Abstract
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is one of the most widely cultivated and economically important vegetables in China. Constructing an effective genetic linkage map and mapping quantitative trait loci (QTLs) related to yield and leafy head morphology is of great importance for molecular breeding of Chinese cabbage. Using two diverse Chinese cabbage inbred lines, ZHB and G291, as parents, an F2 segregating population consisting of 240 individuals was prepared for genetic map construction and phenotype investigation in this study. The two parents are significantly different in both shape and size. Sixteen important agronomic traits of F2 individuals were investigated. A genetic map of 105 intragenic simple sequence repeat (SSR) markers distributed across 10 linkage groups (LGs) was constructed, which was 2034.1 cM in length and had an average inter-locus distance of 21.75 cM. We identified 48 QTLs for the tested important agronomic traits on the studied LGs, with LOD scores of 2.51–12.49, which explained the phenotypic variance of 3.41–26.66%. The QTLs identified in this study will facilitate further genetic analysis and marker-assisted genetic improvement of Chinese cabbage.
1. Introduction
Chinese cabbage (Brassica rapa L. ssp. pekinensis, AA, 2n = 20), which originated in China, is one of the most widely cultivated and economically important vegetables in eastern Asia. After hundreds of years of evolution and breeding, the important agronomic traits related to yield vary greatly among different Chinese cabbage varieties. The inheritance of these yield-related agronomic traits is of great importance for genetic improvement of Chinese cabbage. However, most of these traits are complex quantitative traits, and the expression of the controlling genes is influenced by the internal and/or external environment [1,2]. The genetic base and molecular mechanisms involved in regulating these agronomic traits are yet to be understood.
Genetic linkage maps are effective tools for studying and locating the genetic loci of interesting traits in the genomes of plants. In previous studies, some genetic maps of Chinese cabbage have been constructed using different molecular markers, such as AFLP [3], RFLP [4], STS [5], simple sequence repeat (SSR) [6,7,8,9], InDel [7,8], and SNP [2,10,11], in different genetic populations such as F2 [2,3,6,7,9], F3 [5], RIL [4,8,10], and DH lines [1,11]. A number of quantitative trait loci (QTLs) have been identified in Chinese cabbage recently, including trichome number [12,13,14], flowering time [11,15], flower color [16], anthocyanin accumulation [17], plant morphological traits [7,15,18,19,20,21], orange inner leaves [6,22], seed coat color [3,9,23,24], bolting trait [8], floral stalk length [25], disease resistance [5,26,27,28,29], reproductive fitness traits [4], and yield-related traits [1,30]. For some genetically simple traits such as trichome number [12,13,14], seed coat color [3,9,25], and orange inner leaves [6,22,31], many candidate genes have been identified according to map-based cloning methods, and efficient molecular markers have been developed for marker-assisted selection (MAS). However, for plant morphological and yield-related traits, only a handful of candidate genes and efficient molecular markers have been used for MAS [1].
Intragenic SSRs are more conserved and transferable than extragenic SSRs [32,33,34], especially the expressed sequence tags SSRs (EST-SSRs) found in transcribed sequences. These SSRs are potentially more efficient for QTL mapping, gene targeting, and marker-assisted breeding than genomic-SSRs [35], which have been widely used in genetic linkage map construction in plants [36,37,38,39]. In Chinese cabbage, some EST-SSRs have been used for genetic linkage map construction with other molecular markers [30].
In previous studies, genomic SSRs and EST-SSRs have been identified and analyzed at the whole genome and transcriptome levels in Chinese cabbage [40,41]. In the present study, we aimed to construct a genetic linkage map using intragenic SSRs and map QTLs for important agronomic traits in Chinese cabbage. This study will provide useful information for better understanding of the molecular bases of these complex quantitative traits and molecular breeding in Chinese cabbage.
2. Materials and Methods
2.1. Plant Materials and Trait Measurements
A F2 segregating population was developed by crossing two Chinese cabbage inbred lines, ZHB and G291. The two parents are significantly different in both size and shape. The parents and the F2 generations were planted in the field at the normal sowing time (15 August 2016) in Changqing, Jinan, China, with a row spacing of 50 cm and plant spacing of 50 cm. The plants were harvested in mid-November 2016. A total of 240 F2 individuals were selected randomly for trait measurements and genetic linkage map construction. Sixteen agronomic traits, including plant height (PH), plant width (PW), gross weight (GW), number of non-wrapper leaves (NNL), head weight (HW), head height (HH), head diameter (HD), number of head leaves (NHL), number of all leaves (NAL), maximum leaf length (MLL), maximum leaf width (MLW), petiole length (PEL), petiole width (PEW), petiole thickness (PET), stem length (SL), and stem width (SW), were measured following the descriptions for Brassica by the International Bureau of Plant Genetic Resources (IBPGR, 1990) (Table 1). The mean values and standard deviations for the agronomic parameters and the correlations between agronomic traits were analyzed by SPSS v13.0 for Windows (SPSS Inc., Chicago, IL, USA).

Table 1.
Summary of the agronomic traits and their measurements.
2.2. DNA Isolation and Marker Genotyping
The total DNA of the parental lines and F2 individuals was extracted from young leaves (two weeks old) using the modified CTAB method [42]. DNA quantity and quality were assessed using a NanoDrop ND-1000 Spectrophotometer (Nano-Drop, Wilmington, DE, USA) and electrophoresis in 1.0% agarose gel using 0.5× TBE electrophoresis buffer, respectively. The DNA was diluted to 10 ng/μL and amplified by polymerase chain reaction (PCR). Five hundred SSRs distributed across 10 chromosomes of the Brassica rapa A genome were selected for polymorphic survey between the parental lines according to Shi et al. (2014) and Ding et al. (2015) [40,41]. PCR reactions were performed in a 96-well plate at 95 °C for 5 min, followed by 35 cycles of reaction (95 °C for 30 s, followed by 55–60 °C for 30 s and 72 °C for 30 s), and a final step of 72 °C for 5 min. The appropriate annealing temperatures depend on each primer pair. The PCR products were resolved by 6% denaturing polyacrylamide gel electrophoresis. Codominant polymorphic SSR markers with a single PCR brand selected from the parental polymorphism were used for marker genotyping in the F2 lines.
2.3. Genetic Map Construction and QTL Analysis
We used the QTL Icimapping software V4.1 to construct the genetic map [43]. Redundant markers and markers with a missing rate greater than 20% were deleted using the “BIN” functionality of the software. The chi-square (χ2) test was used to check all the polymorphic SSRs for the goodness of fit against a 1:2:1 segregation ratio (p < 0.01). For genetic map construction, the genotype data of the homozygous alleles from the parent lines ZHB and G291 were recorded as “2” and “0”, respectively. The heterozygous genotypes were recorded as “1”, and all missing data were recorded as “−1”. All markers were grouped at LOD = 2.5 for genetic map construction. The ICIM-ADD mapping method at a LOD threshold of 2.5 was used for QTL mapping with the Icimapping software V4.1 [43].
3. Results
3.1. Construction of the Brassica rapa Linkage Map
A total of 500 intragenic SSR markers, distributed across 10 chromosomes of the Brassica rapa A genome (50 markers per chromosome), were randomly selected for polymorphic survey between the parental lines from the SSRs developed by Shi et al. (2014) and Ding et al. (2015) [40,41]. Among the 500 intragenic SSRs, 133 were polymorphic between ZHB and G291 with a polymorphism rate of 26.6%. Only 105 clearly visible co-dominant polymorphic SSRs were recognized as usable markers for map construction (Table S1). Of the 105 polymorphic SSRs, 62 were located in exons and 43 were located in introns (Table S1).
The polymorphic SSRs were screened on these 240 F2 individuals, and the results showed that 98 markers (93.33%) had the expected 1:2:1 segregation for the parental alleles (p < 0.01), while seven markers (6.67%) were distorted from the expected segregation ratio. Of the seven distorted markers, one was on A01, A06, and A07, respectively, and two were distributed on A03 and A09, respectively. Four distorted markers had a segregation bias in favor of G291, and one in favor of ZHB, while the remaining two distorted markers were in favor of the heterozygous genotype.
A genetic map comprising these 105 polymorphic SSRs was constructed, which was 2034.1 cM in length (Table 2). The SSRs were assigned to 10 linkage groups (LGs), putatively corresponding to the haploid chromosome number of Brassica. rapa. The number of the SSR markers in each of the 10 linkage groups varied from 5 (A05) to 15 (A06) (Table 2). The length of LGs varied from 85.53 cM (A05) to 390.5 cM (A06), and the average linkage group size was 203.41 cM. The average inter-locus distance was 21.75 cM. The smallest marker interval of 2.33 cM was found between A10S24 and A10S23 on A10, while the largest marker interval of 94.1 cM was found on A06 between A06S6 and A06S19 (Table 2).

Table 2.
Features of the EST-SSR-based genetic linkage map of Brassica rapa.
3.2. Investigation and Statistical Analysis of Agronomic Parameters
A total of 240 F2 individuals were randomly selected for trait measurements and genetic linkage map construction. Sixteen important agronomic traits were investigated and analyzed thoroughly (Table 1). The results showed that the parents were significantly different in all the sixteen agricultural traits (Table 3). The plant and leafy head of the paternal parent ‘G291’ were larger and heavier than those of the maternal parent ‘ZHB’, while ‘ZHB’ had longer CL than ‘G291’ (Table 3). The F1 line exhibited strong heterosis, as almost all the tested traits were larger than the parents (Table 3). In the F2 population, all the 16 traits investigated in the study showed a continuous distribution and a wide genetic variation (Table 3, Figure S1). Coefficient of Variation (CV) was used to evaluate the genetic variation of the traits in these 240 F2 lines, and the results showed that head weight, stem length, and gross weight had wider variations than the other traits, with a CV of 33.29%, 30.73%, and 29.52%, respectively, followed by number of non-wrapper leaves with a CV of 23.06% (Table 3). The CV of the other 13 agricultural traits varied from 10.82% to 16.63% (Table 3).

Table 3.
Overview of the phenotypic traits in the parental, F1, and F2 lines used for mapping construction and QTL mapping in Brassica rapa.
Most of the traits showed significant positive correlations with other traits (Table 4). Head weight, as the most important trait representing the yield of Chinese cabbage, showed significant positive correlations with all other traits tested in the study except number of non-wrapper leaves (p < 0.01). Head weight had the largest correlation coefficient with gross weight (with the correlation coefficient of 0.824), followed by that with head diameter and MLW, with correlation coefficients of 0.645 and 0.627, respectively (Table 4).

Table 4.
Correlation co-efficient analysis of the 16 agricultural traits tested in the study.
3.3. QTL Analysis
A total of 48 QTLs on ten chromosomes were detected for the 16 traits of Chinese cabbage. Two (on A04 and A09) to twelve QTLs (on A03) were detected in these ten LGs. The number of the detected QTLs ranged from 0 for stem length and head diameter to 7 for number of non-wrapper leaves, and the confidence interval covered by individual QTLs ranged from 2.33 cM (qMLL-6) to 96.06 cM (qSW-2). The percentage of phenotypic variation (R2) explained by individual QTLs ranged from 3.41% (qPEL-3) to 26.66% (qPEW-2), and the LOD scores of individual QTLs varied from 2.51 (qHH-4) to 12.49 (qPEW-2) (Table 5).

Table 5.
Details of the QTLs identified for the 16 traits of Chinese cabbage.
Two and four QTLs for plant height (qPH-1 and qPH-2) and plant width (qPW-1, qPW-2, qPW-3 and qPW-4) were detected, respectively. Of these QTLs, qPH-1 and qPW-1 showed a relatively higher LOD score and R2, suggesting that these QTLs may be a major QTL for plant height and plant width, respectively. One QTL for gross weight (qGW-1) was detected on A10 between the SSR A10S4-A10S24 with a LOD score of 3.43 and a confidence interval of 4.45 cM, explaining 5.26% of the phenotypic variation (Table 5).
Eight QTLs were identified for leafy head-related traits (head weight, head height and head diameter). Two of these QTLs, qHW-1 on A05 and qHW-2 on A10, were identified for head weight, explaining 6.17% and 5.22% of the phenotypic variation, respectively. Six QTLs for head height (qHH-1 on A01, qHH-2 on A02, qHH-3 on A03, qHH-4 on A05, qHH-5 on A06 and qHH-6 on A10) were detected and the individual QTL explained 4.29–13.39% of the phenotypic variation. qHH-2 on A02 was the major QTL with a comparatively higher LOD of 6.96, explaining 13.39% of the phenotypic variation. No QTL was identified for head diameter in the study (Table 5).
For leaf number-related traits (number of non-wrapper leaves, number of head leaves and number of all leaves), 11 QTLs were detected on 9 LGs (A01, A02, A03, A04, A05, A06, A07, A09 and A10). Seven QTLs were identified for number of non-wrapper leaves, of which three were located on A07, and one was located on A02, A03, A04 and A09, respectively. qNNL-2, qNNL-3 and qNNL-7 were the major QTLs for number of non-wrapper leaves with a relatively higher LOD (5.19, 5.97 and 3.55, respectively) and R2 (10.19%, 9.98% and 13.33%, respectively). Two QTLs were detected for number of head leaves, of which qNHL-1 on A01 between A1S30 and A1S5 was the major QTL, with a LOD score of 3.23 and R2 of 15.13% at the peak position of 58.80 cM. For number of all leaves, two QTLs were detected on A02 and A05 with an R2 of 4.66% and 5.97%, respectively (qNAL-1 and qNAL-2).
A total of 19 QTLs were identified for the five leaf-related traits (maximum leaf length, maximum leaf width, petiole length, petiole width and petiole thickness) on 8 LGs (A02, A03, A04, A06, A07, A08, A09 and A10). For maximum leaf length, 6 QTLs were detected on A02, A03, A04 and A10, of which qMLL-3 and qMLL-1 were the major QTLs with a relatively higher LOD (7.61 and 5.60, respectively) and R2 (12.42% and 10.07, respectively). Two QTLs were identified for a maximum leaf width on A03 and A07, explaining 7.74% and 9.23% of the phenotypic variation, respectively. Three, three, and five QTLs were identified for petiole length, petiole width and petiole thickness, respectively. One major QTL for petiole width was detected on A09 (qPEW-2) with an LOD of 12.49 and R2 of 26.66%. No major QTLs were detected for petiole length and petiole thickness as the R2 of these QTLs were lower than 10% (Table 5).
For the stem-related traits (stem length and stem width), three QTL loci for stem width were identified on A06, A08 and A09. No QTL was detected for stem length in the study.
3.4. Clustering of QTLs
Of the 48 QTLs detected in 10 LGs, many QTLs were found to map in the same QTL region. The QTL region of A02, A03 and A10 showed multiple QTLs for three or more traits (Figure 1). Six QTLs for plant height (qPH-1), head height (qHH-2), number of head leaves (qNHL-2), number of all leaves (qNAL-1), maximum leaf length (qMLL-1) and petiole length (qPEL-1) were mapped in the middle portion of A02 (107.24–121.10 cM) (Figure 1 and Figure S2). Four of these six QTLs, qPH-1, qHH-2, qMLL-1 and qPEL-1, obtained increasing alleles from the parent line “G291”, while qNHL-2 and qNAL-1 obtained increasing alleles from the parent line “ZHB” (Table 5). Four QTLs for plant width (qPW-2), head height (qHH-3), maximum leaf length (qMLL-3) and petiole length (qPEL-2) were mapped nearby A3S20 (132.08 cM) of A03, which derived increasing alleles from the parent line “G291” (Figure 1 and Table 5). The lower part of A03 (172.68–187.61 cM) showed mapping of QTLs for plant width (qPW-3), maximum leaf length (qMLL-3), petiole length (qPEL-3) and petiole thickness (qPET-2). Three (qMLL-3, qPEL-3, and qPET-2) of the four QTLs obtained increasing alleles from the parent line “G291”, except qPW-2. We also observed three QTLs for gross weight (qGW-1), head height (qHH-6), and maximum leaf length (qMLL-6) nearby A10S24 (74.85 cM) of A10, all of which derived increasing alleles from the parent line “G291” (Figure 1 and Table 5).
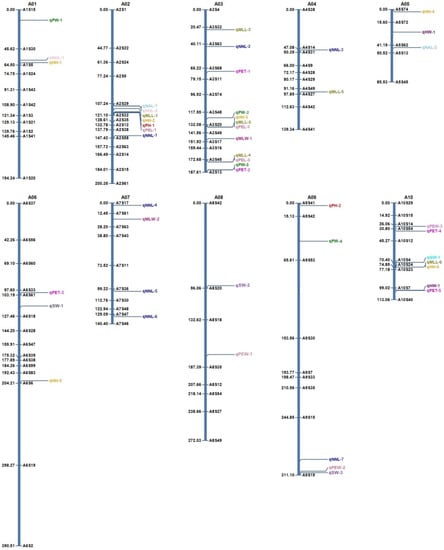
Figure 1.
Brassica rapa genetic linkage map and QTLs for agronomic traits discovered in F2 lines. Genetic distances (cM) are shown on the left side of the linkage group, and the names of the SSRs are shown on the right side of the linkage group.
4. Discussion
4.1. Intragenic SSR-Based Linkage Map Construction in Chinese Cabbage
With the development of high-throughput sequencing technology, the whole genome and transcriptome sequencing of Chinese cabbage have been performed in recent years [46,47], which provides great convenience for developing molecular markers and identifying genetic loci regulating qualitative and quantitative traits in Chinese cabbage. In previous studies, over 140 thousand genomic SSRs and 10 thousand EST-SSRs with a clear physical position have been developed at the whole genome and transcriptome level, respectively [40,41]. In the present study, 500 intragenic SSR markers distributed across 10 chromosomes of the Brassica rapa A genome (50 markers per chromosome) were randomly selected for polymorphic survey between the parental lines. Finally, 105 clearly visible co-dominant polymorphic SSRs were used for genetic map construction and QTLs mapping. This is the first genetic map construction in Chinese cabbage exclusively using distributed intragenic SSR markers. As the physical positions were clear for all the SSRs, it will provide great convenience for further map-based cloning and candidate gene screening for QTLs using this genetic map.
4.2. QTLs for Important Agronomic Traits of Chinese Cabbage
As an economically important leafy vegetable, yield and morphology-related agronomic traits, such as plant height, plant width, leafy head-related traits, leaf number-related traits and central axis-related traits, are important in Chinese cabbage breeding to achieve more attractive plants with higher yield and better architecture according to customers’ demands. In previous studies, QTLs for some yield and morphology-related agronomic traits have been identified using DH, F2, and RIL lines [1,7,15,18,19,20,21,30]. The occurrence of detectable QTL depends on polymorphisms present in the studied population, so different QTLs can be identified in different studies of the same species. Furthermore, as most of these traits were inherited, complex quantitative traits, the QTLs detected under different backgrounds and environments or by different software are usually not consistent. Here, we list the information of QTLs for important agronomic traits identified in previous studies to help us screen efficient candidate QTLs (Table 5).
Plant height and width are important traits, which are associated closely with yield and morphology of Chinese cabbage. Plant width also influences plant space in the field culture. In this study, two QTLs for plant height were identified on A02 and A09. QTLs for plant height have also been detected on A02 [30] and A09 [2,44] in previous studies. QTLs for plant height have been detected on A01, A04, A07, A08 and A10 in other studies [30,44,45]. Three QTLs for plant width are located on the linkage group A03 and A09, on which the QTL for plant width also has been identified in previous studies [2,44,45]. QTLs for plant width have been found on A05, A07 and A10 in previous studies [2,44,45]. One QTL for plant width was also identified on A01 in the study, which may be a new candidate locus for plant width (Table 5).
The leafy head is the main edible part of Chinese cabbage. Leafy head-related traits are the most important traits for breeding and production of Chinese cabbage. QTLs for head weight have been detected on A02, A03, A04, A05, A06, A07, A08, A09, and A10 in previous studies [1,10,30,45]. In this study, two QTLs for head weight were located on A05 and A10, which is consistent with the linkage group for head weight found by Yu et al. (2013) [10] and Liu et al. (2013) [1], respectively. Six QTLs for head height were identified on A01, A02, A03, A05, A06 and A10 in this study. QTLs for head height have been found on A01, A02, A06 and A10 according to Liu et al. (2013) [1], and on A03 according to Ge et al. (2011) [40] and Liu et al. (2015) [45]. No QTL for head height has been identified on A05 in previous studies, which might be a potentially new locus regulating head height (Table 5).
As an important leafy vegetable, leaf number and morphology are important in Chinese cabbage breeding. In this study, three leaf number-related traits and five leaf morphology-related traits were investigated, and 19 QTLs were identified distributed on 8 LGs (A02, A03, A04, A06, A07, A08, A09 and A10). The results indicated that the leaf traits were complex quantitative traits controlled by many genes spreading nearly all the LGs of Chinese cabbage, which is consistent with those reported in previous studies [15,20,21,30,44] (Table 5).
Stem-related traits, especially stem length, are usually used to evaluate the tolerance to bolting of Chinese cabbage. In this study, no QTL was identified for stem length, probably because the parents of the F2 line in this study were both non-resistant bolting varieties.
As the agronomic traits were significantly correlated (Table 4), many co-localized QTLs were identified on A02, A03 and A10 in this study (Figure 1 and Figure S2). The QTL clusters for some agronomic traits were also identified on these LGs in previous studies in Chinese cabbage [1,9,20,21,30]. It indicated that these chromosomes might carry important genes regulating more than one agronomic trait, and could be very useful for the improvement of more than one trait in breeding of Chinese cabbage.
Many QTLs detected in the study have a large region size (>10 cM); there are hundreds of gene in the region, so it is difficult to pick out the candidate genes. Further fine mapping for these QTLs should be conducted by using a high-density genetic map. For the QTLs with an interval size ≤ 10 cM, the gene information was taken from the Brassicaceae Database (http://www.brassicadb.cn/, accessed on 6 February 2022) (Table S2). BraTCPs [48] and BraGRFs [49] genes were reported to be involved in controlling organ size in Chinese cabbage. In the study, 3 BraTCP (Bra032970, Bra012600 and Bra027284) and 1 BraGRF (Bra033281) genes were found in the QTLs regions. Genes involved in the Auxin signaling pathway play important roles in regulating leafy head formation of Chinese cabbage [50]. Here we found that 10 auxin-related genes (Bra026598, Bra026597, Bra026596, Bra032954, Bra019369, Bra019255, Bra027232, Bra034725, Bra008615 and Bra008722) were located in the QTLs regions. For qMLL-6 and qHH-6, only two genes, Bra033221 (SPL8) and Bra033222 (NOT1), were found in the QTL region. These genes may be important candidates for regulating the agronomic traits of Chinese cabbage.
5. Conclusions
In summary, a genetic map comprising 105 EST-SSR markers distributing across 10 LGs were constructed, and a total of 48 QTLs regulating 16 agronomic traits were identified in Chinese cabbage in this study. QTLs consistent with previous studies could be potential candidate QTLs for further genetic analysis and marker-assisted genetic improvement of Chinese cabbage.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8020165/s1, Table S1: Details of the 500 SSRs used in this study. Table S2: Gene information for QTLs with the interval size ≤10 cM. Figure S1: Frequency distribution of agricultural traits for the F2 population derived from a cross between G291 and ZHB. PH: plant height, PW: plant width, GW: gross weight, NNL: number of non-wrapper leaves, HW: head weight, HH: head height, HD: head diameter, NHL: number of head leaves, NAL: number of all leaves, MLL: maximum leaf length, MLW: maximum leaf width, PEL: petiole length, PEW: petiole width, PET: petiole thickness, SL: stem length, and SW: stem width. Figure S2: Brassica rapa genetic linkage map and QTLs for agronomic traits discovered in F2 lines with LOD score.
Author Contributions
H.G., J.L., F.W. and J.G. designed the experiments; X.Y., H.W., Y.C. and J.L. performed the experiments; H.G., J.L. and N.Q. analyzed the data; F.W., H.L. and Y.Z. contributed reagents/materials/analysis tools; J.L. and J.G. wrote the main manuscript text. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Agricultural Science and Technology Innovation Project of SAAS (CXGC2021A10); the Modern Agricultural Industrial Technology System Funding of Shandong Province, China (SDAIT-05); the Key R&D Program of Shandong Province, China (2019GHZ014); Shandong Upgraded Project of “Bohai Granary” Science and Technology Demonstration Engineering (2019BHLC005); Taishan Scholars Program of Shandong Province, China (tsqn201909167); Prospect of Shandong Seed Project, China (2019LZGC0060101); China Agriculture Research System (CARS-23-G13); and the National Natural Science Foundation of China, China (31401869).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Liu, Y.; Zhang, Y.; Xing, J.; Liu, Z.; Feng, H. Mapping quantitative trait loci for yield-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Euphytica 2013, 193, 221–234. [Google Scholar] [CrossRef]
- Huang, L.; Yang, Y.; Zhang, F.; Cao, J. A genome-wide SNP-based genetic map and QTL mapping for agronomic traits in Chinese cabbage. Sci. Rep. 2017, 7, 46305. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhao, Z.; Du, Z.; Yao, Y.; Xu, L.; Tang, G. Genetic characterization and fine mapping of a yellow-seeded gene in Dahuang (a Brassica rapa landrace). Theor. Appl. Genet. 2012, 124, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Dechaine, J.M.; Brock, M.T.; Weinig, C. QTL architecture of reproductive fitness characters in Brassica rapa. BMC Plant Biol. 2014, 14, 66. [Google Scholar] [CrossRef]
- Saito, M.; Kubo, N.; Matsumoto, S.; Suwabe, K.; Tsukada, M.; Hirai, M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 2006, 114, 81. [Google Scholar] [CrossRef]
- Feng, H.; Li, Y.; Liu, Z.; Liu, J. Mapping of or, a gene conferring orange color on the inner leaf of the Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 2012, 29, 235–244. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Ji, X.; Zhang, L.; Liu, Y.; Lv, X.; Feng, H. Identification and validation of a major QTL controlling the presence/absence of leaf lobes in Brassica rapa L. Euphytica 2015, 205, 761–771. [Google Scholar] [CrossRef]
- Liu, Y.; Li, C.; Shi, X.; Feng, H.; Wang, Y. Identification of QTLs with additive, epistatic, and QTL × environment interaction effects for the bolting trait in Brassica rapa L. Euphytica 2016, 210, 427–439. [Google Scholar] [CrossRef]
- Ren, Y.; Wu, J.; Zhao, J.; Hao, L.; Zhang, L. Identification of SSR markers closely linked to the yellow seed coat color gene in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Biol. Open 2017, 6, 278–282. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Zhong, W.; Bai, J.; Liu, P.; He, Y. QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 2013, 8, e76059. [Google Scholar] [CrossRef]
- Liu, J.; Liu, B.; Cheng, F.; Liang, J.; Wang, X.; Wu, J. A high density linkage map facilitates QTL mapping of flowering time in Brassica rapa. Hortic. Plant J. 2016, 2, 217–223. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, Y.; Yuan, Y.; Zhang, X.; Geng, J.; Chen, Y.; Cloutier, S.; McVetty, P.B.E.; Li, G. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol. Biol. 2009, 69, 553–563. [Google Scholar] [CrossRef]
- Ye, L.X.; Hu, F.Y.; Ren, J.; Huang, S.N.; Liu, W.J.; Feng, H.; Liu, Z.Y. Fine mapping and candidate gene analysis of Brtri1, a gene controlling trichome development in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Genet. Mol. Res. 2016, 15, gmr15048924. [Google Scholar] [CrossRef]
- Kawakatsu, Y.; Nakayama, H.; Kaminoyama, K.; Igarashi, K.; Yasugi, M.; Kudoh, H.; Nagano, A.J.; Yano, K.; Kubo, N.; Kimura, S. A GLABRA1 ortholog on LG A9 controls trichome number in the Japanese leafy vegetables Mizuna and Mibuna (Brassica rapa L. subsp. nipposinica L. H. Bailey): Evidence from QTL analysis. J. Plant Res. 2017, 130, 539–550. [Google Scholar] [CrossRef]
- Lou, P.; Zhao, J.; Kim, J.S.; Shen, S.; Del Carpio, D.P.; Song, X.; Jin, M.; Vreugdenhil, D.; Wang, X.; Koornneef, M.; et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2017, 58, 4005–4016. [Google Scholar] [CrossRef]
- Zhang, N.; Chen, L.; Ma, S.; Wang, R.; He, Q.; Tian, M.; Zhang, L. Fine mapping and candidate gene analysis of the white flower gene Brwf in Chinese cabbage (Brassica rapa L.). Sci. Rep. 2020, 10, 6080. [Google Scholar] [CrossRef]
- Guo, N.; Wu, J.; Zheng, S.; Cheng, F.; Liu, B.; Liang, J.; Cui, Y.; Wang, X. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). Mol. Breed. 2015, 35, 113. [Google Scholar] [CrossRef]
- Song, K.; Slocum, M.K.; Osborn, T.C. Molecular marker analysis of genes controlling morphological variation in Brassica rapa (syn. campestris). Theor. Appl. Genet. 1995, 90, 1–10. [Google Scholar] [CrossRef]
- Kubo, N.; Saito, M.; Tsukazaki, H.; Kondo, T.; Matsumoto, S.; Hirai, M. Detection of quantitative trait loci controlling morphological traits in Brassica rapa L. Breed. Sci. 2010, 60, 164–171. [Google Scholar] [CrossRef][Green Version]
- Xiao, D.; Wang, H.; Basnet, R.K.; Zhao, J.; Lin, K.; Hou, X.; Bonnema, G. Genetic dissection of leaf development in Brassica rapa using a genetical genomics approach. Plant Physiol. 2014, 164, 1309–1325. [Google Scholar] [CrossRef]
- Choi, S.R.; Yu, X.; Dhandapani, V.; Li, X.; Wang, Z.; Lee, S.Y.; Heon Oh, S.; Pang, W.; Ramchiary, N.; Hong, C.; et al. Integrated analysis of leaf morphological and color traits in different populations of Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2017, 130, 1617–1634. [Google Scholar] [CrossRef]
- Zou, L.C.; Zheng, Y.; Wang, P.; Zhang, X.; Wang, Y.H.; Liu, Z.Y.; Feng, H. Fine mapping and characterization of the or gene in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Genet. Mol. Res. 2016, 15, gmr.15028370. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, L.; Guo, S.; An, F.; Du, D. Fine mapping and whole-genome resequencing identify the seed coat color gene in Brassica rapa. PLoS ONE 2016, 11, e0166464. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Y.; Sun, J.; Feng, H.; Wang, Y. Identification and validation of major and minor QTLs controlling seed coat color in Brassica rapa L. Breed. Sci. 2019, 69, 47–54. [Google Scholar] [CrossRef]
- Liu, S.; Wang, R.; Zhang, Z.; Li, Q.; Wang, L.; Wang, Y.; Zhao, Z. High-resolution mapping of quantitative trait loci con-trolling main floral stalk length in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2019, 20, 437. [Google Scholar] [CrossRef]
- Kato, T.; Hatakeyama, K.; Fukino, N.; Matsumoto, S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed. Sci. 2013, 63, 116–124. [Google Scholar] [CrossRef]
- Yu, S.; Su, T.; Zhi, S.; Zhang, F.; Wang, W.; Zhang, D.; Zhao, X.; Yu, Y. Construction of a sequence-based bin map and mapping of QTLs for downy mildew resistance at four developmental stages in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 2016, 36, 44. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, X.; Zeng, Q.; Zhang, Z.; Liu, S.; Pei, Y.; Wang, S.; Liu, X.; Xu, W.; Fu, W.; et al. Identification and mapping of a novel Turnip mosaic virus resistance gene TuRBCS01 in Chinese cabbage (Brassica rapa L.). Plant Breed. 2015, 134, 221–225. [Google Scholar] [CrossRef]
- Laila, R.; Park, J.; Khan Robin, A.H.; Natarajan, S.; Kenta Shirasawa, V.H.; Isobe, S.; Kim, H.; Nou, I. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.). BMC Plant Biol. 2019, 19, 13. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, Y.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinesis). Hortic. Environ. Biotechnol. 2011, 52, 494–501. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Ding, Q.; Li, H.; Liu, L.; Wang, F.; Gao, J. Transcriptome Analysis of Orange Head Chinese Cabbage (Brassica rapa L. ssp. pekinensis) and Molecular Marker Development. Int. J. Genom. 2017, 2017, 6835810. [Google Scholar] [CrossRef]
- Eujayl, I.; Sledge, M.K.; Wang, L.; May, G.D.; Chekhovskiy, K.; Zwonitzer, J.C.; Mian, M.A.R. Medicago truncatula ESTSSRs reveal cross-species genetic markers for Medicago spp. Theor. Appl. Genet. 2004, 108, 414–422. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Bernard, M.; Leroy, P.; Feuillet, C.; Sourdille, P. High transferability of bread wheat EST-derived SSRs to other cereals. Theor. Appl. Genet. 2005, 111, 677–687. [Google Scholar] [CrossRef]
- Saha, M.C.; Cooper, J.D.; Rouf Mian, M.A.; Chekhovskiy, K.; May, G.D. Tall fescue genomic SSR markers: Development and transferability across multiple grass species. Theor. Appl. Genet. 2006, 113, 1449–1458. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Shirasawa, K.; Oyama, M.; Hirakawa, H.; Sato, S.; Tabata, S.; Fujioka, T.; Kimizuka-Takagi, C.; Sasamoto, S.; Watanabe, A.; Kato, M.; et al. An EST-SSR Linkage Map of Raphanus sativus and Comparative Genomics of the Brassicaceae. DNA Res. 2011, 18, 221–232. [Google Scholar] [CrossRef]
- Liu, C.; Yuan, D.; Lin, Z. Construction of an EST-SSR-based interspecific transcriptome linkage map of fibre development in cotton. J. Genet. 2014, 93, 689–697. [Google Scholar] [CrossRef]
- El-Rodeny, W.; Kimura, M.; Hirakawa, H.; Sabah, A.; Shirasawa, K.; Sato, S.; Tabata, S.; Sasamoto, S.; Watanabe, A.; Kawashima, K.; et al. Development of EST-SSR markers and construction of a linkage map in faba bean (Vicia faba). Breed. Sci. 2014, 64, 252–263. [Google Scholar] [CrossRef]
- Dhaka, N.; Mukhopadhyay, A.; Paritosh, K.; Gupta, V.; Pental, D.; Pradhan, A.K. Identification of genic SSRs and construction of a SSR-based linkage map in Brassica juncea. Euphytica 2017, 213, 15. [Google Scholar] [CrossRef]
- Shi, J.; Huang, S.; Zhan, J.; Yu, J.; Wang, X.; Hua, W.; Liu, S.; Liu, G.; Wang, H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 2014, 21, 53–68. [Google Scholar] [CrossRef]
- Ding, Q.; Li, J.; Wang, F.; Zhang, Y.; Li, H.; Zhang, J.; Gao, J. Characterization and Development of EST-SSRs by Deep Transcriptome Sequencing in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Int. J. Genom. 2015, 2015, 473028. [Google Scholar] [CrossRef]
- Winnepenninckx, B.; Backeljau, T.; Wachter, R.D. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Zhang, Y. Construction of a Molecular Genetic Map and QTL Mapping for Major Agronomic Traits in Chinese Cabbage (Brassica campestris spp. pekinensis). Doctoral Dissertation, Shenyang Agricultural University, Shenyang, China, 2012. (In Chinese). [Google Scholar]
- Liu, J. Linkage Map Construction and QTL Analysis for Agronomic Traits in Chinese Cabbage. Master’s Dissertation, Tianjing Normal University, Tianjin, China, 2015. (In Chinese). [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Li, H.; Liu, L.; Zhang, Y.; Gao, J.; Wang, X. Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 2012, 99, 299–307. [Google Scholar] [CrossRef]
- Wang, F.; Tan, T.; Zhang, Y.; Li, J.; Li, H.; Li, L.; Liu, L.; Gao, J. Cloning and Functional Analysis of BrTCP24 Gene in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Biotechnol. 2013, 21, 911–919. [Google Scholar] [CrossRef]
- Wang, F.; Qiu, N.; Ding, Q.; Li, J.; Zhang, Y.; Li, H.; Gao, J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2014, 15, 807. [Google Scholar] [CrossRef]
- He, Y.; Xue, W.X.; Sun, Y.D.; Yu, X.H.; Liu, P.L. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 2000, 10, 151–160. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).