Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Host Species and Cultivar Culture Conditions
2.2. Bacterial Strains and Culture Conditions
2.3. Inhibition Activity Assays between Group I and Group II Strains
2.4. Competition Assay between Group I and II Strains In Vitro
2.4.1. Establishment of the Standard Curve
2.4.2. Colony Dynamics of the Two Group Strains In Vitro
2.5. Competition Assay between Group I and II Strains in Melon and Watermelon Cotyledons
2.5.1. Establishment of the Standard Curve for Determination of Colony Number in Melon and Watermelon
2.5.2. Establishment of the Standard Curve
2.5.3. Colony Dynamics of Group I and II Strains in Melon and Watermelon Cotyledons
2.6. Data Processing/Statistical Analysis
3. Results
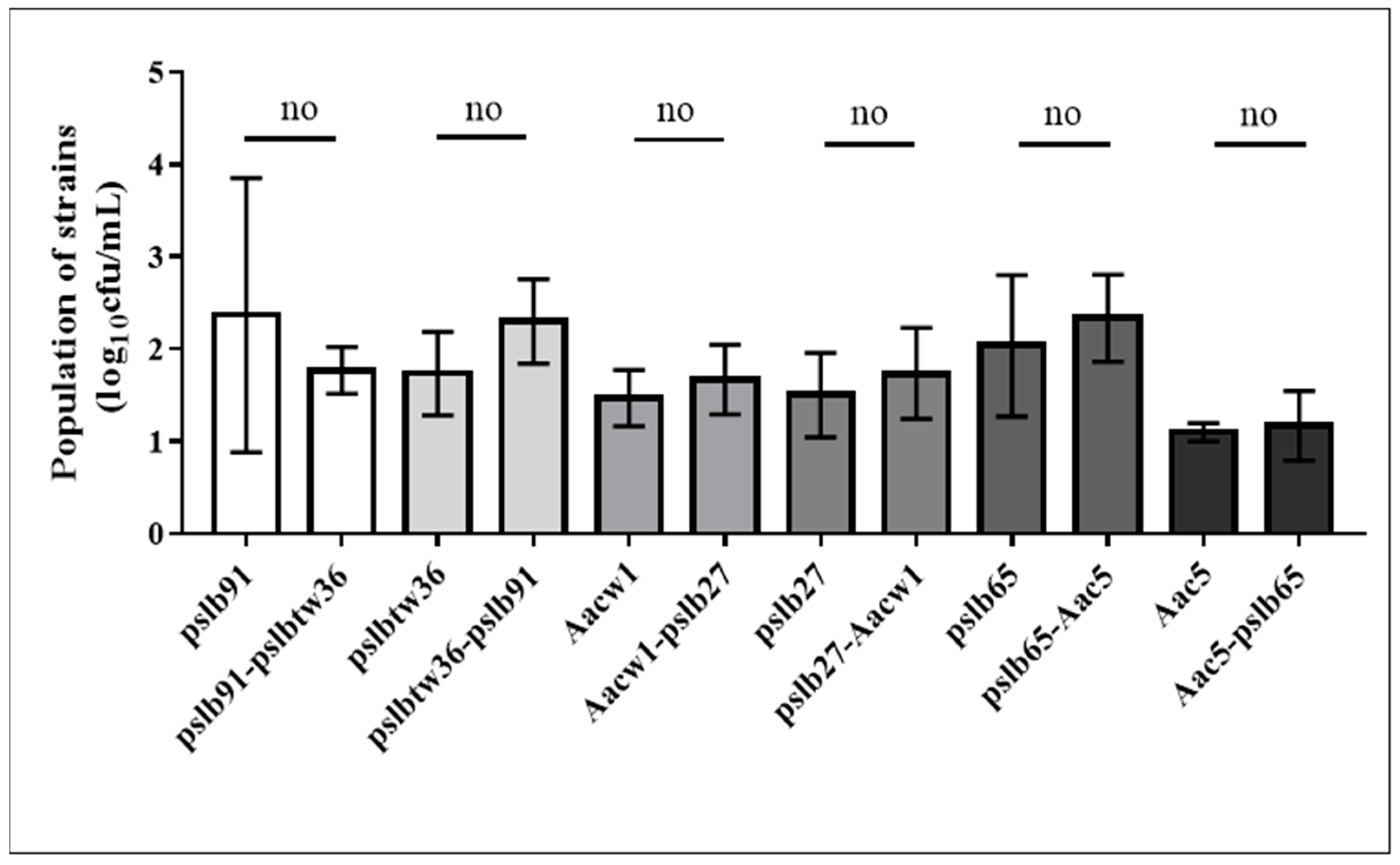
3.1. Cell-Free Culture Supernatants from A. Citrulli Group I or Group II Strains Do Not Inhibit Each Other’s Growth in Culture
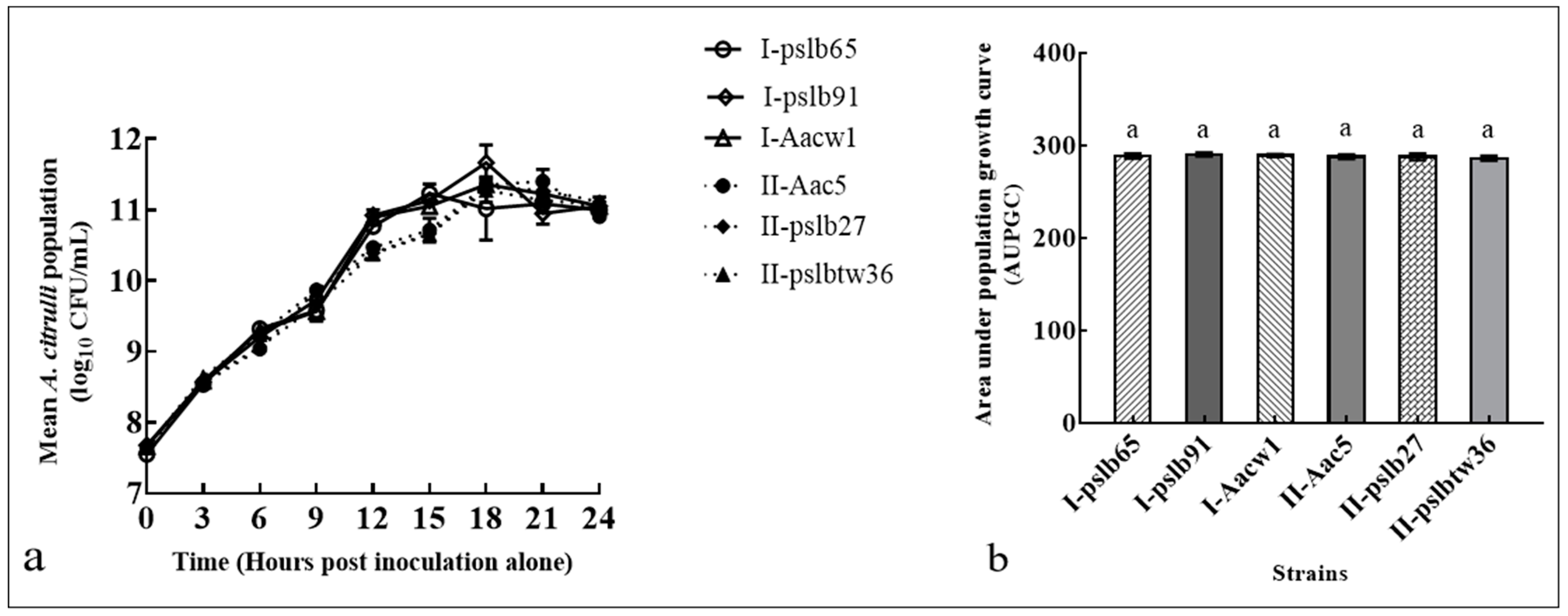
3.2. Colony Dynamics of the Two Group Strains When Cultured Separately In Vitro
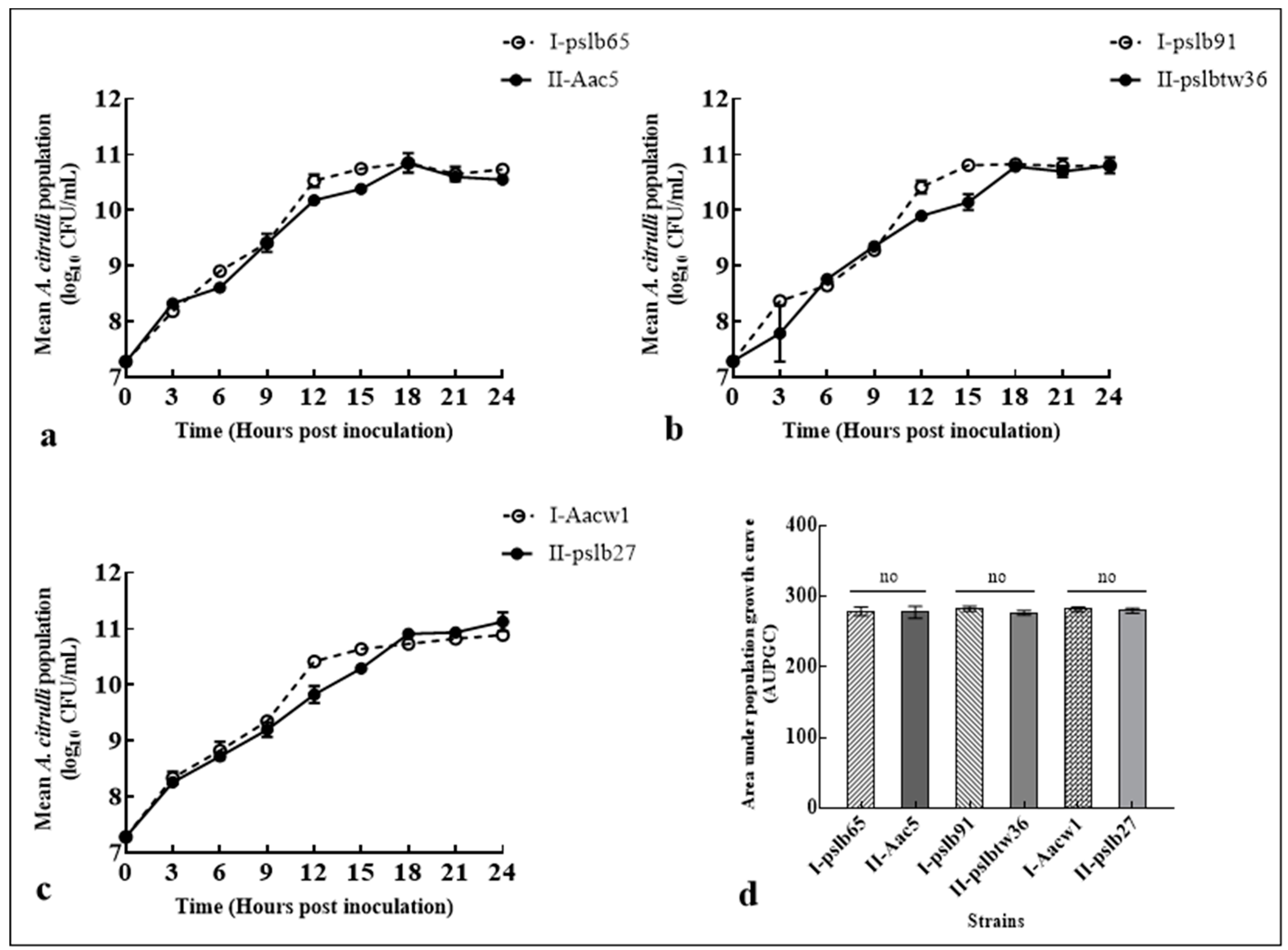
3.3. Colony Dynamics of Group I or Group II Strains in Mixed Culture In Vitro
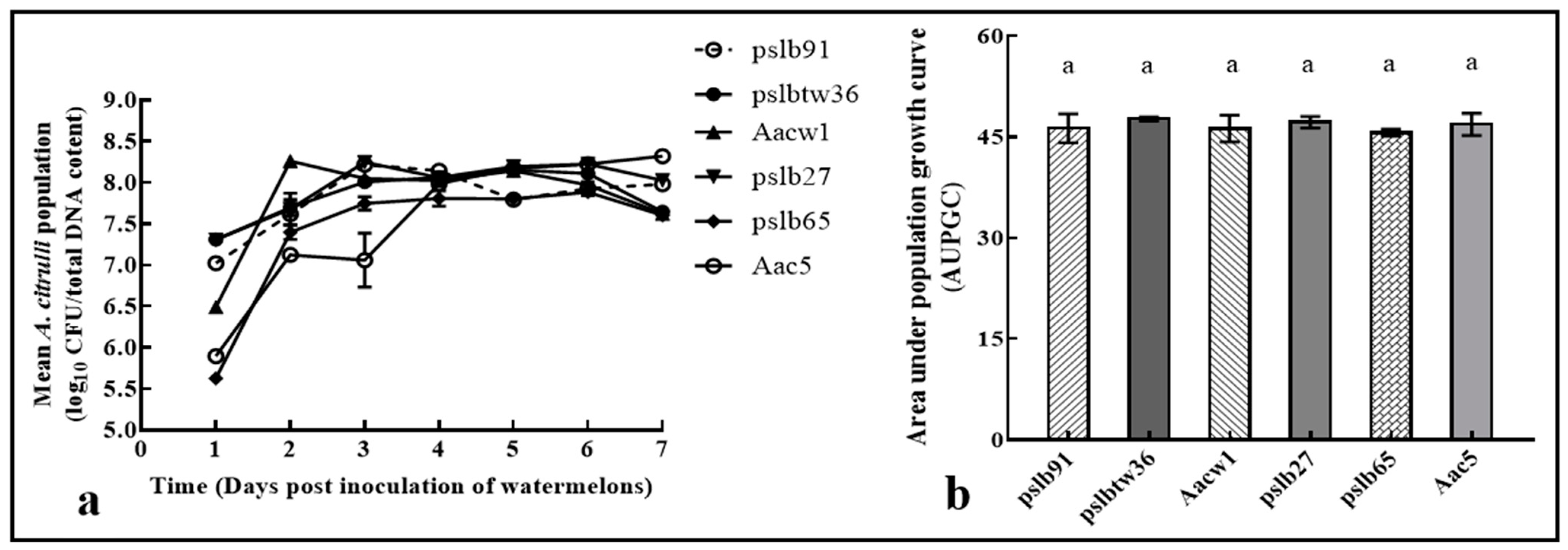
3.4. Colony Dynamics of Group I and Group II Strains Individually Inoculated into Melon and Watermelon Cotyledons
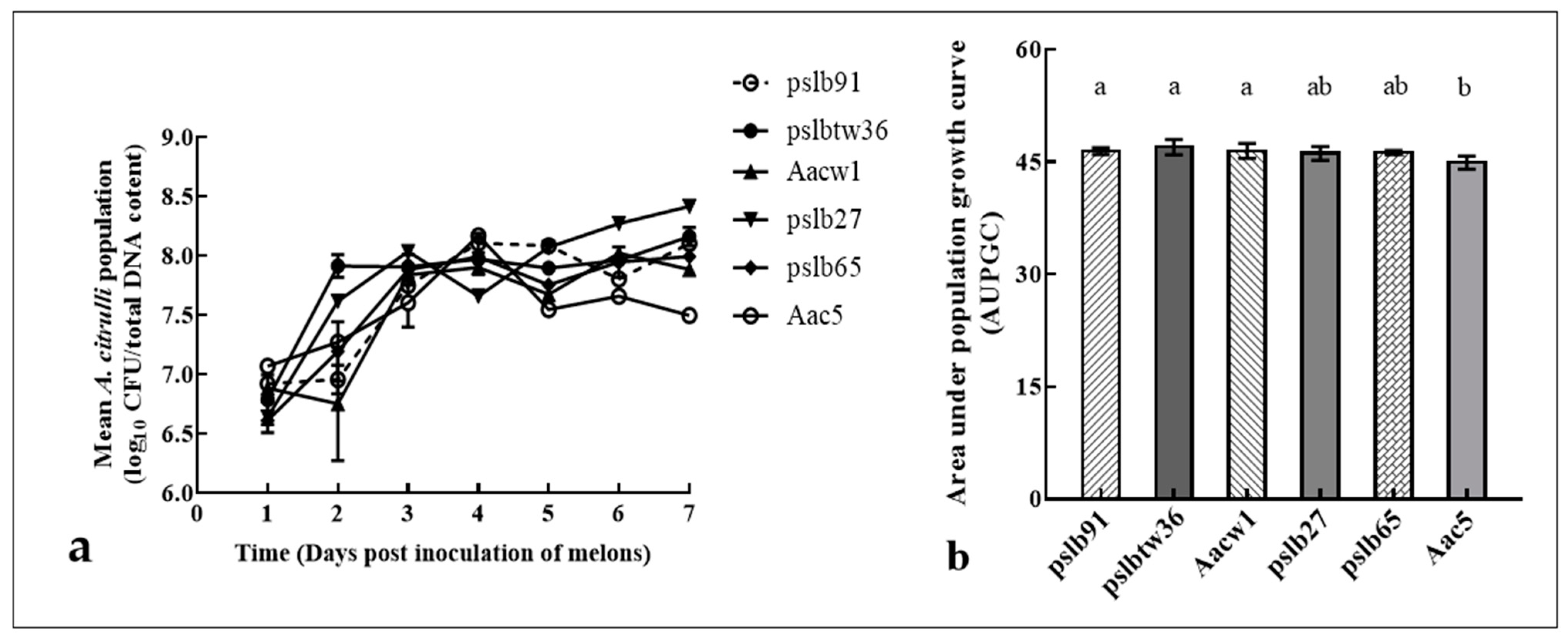
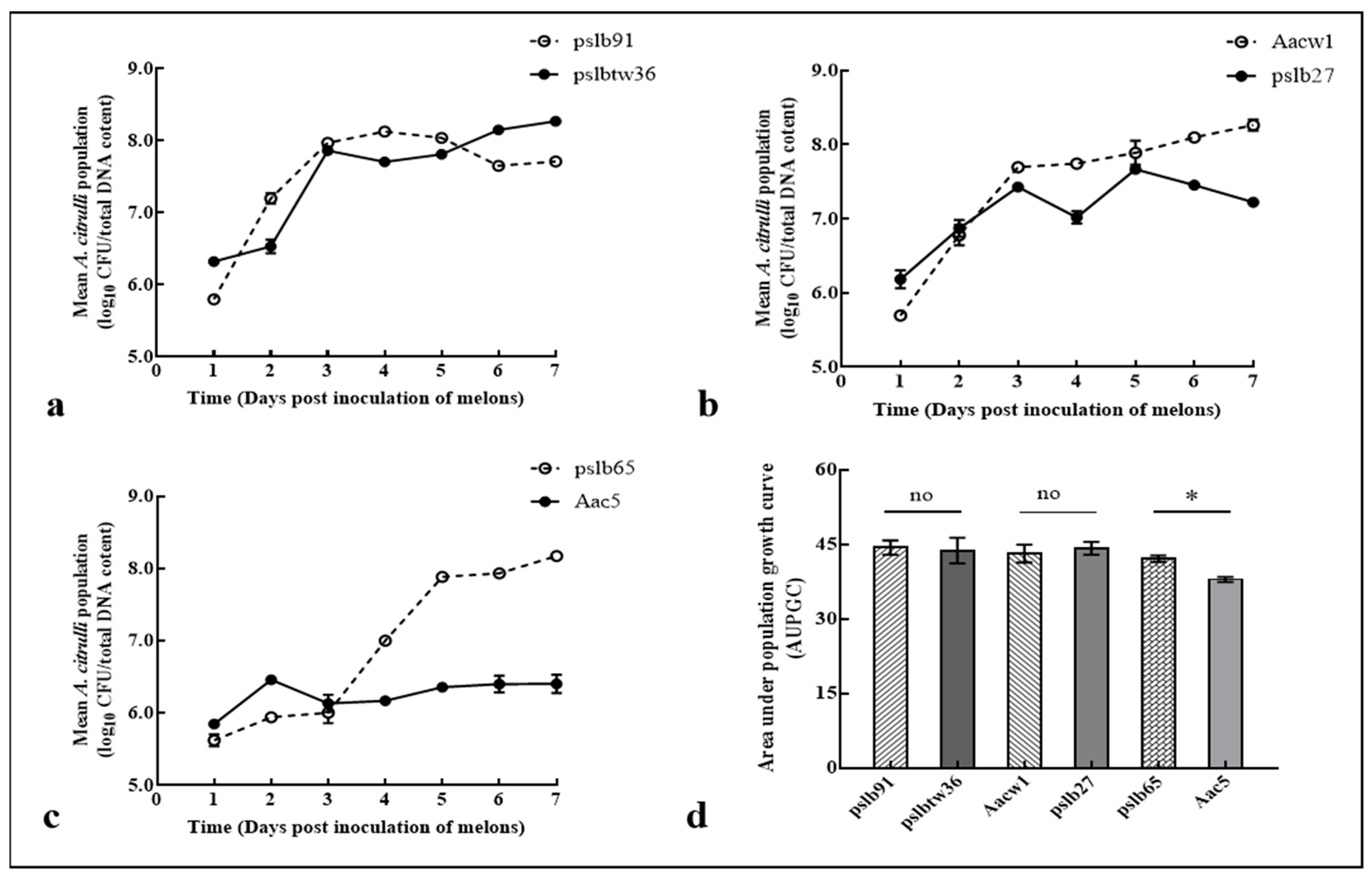
3.5. Colony Dynamics of Group I and Group II Strains after Mixed Inoculation into Melon and Watermelon Cotyledons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schaad, N.W.; Postnikova, E.; Sechler, A.; Claflin, L.E.; Vidaver, A.K.; Jones, J.B.; Agarkova, I.; Ignatov, A.; Dickstein, E.; Ramundo, B.A. Reclassification of subspecies of Acidovorax avenae as A. avenae (Manns 1905) emend; A. cattleyae (Pavarino, 1911) comb. nov., A. citrulli (Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 2008, 31, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Latin, R.X.; Rane, K.K. Bacterial fruit blotch of watermelon in Indiana. Plant Dis. 1990, 74, 331. [Google Scholar] [CrossRef]
- Walcott, R.R.; Fessehaie, A.; Castro, A.C. Differences in pathogenicity between two genetically distinct groups of Acidovorax avenae subsp. citrulli on cucurbit hosts. J. Phytopathol. 2004, 152, 277–285. [Google Scholar] [CrossRef]
- Burdman, S.; Kots, N.; Kritzman, G.; Kopelowitz, J. Molecular, physiological, and host-range characterization of Acidovorax avenae subsp. citrulli isolates from watermelon and melon in Israel. Plant Dis. 2005, 89, 1339–1347. [Google Scholar] [CrossRef]
- Ren, Y.Z.; Li, H.; Li, G.Y.; Wang, Q.Y.; Li, J.Q. First report of Acidovorax avenae subsp. citrulli infecting edible seed watermelon (Citrullus lanatus var. lanatus) in China. Plant Dis. 2006, 90, 1112. [Google Scholar] [CrossRef] [PubMed]
- Palkovics, L.; Petróczy, M.; Kertész, B.; Németh, J.; Bársony, C.; Mike, Z.; Hevesi, M. First report of bacterial fruit blotch of watermelon caused by Acidovorax avenae subsp. citrulli in Hungary. Plant Dis. 2008, 92, 834–835. [Google Scholar] [CrossRef] [PubMed]
- Amadi, J.E.; Adebola, M.O.; Eze, C.S. Isolation and identification of a bacterial blotch organism from watermelon (Citrullis lanatus (Thunb.) Matsum. And Nakai). Afr. J. Agr. Res. 2009, 4, 1291–1294. [Google Scholar] [CrossRef]
- Eckshtain-Levi, N.; Shkedy, D.; Gershovits, M.; Da Silva, G.M.; Tamir-Ariel, D.; Walcott, R.; Pupko, T.; Burdman, S. Insights from the genome sequence of Acidovorax citrulli M6, a group I strain of the causal agent of bacterial fruit blotch of cucurbits. Front. Microbiol. 2016, 7, 430. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Randhawa, P.; Bonde, M.; Schaad, N.W. Evaluation of seed treatments for the eradication of Acidovorax avenae subsp. citrulli from melon and watermelon seeds. Can. J. Plant Pathol. 2009, 31, 180–185. [Google Scholar] [CrossRef]
- Yan, S.; Yang, Y.; Wang, T.; Zhao, T.; Schaad, N.W. Genetic diversity analysis of Acidovorax citrulli in China. Eur. J. Plant Pathol. 2013, 136, 171–181. [Google Scholar] [CrossRef]
- Yang, Y.W.; Zhao, M.; Zhang, L.Q.; Qiao, P.; Bai, X.; Zhang, X.X.; Walcott, R.R.; Guan, W.; Zhao, T.C. Development of a multiplex PCR assay based on the pilA gene sequences to detect different types of Acidovorax citrulli. J. Microbiol. Methods 2019, 158, 93–98. [Google Scholar] [CrossRef] [PubMed]
- O’brien, R.G.; Martin, H.L. Bacterial blotch of melons caused by strains of Acidovorax avenae subsp. citrulli. Anim. Prod. Sci. 1999, 39, 479–485. [Google Scholar] [CrossRef]
- Zivanovic, M.; Walcott, R.R. Further characterization of genetically distinct groups of Acidovorax citrulli strains. Phytopathology 2017, 107, 29–35. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Zhao, M.; Yang, Y.W.; Guan, W.; Ji, W.Q.; Bai, Q.R.; Gao, J.; Walcott, R.R.; Zhao, T.C. Transcriptomic analysis of early interaction between melon seedlings and Acidovorax citrulli strains. Acta Phytophy. Sin. 2017, 47, 776–789. [Google Scholar] [CrossRef]
- Yan, L.; Hu, B.; Chen, G.; Zhao, M.; Walcott, R.R. Further Evidence of Cucurbit Host Specificity among Acidovorax citrulli Groups Based on a Detached Melon Fruit Pathogenicity Assay. Phytopathology 2017, 107, 1305–1311. [Google Scholar] [CrossRef][Green Version]
- Zhao, M.; Dutta, B.; Luo, X.; Burdman, S.; Walcott, R. Genetically Distinct Acidovorax citrulli Strains Display Cucurbit Fruit Preference Under Field Conditions. Phytopathology 2020, 110, 973–980. [Google Scholar] [CrossRef]
- Hui, W.G.; Zhao, T.C.; Schaad, N.W.; Sun, F.Z.; Wang, J.R. Establishment of rapid detection method for the pathogen of Hami Melon fruit blotch. Acta Phytophy. Sin. 2007, 40, 2495–2501. [Google Scholar]
- Bahar, O.; Efrat, M.; Hadar, E.; Dutta, B.; Walcott, R.R.; Burdman, S. New subspecies-specific polymerase chain reaction-based assay for the detection of Acidovorax avenae subsp. citrulli. Plant Pathol. 2008, 57, 754–763. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Hu, B.; Liu, F. Reliable and sensitive detection of Acidovorax citrulli in cucurbit seed using a padlock-probe-based assay. Plant Dis. 2013, 97, 961–966. [Google Scholar] [CrossRef]
- Zhao, T.; Feng, J.; Sechler, A.; Randhawa, P.; Li, J.; Schaad, N.W. An improved assay for detection of Acidovorax citrulli in watermelon and melon seed. Seed Sci. Technol. 2009, 37, 337–349. [Google Scholar] [CrossRef]
- Ha, Y.; Fessehaie, A.; Ling, K.S.; Wechter, W.P.; Keinath, A.P.; Walcott, R.R. Simultaneous detection of Acidovorax avenae subsp. citrulli and Didymella bryoniae in cucurbit seedlots using magnetic capture hybridization and real-time polymerase chain reaction. Phytopathology 2009, 99, 666–678. [Google Scholar] [CrossRef][Green Version]
- Walcott, R.R.; Castro, A.C.; Fessehaie, A.; Ling, K. Progress towards a commercial PCR-based seed assay for Acidovorax avenae subsp. citrulli. Seed Sci. Technol. 2006, 34, 101–116. [Google Scholar] [CrossRef]
- Zhao, W.; Lu, J.; Ma, W.; Xu, C.; Kuang, H.; Zhu, S. Rapid on-site detection of Acidovorax avenae subsp citrulli by gold-labeled DNA strip sensor. Biosens. Bioelectron. 2011, 26, 4241–4244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tian, Q.; Zhu, S.F.; Zhao, W.J.; Liu, F.Q. Rapid on-site detection of Acidovorax citrulli by cross-priming amplification. Mol. Cell Probes. 2012, 26, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Zhou, Q.; Li, B.; Liu, B.P.; Wu, G.X.; Ibrahim, M.; Xie, G.L.; Li, H.Y.; Sun, G.C. Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovorax oryzae and Acidovorax citrulli. BMC Microbiol. 2012, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Lin, Z.Y.; Ma, Y.M.; Gao, B.D.; Liu, H.Q.; Zhao, T.C. Rapid discrimination between groups I and II of Acidovorax citrulli using a primer pair specific to a pilL Gene. J. Phytopathol. 2016, 164, 558–562. [Google Scholar] [CrossRef]
- Ji, W.; Zhao, M.; Fei, N.; Yang, L.; Qiao, P.; Walcott, R.; Yang, Y.; Zhao, T. Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen. Int. J. Mol. Sci. 2022, 23, 9144. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab Clin. Med. 1954, 44, 301–307. [Google Scholar] [CrossRef]
- Fei, N.; Ji, W.; Yang, L.; Yu, C.; Qiao, P.; Yan, J.; Guan, W.; Yang, Y.; Zhao, T. Hcp of the Type VI Secretion System (T6SS) in Acidovorax citrulli Group II Strain Aac5 Has a Dual Role as a Core Structural Protein and an Effector Protein in Colonization, Growth Ability, Competition, Biofilm Formation, and Ferric Iron Absorption. Int. J. Mol. Sci. 2022, 23, 9632. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Niu, P.; Xie, J.; Jiang, W.; Huang, Y.; Bie, Z. Screening Suitable Reference Genes for Normalization in Reverse Transcription Quantitative Real-Time PCR Analysis in Melon. PLoS ONE 2014, 27, e87197. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Gao, L.; Zhao, S.; Jiang, W.; Huang, Y.; Bie, Z. Identification of Suitable Reference Genes for Gene Expression Normalization in qRT-PCR Analysis in Watermelon. PLoS ONE 2014, 28, e90612. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the (2-ΔΔCT) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Baltrus, D.A.; Nishimura, M.T.; Dougherty, K.M.; Biswas, S.; Mukhtar, M.S.; Vicente, J.; Holub, E.B.; Dangl, J.L. The Molecular Basis of Host Specialization in Bean Pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 2012, 25, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Eckshtain-Levi, N.; Munitz, T.; Živanović, M.; Traore, S.M.; Spröer, C.; Zhao, B.; Welbaum, G.; Walcott, R.; Sikorski, J.; Burdman, S. Comparative analysis of type III secreted effector genes reflects divergence of Acidovorax citrulli strains into three distinct lineages. Phytopathology 2014, 104, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Da Silva, G.M.; Wagner, N.; Shkedy, D.; Zhao, M.; Pizarro, L.; Bar, M.; Walcott, R.; Sessa, G.; et al. Show me your secret(ed) weapons: A multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus. Mol. Plant Pathol. 2020, 21, 17–37. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Qiao, P.; Wang, T.; Ji, W.; Fei, N.; Zhang, L.; Guan, W.; Zhao, T. Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains. Horticulturae 2022, 8, 1173. https://doi.org/10.3390/horticulturae8121173
Yang Y, Qiao P, Wang T, Ji W, Fei N, Zhang L, Guan W, Zhao T. Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains. Horticulturae. 2022; 8(12):1173. https://doi.org/10.3390/horticulturae8121173
Chicago/Turabian StyleYang, Yuwen, Pei Qiao, Tielin Wang, Weiqin Ji, Nuoya Fei, Liqun Zhang, Wei Guan, and Tingchang Zhao. 2022. "Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains" Horticulturae 8, no. 12: 1173. https://doi.org/10.3390/horticulturae8121173
APA StyleYang, Y., Qiao, P., Wang, T., Ji, W., Fei, N., Zhang, L., Guan, W., & Zhao, T. (2022). Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains. Horticulturae, 8(12), 1173. https://doi.org/10.3390/horticulturae8121173




