Marker-Assisted Pyramiding of Genes for Multilocular Ovaries, Self-Compatibility, and Clubroot Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Marker Development, Validation, and Phenotype Genetic Analysis
2.3. Crossing Program and MAS for Gene Pyramiding
2.4. Phenotypic Characterization
3. Results
3.1. Marker Development and Genetic Analysis for Multilocular and Self-Compatibility Traits
3.2. Polymorphic Marker Screening for Foreground Selection
3.3. Polymorphic Markers Screening for Background Selection of the Whole Genome
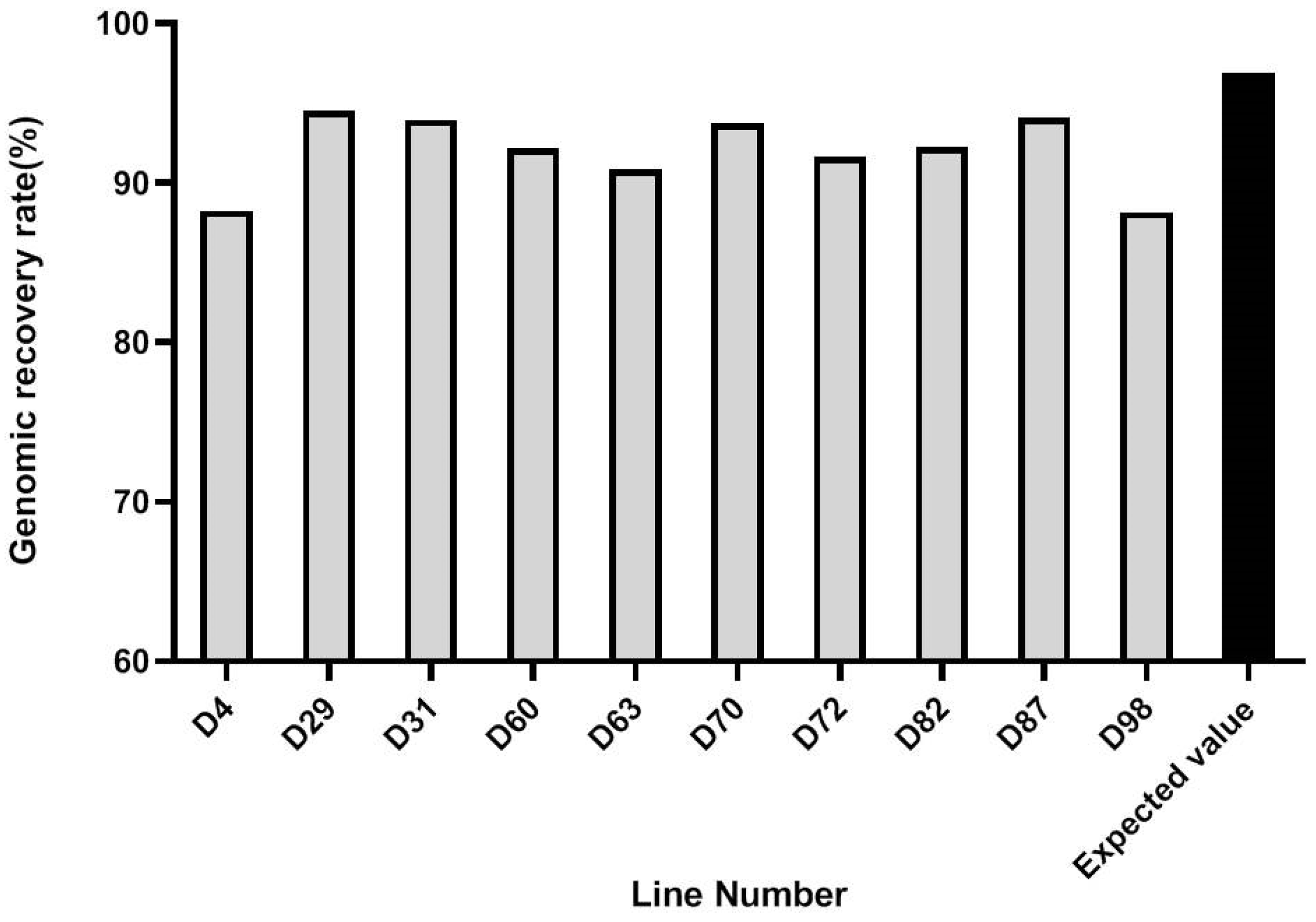
3.4. Marker-Assisted Pyramiding of Multilocular, Self-Compatibility, and CRb Genes
3.5. Phenotypic Evaluation of a Pyramided Line
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagaharu, U. Genome analysis in Brassica carinata with special reference to the experimental formation of Brassica napus, a peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Zhao, J.; Wang, X.; Deng, B.; Lou, P.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Somnath, R.; Sinhamahapatra, S.P. Relationship between seed yield and yield components in bilocular and tetralocular yellow sarson (Brassica rapa). Indian J. Agric. Sci. 2011, 81, 643–647. [Google Scholar]
- Yadava, S.K.; Paritosh, K.; Panjabi-Massand, P.; Gupta, V.; Chandra, A.; Sodhi, Y.S.; Pradhan, A.K.; Pental, D. Tetralocular ovary and high silique width in yellow sarson lines of Brassica rapa (subspecies trilocularis) are due to a mutation in Bra034340 gene, a homologue of CLAVATA3 in Arabidopsis. Theor. Appl. Genet. 2014, 127, 2359–2369. [Google Scholar] [CrossRef]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005, 56, 467–489. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Nishio, T. Commonalities and differences between Brassica and Arabidopsis self-incompatibility. Hortic. Res. 2014, 1, 14054. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.J. Self-incompatibility systems in angiosperms: III. Crucif. Hered. 1955, 9, 53–68. [Google Scholar] [CrossRef] [Green Version]
- Nasrallah, J.B.; Nishio, T.; Nasrallah, M.E. The Self-incompatibility genes of Brassica: Expression and use in genetic ablation of floral tissues. Plant Mol. Biol. 1991, 42, 393–422. [Google Scholar] [CrossRef]
- Watanabe, M.; Takasaki, T.; Toriyama, K.; Yamakawa, S.; Isogai, A.; Suzuki, A.; Hinata, K. A high degree of homology exists between the protein encoded by SLG and the S receptor domain encoded by SRK in self-incompatible Brassica campestris L. Plant Cell Physiol. 1994, 35, 1221–1229. [Google Scholar] [CrossRef]
- Stein, J.C.; Howlett, B.; Boyes, D.C.; Nasrallah, M.E.; Nasrallah, J.B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 1991, 88, 8816–8820. [Google Scholar] [CrossRef] [Green Version]
- Hatakeyama, K.; Takasaki, T.; Watanabe, M.; Hinata, K. Molecular characterization of S locus genes, SLG and SRK, in a pollen-recessive self-incompatibility haplotype of Brassica rapa L. Genetics 1998, 149, 1587–1597. [Google Scholar] [CrossRef]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The male determinant of self-incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef]
- Suzuki, G.; Kai, N.; Hirose, T.; Fukui, K.; Nishio, T.; Takayama, S.; Isogai, A.; Watanabe, M.; Hinata, K. Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S(9) haplotype of Brassica campestris (syn. rapa). Genetics 1999, 153, 391–400. [Google Scholar] [CrossRef]
- Kachroo, A.; Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001, 293, 1824–1826. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shimosato, H.; Shiba, H.; Funato, M.; Che, F.S.; Watanabe, M.; Iwano, M.; Isogai, A. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 2001, 413, 534–538. [Google Scholar] [CrossRef]
- Hiroko, S.; Yokota, N.; Shiba, H.; Iwano, M.; Entani, T.; Che, F.S.; Watanabe, M.; Isogai, A.; Takayama, S. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 2007, 19, 107–117. [Google Scholar]
- Dixon, G.R. The occurrence and economic impact of plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Jing, W.; Yun, H.; Li, X.; Li, H. Research progress in clubroot of crucifers. Plant Prot. 2011, 37, 153–158. [Google Scholar]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Special Issue: Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated Control of Clubroot. J. Plant Growth Regul. 2009, 28, 289–303. [Google Scholar] [CrossRef]
- Kageyama, K.; Asano, T. Life Cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Mehraj, H.; Akter, A.; Miyaji, N.; Miyazaki, J.; Shea, D.J.; Fujimoto, R.; Doullah, M.A.-U. Genetics of clubroot and fusarium wilt disease resistance in Brassica vegetables: The Application of marker assisted breeding for disease resistance. Plants 2020, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage. Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Matsumoto, S.; Hirai, M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 2003, 107, 997–1002. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Hirai, M.; Harada, T.; Kubo, N.; Tsukada, M.; Suwabe, K.; Matsumoto, S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 2004, 108, 639–643. [Google Scholar] [CrossRef]
- Saito, M.; Kubo, N.; Matsumoto, S.; Suwabe, K.; Tsukada, M.; Hirai, M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 2006, 114, 81–91. [Google Scholar] [CrossRef]
- Piao, Z.Y.; Deng, Y.Q.; Choi, S.R.; Park, Y.J.; Lim, Y.P. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2004, 108, 1458–1465. [Google Scholar] [CrossRef]
- Chen, J.; Jing, J.; Zhan, Z.; Zhang, T.; Zhang, C.; Piao, Z. Identification of novel QTLs for isolate-specific partial resistance to plasmodiophora brassicae in Brassica rapa. PLoS ONE 2013, 8, e85307. [Google Scholar] [CrossRef] [Green Version]
- Pang, W.; Fu, P.; Li, X.; Zhan, Z.; Yu, S.; Piao, Z. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 2018, 9, 653. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Zhang, X.; Huang, Z.; Chu, M.; Song, T.; Falk, K.C.; Deora, A.; Chen, Q.; Yan, Z.; Mcgregor, L. Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS ONE 2016, 11, e0153218. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, X.; Peng, G.; Falk, K.C.; Strelkov, S.E.; Gossen, B.D. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017, 7, 4516. [Google Scholar] [CrossRef] [PubMed]
- Zhen, H.; Peng, G.; Liu, X.; Abhinandan, D.; Falk, K.C.; Gossen, B.D.; Mcdonald, M.R.; Yu, F. Fine mapping of a clubroot resistance gene in Chinese Cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 2017, 8, 1448. [Google Scholar]
- Yamanaka, N.; Hossain Md, M. Pyramiding three rust-resistance genes confers a high level of resistance in soybean (Glycine max). Plant Breed. 2019, 138, 686–695. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Liu, Y.; Dai, H.; He, J.; Kang, H.; Pan, G.; Huang, J.; Qiu, Z.; Wang, Q.; et al. Marker assisted pyramiding of two brown planthopper resistance genes, Bph3 and Bph27 (t), into elite rice cultivars. Rice 2016, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Yang, Q.; Huang, M.; Guo, T.; Liu, Y.; Wang, J.; Yang, G.; Zhou, J.; Yang, J.; Zhu, X.; et al. Improvement of rice blast resistance by developing monogenic lines, two-gene pyramids and three-gene pyramid through MAS. Rice 2019, 12, 78. [Google Scholar] [CrossRef]
- Shah, N.; Li, Q.; Xu, Q.; Liu, J.; Huang, F.; Zhan, Z.; Qin, P.; Zhou, X.; Yu, W.; Zhu, L.; et al. CRb and PbBa8.1 synergically increases resistant genes expression upon infection of Plamodiophora brassicae in Brassica napus. Genes 2020, 11, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, E.; Ueno, H.; Aruga, D.; Sakamoto, K.; Hayashida, N. Accumulation of three clubroot resistance genes through marker-assisted selection in Chinese cabbage (Brassica rapa ssp. pekinensis). J. Jpn. Soc. Hortic. Sci. 2012, 81, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Tomita, H.; Shimizu, M.; Doullah, M.A.U.; Fujimoto, R.; Okazaki, K. Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Mol. Breed. 2013, 32, 889–900. [Google Scholar] [CrossRef]
- Tyagi, S.; Mir, R.R.; Kaur, H.; Chhuneja, P.; Ramesh, B.; Balyan, H.S.; Gupta, P.K. Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.). Mol. Breed. 2014, 34, 167–175. [Google Scholar] [CrossRef]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and –susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ramchiary, N.; Choi, S.R.; VanNguyen, D.; Hossain, M.J.; Yang, H.; Lim, Y.P. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 2010, 53, 939–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L.ssp. pekinensis). Hortic. Environ. Biotechnol. 2011, 52, 494–501. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Z.; Zhang, C.; Pang, W.; Choi, S.R.; Lim, Y.P.; Piao, Z. Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa. Mol. Breed. 2014, 34, 1173–1183. [Google Scholar] [CrossRef]
- Li, X.; Ramchiary, N.; Dhandapani, V.; Choi, S.R.; Hur, Y.; Nou, I.-S.; Yoon, M.K.; Lim, Y.P. Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Res. 2013, 20, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapa L. Breed. Sci. 2011, 61, 160–167. [Google Scholar] [CrossRef]
- Williams, P.H. A system for the determination of races of Plasmodiophora brassicae that infect Cabbage and Rutabaga. Phytopathology 1966, 56, 624–626. [Google Scholar]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Nasrallah, J.B. Plant mating systems: Self-incompatibility and evolutionary transitions to self-fertility in the mustard family. Curr. Opin. Genet. Dev. 2017, 47, 54–60. [Google Scholar] [CrossRef]
- Xu, P.; Lv, Z.; Zhang, X.; Wang, X.; Pu, Y.; Wang, H.; Yi, B.; Wen, J.; Ma, C.; Tu, J.; et al. Identification of molecular markers linked to trilocular gene (mc1) in Brassica juncea L. Mol. Breed. 2014, 33, 425–434. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, V.K.; Singh, S.P.; Pandian, R.T.P.; Ellur, R.K.; Singh, D. Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012, 2012, pls029. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalingam, J.; Raveendra, C.; Savitha, P.; Vidya, V.; Chaithra, T.L.; Velprabakaran, S.; Saraswathi, R.; Ramanathan, A.; Pillai, M.P.A.; Arumugachamy, S.; et al. Gene Pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 2020, 11, 591457. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Chiu, C.H.; Yap, R.; Tseng, Y.C.; Wu, Y.P. Pyramiding bacterial blight resistance genes in Tainung82 for broad-spectrum resistance using marker-assisted selection. Int. J. Mol. Sci. 2020, 21, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balachiranjeevi, C.; Bhaskar, N.S.; Abhilash, V.; Akanksha, S.; Viraktamath, B.C.; Madhav, M.S.; Hariprasad, A.S.; Laha, G.S.; Prasad, M.S.; Balachandran, S.M.; et al. Marker-assisted introgression of bacterial blight and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice. Mol. Breed. 2015, 35, 151. [Google Scholar] [CrossRef]





| Gene | Chromosomal Location | Markers | Type of Marker | Character |
|---|---|---|---|---|
| Bra034340 | A4 | sau_um190 | Based on PCR | Co-dominant |
| cun_146a | Based on PCR | Co-dominant | ||
| Teo-1 | SSR | Co-dominant | ||
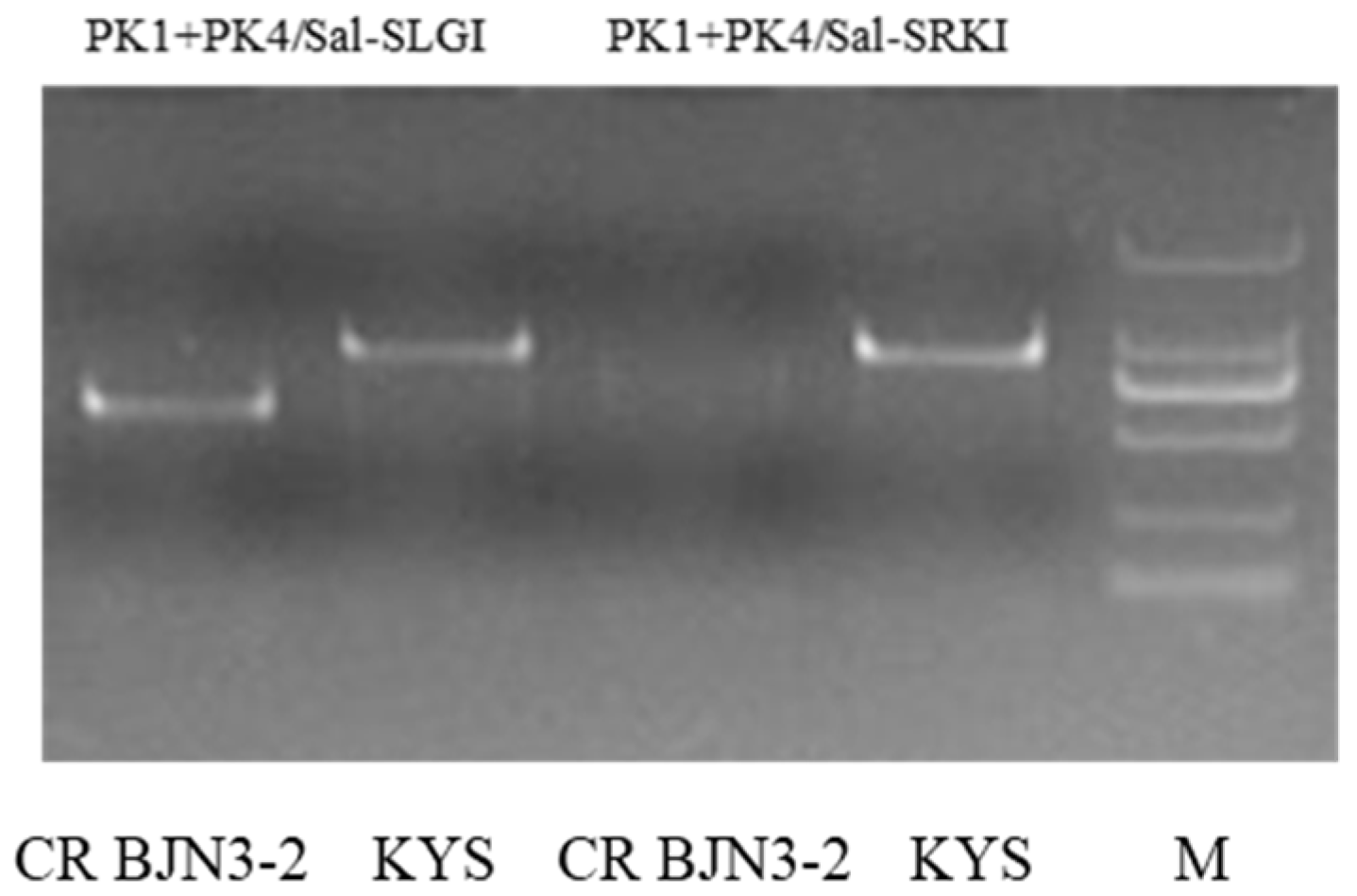
| S-Locus | A7 | PK1+PK4/Sal-SLGI | Based on PCR | Co-dominant |
| SCF-6 | SSR | Co-dominant | ||
| SC-12 | SSR | Co-dominant | ||
| CRb | A3 | TCR74 | SSR | Co-dominant |
| TCR79 | SSR | Co-dominant |
| Material | Level 0 | Level 1 | Level 2 | Level 3 | Disease INCIDENCE (%) | Disease Index (DI) |
|---|---|---|---|---|---|---|
| CR BJN3-2 | 30 | 3 | 6 | 5 | 31.8 | 23.5 |
| BJN3-2 | 50 | 100 | 100 | |||
| 552-39 | 48 | 2 | 4 | 4 |
| Material | SCI |
|---|---|
| CR BJN3-2 | 0.48 ± 0.02 |
| KYS | 1.37 ± 0.29 |
| 552-39 | 1.15 ± 0.18 |
| Agronomic Traits | 552-39 | CR BJN3-2 |
|---|---|---|
| Plant height (cm) | 35.3 ± 0.58 | 34.5 ± 4.95 |
| Plant width (cm) | 49.0 ± 7.00 | 48.0 ± 0.00 |
| Plant weight (kg) | 2.23 ± 0.29 | 2.17 ± 0.15 |
| Number of outer leaves | 25.7 ± 3.06 | 19.5 ± 2.12 |
| Leaf length (cm) | 32.0 ± 1.73 | 34.0 ± 2.12 |
| Leaf width (cm) | 19.8 ± 2.75 | 21.0 ± 0.00 |
| Petiole length (cm) | 18.5 ± 1.00 | 17.8 ± 1.06 |
| Petiole width (cm) | 5.3 ± 0.58 | 5.5 ± 0.71 |
| Petiole color | Green | Green |
| Head weight (kg) | 1.16 ± 0.17 | 1.09 ± 0.08 |
| Head shape | Folded | Folded |
| Head solidity | Compaction | Compaction |
| Head length (cm) | 27.7 ± 2.84 | 23.9 ± 0.21 |
| Head width (cm) | 12.8 ± 2.47 | 10.7 ± 0.35 |
| Head color | Pale Yellow | Pale Yellow |
| Stem length (cm) | 2.83 ± 1.26 | 3.25 ± 0.35 |
| Stem width (cm) | 2.30 ± 0.75 | 2.90 ± 0.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Zhao, H.; Ma, Y.; Jiang, M.; Zhan, Z.; Li, X.; Piao, Z. Marker-Assisted Pyramiding of Genes for Multilocular Ovaries, Self-Compatibility, and Clubroot Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2022, 8, 139. https://doi.org/10.3390/horticulturae8020139
Zheng J, Zhao H, Ma Y, Jiang M, Zhan Z, Li X, Piao Z. Marker-Assisted Pyramiding of Genes for Multilocular Ovaries, Self-Compatibility, and Clubroot Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae. 2022; 8(2):139. https://doi.org/10.3390/horticulturae8020139
Chicago/Turabian StyleZheng, Jingyi, Huicai Zhao, Yingmei Ma, Mingliang Jiang, Zongxiang Zhan, Xiaonan Li, and Zhongyun Piao. 2022. "Marker-Assisted Pyramiding of Genes for Multilocular Ovaries, Self-Compatibility, and Clubroot Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)" Horticulturae 8, no. 2: 139. https://doi.org/10.3390/horticulturae8020139
APA StyleZheng, J., Zhao, H., Ma, Y., Jiang, M., Zhan, Z., Li, X., & Piao, Z. (2022). Marker-Assisted Pyramiding of Genes for Multilocular Ovaries, Self-Compatibility, and Clubroot Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae, 8(2), 139. https://doi.org/10.3390/horticulturae8020139






