso-Brasileiro de Horticultura, Lisbon, Portugal, 1–4 November 2017; pp. 510–516. Available online: https://aph.aphorticultura.pt/wp-content/uploads/2019/10/the_effect_of_topography_on.pdf (accessed on 6 March 2022).
- Jasse, A.; Berry, A.; Aleixandre-Tudo, J.L.; Poblete-Echeverría, C. Intra-block spatial and temporal variability of plant water status and its effect on grape and wine parameters. Agric. Water Manag. 2020, 246, 106696. [Google Scholar] [CrossRef]
- Taylor, J.; Dresser, J.; Hickey, C.; Nuske, S.; Bates, T. Considerations on spatial crop load mapping. Aust. J. Grape Wine Res. 2018, 25, 144–155. [Google Scholar] [CrossRef]
- Carrillo, E.; Matese, A.; Rousseau, J.; Tisseyre, B. Use of multi-spectral airborne imagery to improve yield sampling in viticulture. Precis. Agric. 2015, 17, 74–92. [Google Scholar] [CrossRef]
- Seng, K.P.; Ang, L.-M.; Schmidtke, L.M.; Rogiers, S.Y. Computer Vision and Machine Learning for Viticulture Technology. IEEE Access 2018, 6, 67494–67510. [Google Scholar] [CrossRef]
- Victorino, G.F.; Braga, R.; Santos-Victor, J.; Lopes, C.M. Yield components detection and image-based indicators for non-invasive grapevine yield prediction at different phenological phases. OENO One 2020, 54, 833–848. [Google Scholar] [CrossRef]
- Diago, M.P.; Tardáguila, J.; Aleixos, N.; Millán, B.; Prats-Montalban, J.M.; Cubero, S.; Blasco, J. Assessment of cluster yield components by image analysis. J. Sci. Food Agric. 2014, 95, 1274–1282. [Google Scholar] [CrossRef] [Green Version]
- Lopes, C.M.; Graça, J.; Sastre, J.; Reyes, M.; Guzmán, R.; Braga, R.; Monteiro, A.; Pinto, P.A. Vineyard Yeld Estimation by VINBOT Robot—Preliminary Results with the White Variety Viosinho. In Proceedings 11th International Terroir Congres; Jones, G., Doran, N., Eds.; Southern Oregon University: Ashland, OR, USA, 2016; pp. 458–463. [Google Scholar]
- Milella, A.; Marani, R.; Petitti, A.; Reina, G. In-field high throughput grapevine phenotyping with a consumer-grade depth camera. Comput. Electron. Agric. 2018, 156, 293–306. [Google Scholar] [CrossRef]
- Hacking, C.; Poona, N.; Manzan, N.; Poblete-Echeverría, C. Investigating 2-D and 3-D Proximal Remote Sensing Techniques for Vineyard Yield Estimation. Sensors 2019, 19, 3652. [Google Scholar] [CrossRef] [Green Version]
- Aquino, A.; Barrio, I.; Diago, M.-P.; Millan, B.; Tardaguila, J. vitisBerry: An Android-smartphone application to early evaluate the number of grapevine berries by means of image analysis. Comput. Electron. Agric. 2018, 148, 19–28. [Google Scholar] [CrossRef]
- Liu, S.; Zeng, X.; Whitty, M. A vision-based robust grape berry counting algorithm for fast calibration-free bunch weight estimation in the field. Comput. Electron. Agric. 2020, 173, 105360. [Google Scholar] [CrossRef]
- Aquino, A.; Millan, B.; Diago, M.-P.; Tardaguila, J. Automated early yield prediction in vineyards from on-the-go image acquisition. Comput. Electron. Agric. 2018, 144, 26–36. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Sagan, V.; Sidike, P.; Kwasniewski, M.T. Dual Activation Function-Based Extreme Learning Machine (ELM) for Estimating Grapevine Berry Yield and Quality. Remote Sens. 2019, 11, 740. [Google Scholar] [CrossRef] [Green Version]
- Zabawa, L.; Kicherer, A.; Klingbeil, L.; Milioto, A.; Topfer, R.; Kuhlmann, H.; Roscher, R. Detection of Single Grapevine Berries in Images Using Fully Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Long Beach, CA, USA, 16–17 June 2019; 2019; pp. 2571–2579. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zou, L.; Ren, C.; Ren, F.; Wang, Y.; Fan, P.; Li, S.; Liang, Z. VvSWEET10 mediates sugar accumulation in grapes. Genes 2019, 10, 255. [Google Scholar] [CrossRef] [Green Version]
- Tello, J.; Aguirrezábal, R.; Hernáiz, S.; Larreina, B.; Montemayor, M.; Vaquero, E.; Ibáñez, J. Multicultivar and multivariate study of the natural variation for grapevine bunch compactness. Aust. J. Grape Wine Res. 2015, 21, 277–289. [Google Scholar] [CrossRef]
- OIV Descriptor List for Grape Varieties and Vitis Species (2nd Edition). Available online: http://www.oiv.int/en/technical-standards-and-documents/description-of-grape-varieties/oiv-descriptor-list-for-grape-varieties-and-vitis-species-2nd-edition (accessed on 6 March 2022).
- Font, D.; Tresanchez, M.; Martínez, D.; Moreno, J.; Clotet, E.; Palacín, J. Vineyard Yield Estimation Based on the Analysis of High Resolution Images Obtained with Artificial Illumination at Night. Sensors 2015, 15, 8284–8301. [Google Scholar] [CrossRef] [Green Version]
- Herrero-Huerta, M.; González-Aguilera, D.; Rodríguez-Gonzálvez, P.; Hernández-López, D. Vineyard yield estimation by automatic 3D bunch modelling in field conditions. Comput. Electron. Agric. 2015, 110, 17–26. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Toscano, P.; Cinat, P.; Berton, A.; Matese, A. A Low-Cost and Unsupervised Image Recognition Methodology for Yield Estimation in a Vineyard. Front. Plant Sci. 2019, 10, 559. [Google Scholar] [CrossRef] [Green Version]
- Nuske, S.; Gupta, K.; Narasimhan, S.; Singh, S. Modeling and calibrating visual yield estimates in vineyards. In Field and Service Robotics; Springer: Berlin/Heidelberg, Germany, 2014; Volume 92, pp. 343–356. [Google Scholar] [CrossRef] [Green Version]
- Tello, J.; Cubero, S.; Blasco, J.; Tardáguila, J.; Aleixos, N.; Ibáñez, J. Application of 2D and 3D image technologies to characterise morphological attributes of grapevine clusters. J. Sci. Food Agric. 2016, 96, 4575–4583. [Google Scholar] [CrossRef] [Green Version]
- Cubero, S.; Diago, M.; Blasco, J.; Tardaguila, J.; Prats-Montalbán, J.; Ibáñez, J.; Tello, J.; Aleixos, N. A new method for assessment of bunch compactness using automated image analysis. Aust. J. Grape Wine Res. 2015, 21, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Smart, R.E.; Turkington, C.R.; Evans, J.C. Grapevine Response to Furrow and Trickle Irrigation. Am. J. Enol. Vitic. 1974, 25, 62–66. [Google Scholar]
- Teixeira, G.; Monteiro, A.; Santos, C.; Lopes, C. Leaf morphoanatomy traits in white grapevine cultivars with distinct geographical origin. Ciência Técnica Vitivinícola 2018, 33, 90–101. [Google Scholar] [CrossRef] [Green Version]
- Duda, R.O.; Hart, P.E. Use of the Hough transformation to detect lines and curves in pictures. Commun. ACM 1972, 15, 11–15. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Tree-based Methods. In Modern Applied Statistics with S-Plus; Springer: New York, NY, USA, 1997; pp. 413–430. [Google Scholar] [CrossRef]
- Nuske, S.; Achar, S.; Gupta, K.; Narasimhan, S.; Singh, S. Visual Yield Estimation in Vineyards: Experiments with Different Varietals and Calibration Procedures; CMURI-TR-11-39; Carnegie Mellon University: Pittsburgh, PA, USA, 2011; p. 27. [Google Scholar]
- Ivorra, E.; Sánchez, A.; Camarasa, J.; Diago, M.; Tardaguila, J. Assessment of grape cluster yield components based on 3D descriptors using stereo vision. Food Control 2015, 50, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Hacking, C.; Poona, N.; Poblete-Echeverria, C. Vineyard yield estimation using 2-D proximal sensing: A multitemporal approach. OENO One 2020, 54, 793–812. [Google Scholar] [CrossRef]
- Mirbod, O.; Yoder, L.; Nuske, S. Automated measurement of berry size in images. IFAC-Pap. OnLine 2016, 49, 79–84. [Google Scholar] [CrossRef]
- Roscher, R.; Herzog, K.; Kunkel, A.; Kicherer, A.; Töpfer, R.; Förstner, W. Automated image analysis framework for high-throughput determination of grapevine berry sizes using conditional random fields. Comput. Electron. Agric. 2014, 100, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Aquino, A.; Diago, M.P.; Millán, B.; Tardaguila, J. A new methodology for estimating the grapevine-berry number per cluster using image analysis. Biosyst. Eng. 2017, 156, 80–95. [Google Scholar] [CrossRef]
- Clingeleffer, P. Crop Development, Crop Estimation and Crop Control to Secure Quality and Production of Major Wine Grape Varieties: A National Approach. Final Rep. Grape Wine Res. Dev. Corp. 2001, 148. Available online: http://hdl.handle.net/102.100.100/201731?index=1 (accessed on 6 March 2022).
- Kierdorf, J.; Weber, I.; Kicherer, A.; Zabawa, L.; Drees, L.; Roscher, R. Behind the Leaves—Estimation of Occluded Grapevine Berries with Conditional Generative Adversarial Networks. arXiv 2021, arXiv:2105.10325. Available online: http://rs.ipb.uni-bonn.de/wp-content/papercite-data/pdf/kierdorf2021behind.pdf (accessed on 6 March 2022).
- Parr, B.; Legg, M.; Bradley, S.; Alam, F. Occluded Grape Cluster Detection and Vine Canopy Visualisation Using an Ultrasonic Phased Array. Sensors 2021, 21, 2182. [Google Scholar] [CrossRef] [PubMed]


| Cultivar | Vine Age | Site | N° of Bunches | Vine Spacing Row × Interrow; m) | Training System | Pruning System | Irrigation |
|---|---|---|---|---|---|---|---|
| Arinto | 13 | PT | 48 | 1.0 × 2.5 | VSP | Unilateral Royat | Deficit drip |
| Cabernet | 18 | SA | 48 | 2.0 × 2.5 | VSP | Bilateral Royat | Drip |
| Encruzado | 13 | PT | 48 | 1.0 × 2.5 | VSP | Unilateral Royat | Deficit drip |
| Syrah | 20 | PT | 48 | 1.3 × 2.5 | VSP | Bilateral Royat | Rainfed |
| Cultivar | Bw (g) | Tb (n°) | BA (cm2) | BP (cm) | Vb (n°) | bA (cm2) | CI |
|---|---|---|---|---|---|---|---|
| Arinto | 423.5 ± 26.9 | 293.6 ± 18.9 | 107.8 ± 5.5 | 62.3 ± 2.3 | 108.0 ± 5.4 | 1.6 ± 0.1 | 7 |
| Cabernet | 160.6 ± 10.4 | 152.5 ± 9.2 | 85.7 ± 4.2 | 84.8 ± 4.5 | 81.2 ± 3.7 | 1.3 ± 0.1 | 7 |
| Encruzado | 173.0 ± 15.4 | 82.3 ± 7.1 | 60.0 ± 3.9 | 43.2 ± 1.7 | 46.5 ± 2.7 | 2.3 ± 0.1 | 7 |
| Syrah | 72.7 ± 6.5 | 71.4 ± 5.9 | 50.6 ± 4.2 | 68.7 ± 4.1 | 46.8 ± 2.9 | 1.2 ± 0.1 | 3 |
| Predictor | Cultivar | Equation | R2 | RMSPE (%) |
|---|---|---|---|---|
| BA | Arinto | Bw = 4.388 × BA − 56.059 | 0.81 *** | 18.2 |
| Cabernet | Bw = 2.238 × BA − 34.308 | 0.90 *** | 12.7 | |
| Encruzado | Bw = 3.632 × BA − 44.300 | 0.74 *** | 31.3 | |
| Syrah | Bw = 1.222 × BA + 9.493 | 0.86 *** | 19.7 | |
| Combined | Bw = 3.897 × BA − 91.060 | 0.71 *** | 45.0 | |
| Vb | Arinto | Bw = 4.427 × Vb − 65.395 | 0.86 *** | 15.4 |
| Cabernet | Bw = 2.085 × Vb − 12.593 | 0.72 *** | 21.1 | |
| Encruzado | Bw = 5.257 × Vb − 77.639 | 0.86 *** | 23.0 | |
| Syrah | Bw = 2.055 × Vb − 18.919 | 0.73 *** | 27.6 | |
| Combined | Bw = 4.066 × Vb − 81.125 | 0.77 *** | 39.8 | |
| BP | Arinto | Bw = 8.838 × BP − 143.440 | 0.55 *** | 28.0 |
| Cabernet | Bw = 1.220 × BP + 53.987 | 0.25 *** | 34.8 | |
| Encruzado | Bw = 6.603 × BP − 105.400 | 0.19 * | 54.7 | |
| Syrah | Bw = 1.215 × BP − 7.857 | 0.56 *** | 35.1 | |
| Combined | Bw = 1.524 × BP + 111.900 | 0.04 * | 81.6 |
| Step | Variable | Selection | AIC | R2 | Adj-R2 |
|---|---|---|---|---|---|
| 1 | Vb | Added | 1394.5 | 0.78 | 0.77 |
| 2 | BA_BP | Added | 1319.8 | 0.88 | 0.88 |
| 3 | BA_avg(bA) | Added | 1308.0 | 0.90 | 0.89 |
| 4 | BA | Added | 1294.6 | 0.91 | 0.91 |
| 5 | BP | Added | 1286.6 | 0.92 | 0.91 |
| 6 | BA_BP | Removed | 1285.1 | 0.92 | 0.91 |
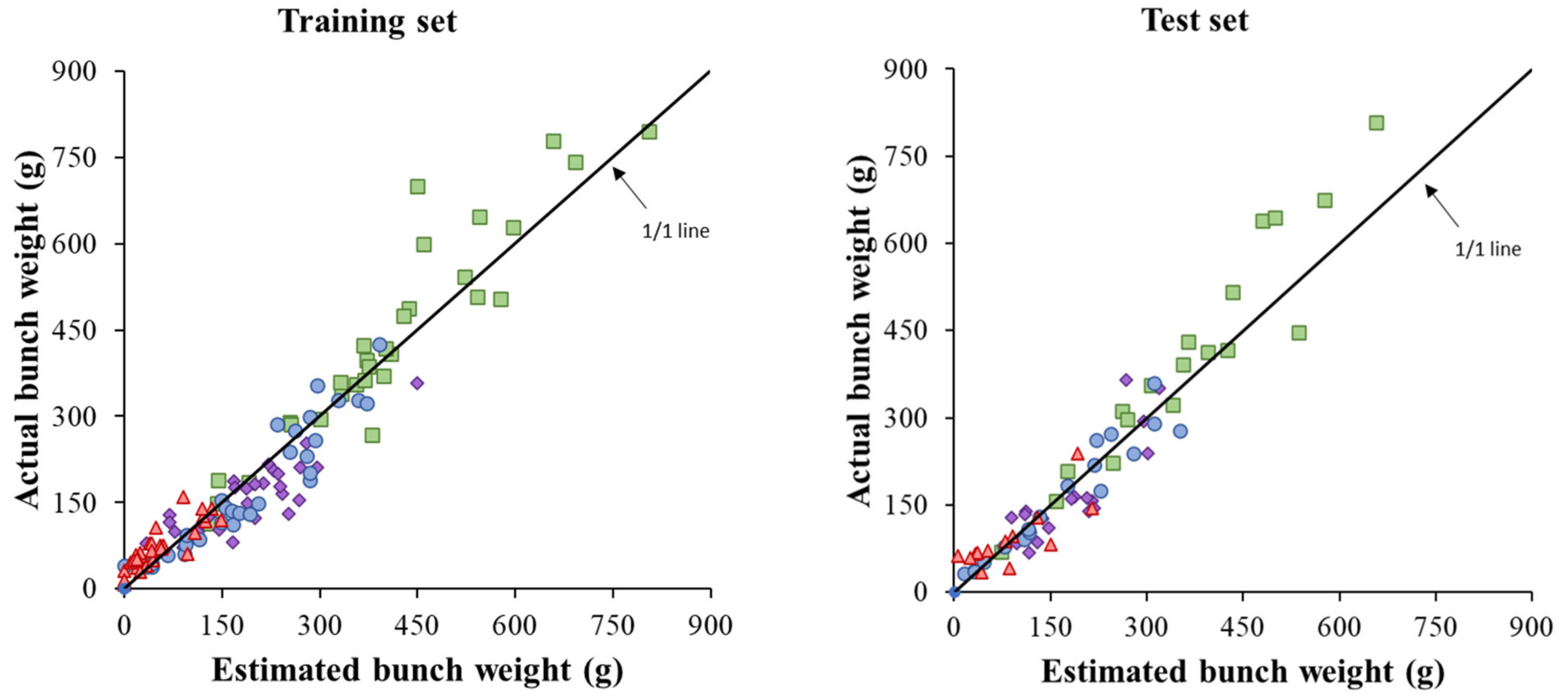
| Data Set | Linear Regression | R2 | RMSPE (%) | Bias (%) | EF | |
|---|---|---|---|---|---|---|
| 1 Slope | 2 Intercept | |||||
| Arinto | 1.202 * | −30.520 n.s. | 0.91 *** | 19.0 | 10.6 | 0.83 |
| Cabernet | 0.996 n.s. | −15.577 n.s. | 0.72 *** | 27.8 | −9.6 | 0.69 |
| Encruzado | 0.924 n.s. | −8.719 n.s. | 0.92 *** | 18.3 | −2.4 | 0.91 |
| Syrah | 0.599 *** | −39.822 *** | 0.70 *** | 60.5 | 23.0 | 0.28 |
| Combined | 1.073 n.s. | −4.184 n.s. | 0.91 *** | 25.9 | 4.9 | 0.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Victorino, G.; Poblete-Echeverría, C.; Lopes, C.M. A Multicultivar Approach for Grape Bunch Weight Estimation Using Image Analysis. Horticulturae 2022, 8, 233. https://doi.org/10.3390/horticulturae8030233
Victorino G, Poblete-Echeverría C, Lopes CM. A Multicultivar Approach for Grape Bunch Weight Estimation Using Image Analysis. Horticulturae. 2022; 8(3):233. https://doi.org/10.3390/horticulturae8030233
Chicago/Turabian StyleVictorino, Gonçalo, Carlos Poblete-Echeverría, and Carlos M. Lopes. 2022. "A Multicultivar Approach for Grape Bunch Weight Estimation Using Image Analysis" Horticulturae 8, no. 3: 233. https://doi.org/10.3390/horticulturae8030233
APA StyleVictorino, G., Poblete-Echeverría, C., & Lopes, C. M. (2022). A Multicultivar Approach for Grape Bunch Weight Estimation Using Image Analysis. Horticulturae, 8(3), 233. https://doi.org/10.3390/horticulturae8030233








