Abstract
Pectins are major components of cell walls in plants. Pectin methylesterases (PMEs) and pectin methylesterase inhibitors (PMEIs) play crucial roles in pectin synthesis and metabolism. Overall, 28 putative DkPMEs and 29 putative DkPMEIs were identified from the D. kaki genome. According to phylogenetic analysis, DkPME/DkPMEI proteins can be classified into four and five clades, respectively. Motif and gene structure analysis showed that DkPME/DkPMEI are highly conserved in the same clades, which indicates that the function of these DkPME/DkPMEI were similar. Besides, DkPME/DkPMEI genes were distributed unevenly on their corresponding chromosomes. Synteny analysis showed that PME or PMEI gene usually matched with more than one DkPME/DkPMEI in D. oleifera, D. lotus, and A. thaliana, implying that the function of these genes in D. kaki may be diverse. Expression analysis showed that DkPME/DkPMEI from the same clade exhibited diverse expression patterns, indicating that these genes might have diverse functions. Functional protein–protein interaction network analysis showed that DkPMEI21 and DkPMEI15 were core nodes and were, respectively, positive and negative regulators for carbohydrate metabolism, stress responses, and sugar signaling. This study provides a theoretical basis for the functional characteristics, evolutionary relationship, and role of these gene families in developing persimmon fruit.
1. Introduction
Plant cell wall, synthesized from cellulose, hemicellulose, pectin, and several glycosylated proteins, plays multiple roles in physiological and developmental processes [1]. Pectin is a significant component of cell walls and mainly contains homogalacturonan (HG), xylogalacturonan (XGA), rhamnogalacturonan-I (RG-I), and rhamnogalacturonan-II (RG-II) [2,3]. The richest form of pectic synthesized in the Golgi apparatus is a highly methyl-esterified state. It is delivered to cell walls and de-methylesterified by pectin methylesterase (PME) [4,5]. Pectin methylesterase inhibitor (PMEI) inhibits PME activity by forming a non-covalent at a 1:1 ratio in plants [6]. PME and PMEI proteins play crucial roles in pectin synthesis and metabolism [7,8].
PME proteins are present widely in higher plants and some microorganisms [8]. For the protein structures, Type-I PME and Type-II PME proteins share the PME domains, and Type-I PME proteins also contains the PRO-region, an additional N-terminal region similar to the PMEI domain [9]. Recently, PME genes have been discovered in various plant species, such as 66 PME genes, which were identified from Arabidopsis thaliana [10], 53 from Citrus sinensis [11], 61 from Glycine max [12], 54 from Fragaria vesca [12], and 56 from Solanum tuberosum [13]. The PME genes are tightly linked to various functions of plants, such as fruit maturity and softening [14,15], seed germination and development [16], pollen development [17,18], stem elongation [19], root hair formation [19], and stress response [20,21].
PMEI proteins belong to a large multigene family, containing a conserved PMEI domain (PF04043) [22,23]. The first PMEI protein was discovered in kiwi fruit and later identified in multiple plants [7]. Until now, 78 PMEI genes have been identified from Arabidopsis thaliana [24], 42 from Pyrus bretschneideri [25], 49 from Oryza sativa [26], 51 from Camellia sinensis [27], 55 from Sorghum bicolor [28], 83 from Linum usitatissimum [29], 95 from Brassica oleracea [30], and 100 from Brassica campestris [31]. PMEI family genes play multiple roles in plant growth, development, and stress response [32]. For example, AtPMEI4 promotes root growth of A. thaliana [33]. AtPMEI5 overexpression affected HG methyl esterification by repressing the PME activity and accelerating seed germination [34]. In rice, OsPMEI28 overexpression suppressed the PME activity, impacting the growth process, resulting in dwarfed phenotypes [35]. Overexpression of Camellia sinensis PMEI2/4 in Arabidopsis decreased PME activity but increased sugar content, promoting early flowering phenotypes [27].
Persimmon (Diospyros kaki) belongs to the family Ebenaceae and is widely distributed in the subtropics and tropics of East Asia [36]. An abundant tannin content characterizes it, and the high soluble tannin content leads to the astringency of persimmon fruit [37]. Previous studies have shown that the interaction of pectin and tannin might be essential in reducing astringency [38,39]. Methanol, which is produced by PMEs catalyzing the pectin de-methyl esterification, might be associated with the loss of astringency [40]. Based on the critical role of PME and PMEI genes in regulating pectin de-methylesterifcation [7], these genes might play essential roles in persimmon de-astringency. However, studies of the PME and PMEI gene families in persimmon are still limited.
The availability of the persimmon genome has facilitated the identification of PME and PMEI gene families. The present study reported a comprehensive analysis to examine their conserved protein motif, gene structure, phylogenetic and evolutionary relationship, chromosome location, collinearity and promoter, protein interaction, and in silico expression analysis of PME and PMEI genes during fruit development. This study provides a theoretical basis for the functional characteristics, evolutionary relationship, and the role of DkPME and DkPMEI genes in developing persimmon fruit.
2. Materials and Methods
2.1. Identification of PME and PMEI Genes
The most updated Hidden Markov Model (HMM) of the PME domain (PF01095) and PMEI domain (PF04043) were downloaded from the Pfam to identify PME and PMEI genes in the persimmon genome (unpublished) using HMMER 3.3 [41]. The presence and completeness of the PME and PMEI domains were evaluated using SMART tools with an E-value < 0.1 [42]. The gene length, molecular weights, and isoelectric point of putative PME and PMEI genes were predicted using the online ExPASy program (https://www.expasy.org/, accessed on 6 April 2022) [43].
2.2. Phylogenetic Analysis and Interaction Network Construction
PME and PMEI proteins sequences from persimmon and arabidopsis were used to investigate the phylogenetic relationships; among them, AtPME and AtPMEI proteins were downloaded from Tair (https://www.arabidopsis.org/index.jsp, accessed on 10 April 2022). Using Clustal X2 at the default parameters, multiple sequence alignments were aligned, and neighbor-joining (NJ) phylogenetic trees were constructed with 1000 bootstrap replicates using MEGA5.0 [44,45]. DkPMEs and DkPMEIs were classified according to their phylogenetic relationship with corresponding A. thaliana PME and PMEI genes. We analyzed the interaction network of DkPME and DkPMEI proteins using a model plant A. thaliana on the STRING protein interaction database (http://string–db.org/, accessed on 13 April 2022) [46].
2.3. Motif Prediction and Gene Structure Analysis of PME and PMEI Genes
The online MEME tool (http://meme.ebi.edu.au/, accessed on 15 April 2022) was used to identify unknown conserved motifs shared among persimmon PME and PMEI proteins [47]. The parameters were used as follows: a maximum number of motifs: 15 (DkPME) and 10 (DkPMEI); 6 ≤ optimum motif width ≤ 50. Gene Structure Display Server online software (http://gsds.cbi.pku.edu.cn/index.php, accessed on 20 April 2022) was used to display the structures of PME and PMEI genes. The motif and gene structure with the phylogenetic tree was shown using the TBtools [48].
2.4. Chromosomal Distribution, Gene Duplication, and Kaks Calculation Analysis
The positions of all PME and PMEI genes from the persimmon genome were extracted from the GFF3 file using TBtools [48]. The collinear relationship of PME and PMEI genes of persimmon and arabidopsis and gene duplication events of PME and PMEI genes were identified using the Multiple Collinearity Scan Toolkit (MCScanX) with default parameters [49]. The chromosomal locations and duplication events were visualized using TBtools [48]. The number of non-synonymous (Ka) and synonymous (Ks) substitutions of paralogous DkPME and DkPMEI gene pairs were computed using TBtools [48], and the divergence time of the duplication events was calculated using the formula T = Ks/(2 × 1.5 × 10−8) × 10−6 million years ago (MYA).
2.5. Promoter Cis-Regulatory Elements Analysis
The upstream 2000 bp sequences of the DkPME and DkPMEI promoter regions were cut and extracted using TBtools [48]. PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 22 April 2022) searched for plant growth and development, hormone-responsive, and abiotic stress cis-acting elements in the promoter sequences [50].
2.6. In Silico Expression Analysis of PME and PMEI Genes
RNA-seq data were available in the NCBI SRA under the Bioproject ID PRJNA771936. Five different stages (T1 = 70, T2 = 100, T3 = 120, T4 = 140, T5 = 160 days after flower (DAF)) of persimmon fruit were sampled, and three independent biological replicates were used. The expression levels of DkPME and DkPMEI genes were calculated as fragments per kilobase million (FPKM). The heat maps were created using the Heatmap tool in Hiplot (https://hiplot.com.cn, accessed on 24 April 2022) based on the transformed data of log2 (FPKM + 1) values [51].
3. Results
3.1. Identification and Phylogenetic Analysis of PME and PMEI Genes in D. kaki
After removing the redundant sequences, 28 putative DkPME genes and 29 putative DkPMEI genes were identified from the D. kaki genome. These genes were named DkPME1 to DkPME28 and DkPMEI1 to DkPMEI29 based on their physical chromosome location. Overall, 14 DkPME contained both PMEI and PME domain (Type-II PME) among these genes. As shown in Table S1, the length of DkPME proteins varied from 253 aa (DkPME9) to 1164 aa (DkPME21), the molecular weights (MW) ranged from 28,529.85 (DkPME9) to 126,409.97 (DkPME21) Da, and the theoretical isoelectric points (pIs) were changed from 5.75 (DkPME22) to 9.38 (DkPME19). The MW of DkPMEI was extended from 6762.71 (DkPMEI17) to 103,158.13 (DkPMEI29) Da, and the encoded amino acids were ranged from 66 (DkPMEI17) to 918 (DkPMEI29) aa, and the pI varied from 4.25 (DkPMEI17) to 11.07 (DkPMEI14) (Table S1).
NJ phylogenetic trees were constructed to verify the evolutionary relationship using the protein sequences of 66 published AtPMEs, 78 AtPMEIs, and putative DkPMEs and DkPMEIs (Figure 1). According to sequence similarity, the PMEs and PMEIs families in D. kaki were divided into 4 and 5 clades, respectively. The Type-II DkPME PMEs were clustered into Clade II, III, and IV, while Clade I only contained Type-I DkPMEs. The largest subfamily of PME was Clade IV which had 14 DkPME proteins, while Clade I, II, and III held 12, 1, and 1 DkPME genes, respectively (Figure 1a). As for the DkPMEI family, CladeI contained 21 proteins, while Group III and IV only contained 1 PMEI protein (Figure 1b).
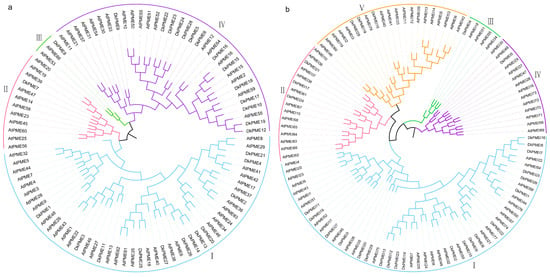
Figure 1.
Phylogeny of the PME/PMEIs in D. kaki. (a) NJ phylogenetic tree of the DkPMEs; (b) NJ phylogenetic tree of the DkPMEIs.
3.2. Conserved Motifs and Structural Analysis
To analyze the structural characteristics of the DkPME and DkPMEI proteins, a total of 15 and 10 conserved motifs of DkPME and DkPMEI proteins were predicted using MEME online software. Of the DkPMEs, motifs 3, 4, 6, 7, and 9 were annotated as PME domains; these motifs were exhibited in 26, 27, 26, 26, and 27 DkPME proteins, respectively, indicating that these PME-related motifs were essential and highly conserved. Motif 13 contained the PMEI domain in DkPME proteins and was presented in 12 Clade I DkPME proteins (Figure 2a). For the DkPMEIs, motifs 1, 2, and 3 were annotated as PMEI domain and were presented in 20, 17, and 24 PMEI proteins, respectively, revealing that PMEI-related motifs were highly conserved in DkPMEI proteins (Figure 3a).
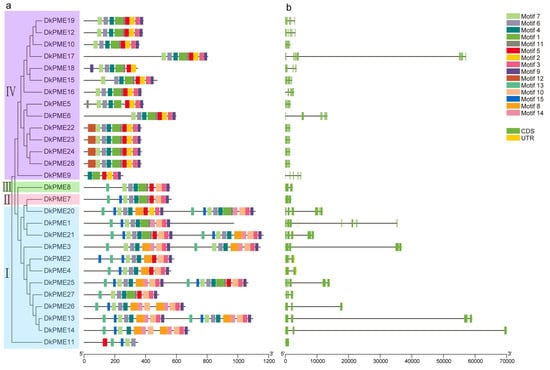
Figure 2.
Conserved motif and gene structure analyses of PME genes in D. kaki. (a) Conserved motif distribution. (b) Exon–intron structure of DkPME genes.
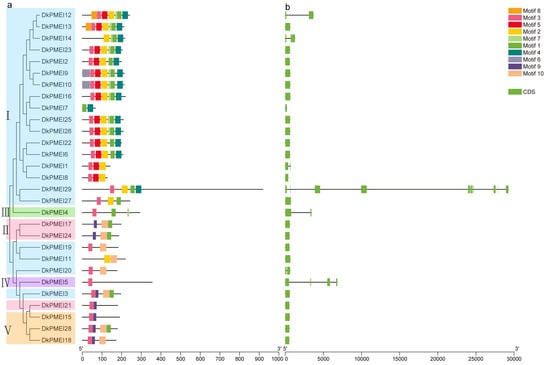
Figure 3.
Conserved motif and gene structure analyses of PMEI genes in D. kaki. (a) Conserved motif distribution. (b) Exon–intron structure of DkPMEI genes.
The DkPME and DkPMEI proteins in the same clade possessed the same conserved motifs, indicating functional similarity in paralogous gene pairs or genes in the same clades. For example, motif 2 of DkPME was prominent in Clade IV, and motifs 14 and 15 were only observed in Clade I (Figure 2a). Motifs 2 and 5 were unique to most of the DkPMEIs in Clade I, while motif 9 was mainly present in Clade V (Figure 3a). The conservation of conserved motifs of DkPME and DkPMEI members in the same clade supported the results of the phylogenetic analysis.
The exon–intron distribution of DkPME and DkPMEI genes was created based on the coding sequence. The DkPME gene structure is relatively conserved, with most having about 3–4 introns, while two genes of Clade IV have only 6 and 9 introns. The exon numbers of Clade IV genes (Type-II PME) were somewhat more extensive than that of Clade I members (Type-I PME), but the intron sizes of Clade IV genes were relatively smaller. Furthermore, two Clade I members had noncoding regions (3′UTR and 5′UTR) (Figure 2a). The number of exons in DkPMEI genes varied from 1 to 10. The introns were detected from only 7 out of 29 DkPMEI genes. Among them, 4 DkPMEI genes belonged to Clade I. Interestingly, most DkPMEI genes lacked introns and noncoding regions (Figure 3a). The gene structures analysis of DkPME and DkPMEI genes also supported the reliability of the phylogenetic classification.
3.3. Chromosomal Locations, Gene Duplication, and Ka/Ks of PME and PMEI Genes
Chromosomal localization analysis showed that DkPME and DkPMEI genes were distributed unevenly on their corresponding chromosomes (Table S1, Figure 4 and Figure 5). DkPME and DkPMEI genes were identified in 13 and 9 of 15 D. kaki chromosomes, respectively (Table S1, Figure 4 and Figure 5). For the DkPMEs, there were three gene clusters (DkPME3-DkPME4, DkPME5-DkPME6-DkPME7, DkPME22-DkPME23-DkPME24), respectively, located on chromosomes 1, 1, 13 (Figure 4). Of the DkPMEIs, there were four gene clusters (DkPMEI6-DkPMEI7-DkPMEI8, DkPMEI9-DkPMEI10, DkPMEI13-DkPMEI14, and DkPMEI25-DkPMEI26) located on chromosomes 3, 5, 6 and 14, respectively (Figure 4).
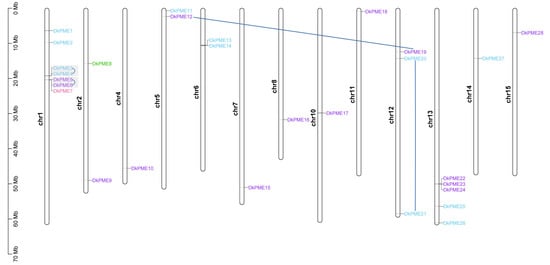
Figure 4.
Chromosome location of DkPME genes in D. kaki. Genes of different groups are expressed in different colors. Darkblue lines showed fragment duplication events, and gray boxes presented tandem duplication events.
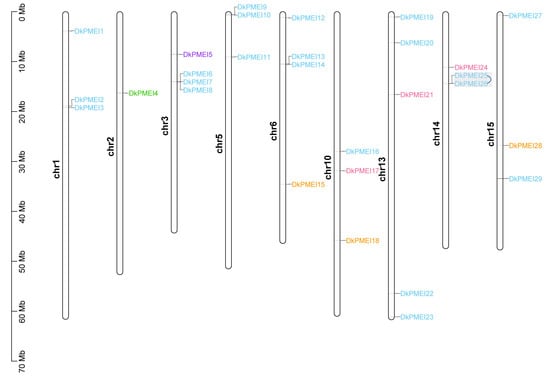
Figure 5.
Chromosome location of DkPMEI genes in D. kaki. Genes of different groups are expressed in different colors. Gray boxes presented tandem duplication events.
The duplication analysis showed that 2 DkPME gene pairs were identified as segment duplication and 2 tandem duplication events. Two DkPME gene clusters were formed by tandem duplication located on chromosome 1. However, only one DkPMEI gene pair originated in tandem duplication and is located on chromosome 14. The Ka/Ks ratios of all tandem duplicated and segment duplicated DkPME/DkPMEI gene pairs were <1, indicating that these DkPME/DkPMEI genes evolved under purifying selection to reduce deleterious mutations after replication. The differentiation time of DkPME genes was primarily between 26.599 and 49.717 million years ago (MYA), and the differentiation time of the DkPMEI gene pair was 39.777 MYA (Table S2).
3.4. Synteny Analysis of PME and PMEI Genes
We analyzed the syntenic relationship between the PME and PMEI genes in D. kaki, its closely related species Diospyros oleifera and Diospyros lotus, and model plants A. thaliana (Figure 6). The results showed that the PME genes in D. kaki had the most homologous gene pairs with PME genes in D. oleifera (30), followed by D. lotus (22) and A. thaliana (21) (Figure 6a–c). However, the PMEI genes in D. kaki had more homologous gene pairs with PMEI genes in D. lotus (35) and D. oleifera (33), followed by A. thaliana (21) (Figure 6d–f). Notably, one PME or PMEI gene usually matched with more than one PME or PMEI in D. oleifera, D. lotus, and A. thaliana, implying that the function of certain DkPME or DkPMEI genes in D. kaki may be diverse.
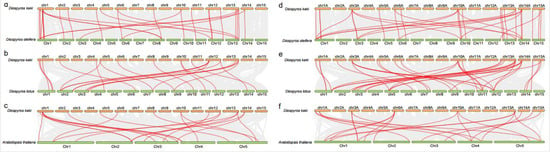
Figure 6.
Synteny analysis of PME and PMEI genes between D. kaki, D. oleifera, and D. lotus, and A. thaliana. (a) PME genes between D. kaki and D. oleifera. (b) PME genes between D. kaki and D. lotus. (c) PME genes between D. kaki and A. thaliana. (d) PMEI genes between D. kaki and D. oleifera. (e) PMEI genes between D. kaki and D. lotus. (f) PMEI genes between D. kaki and A. thaliana.
3.5. Cis-Element Analysis of PME and PMEI Genes
To investigate the gene characteristics and function of DkPME and DkPMEI genes, cis-regulatory elements in the promoters were predicted using PlantCARE. Briefly, multiple light-related elements were identified, namely Box 4, G-box, GT1-motif, TCT-motif, GATA-motif, and so on. Other cis-elements could be categorized into plant growth and development, hormone-responsive, and abiotic stress. Eight cis-elements were involved in plant growth and development, including circadian, MSA-box, CAT-box, RY-element, etc. Ten cis-elements are related to hormone-responsive, i.e., ABRE, TATC-box, CGTCA-motif, AuxRR-core, and P-box. Six cis-elements participated in abiotic stress (e.g., LTR, ARE, and MBS). In addition, the MBSI cis-element was involved in flavonoid biosynthetic genes regulation and was identified in the promoter of DkPMEI12, DkPMEI17, DkPMEI22, and DkPMEI29 (Figure 7 and Table S3).
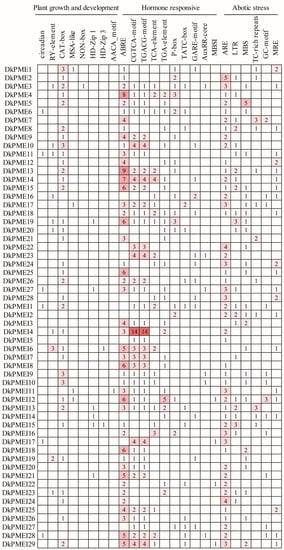
Figure 7.
Cis-acting element in the promoter region of DkPME and DkPMEI genes.
3.6. Expression Patterns of DkPME and DkPMEI Genes during Fruit Development
The expression levels of DkPME and DkPMEI genes during fruit development were evaluated using high-throughput RNA-seq data (Figure 8). Overall, 22 DkPME and 23 DkPMEI genes were expressed in persimmon during fruit development. Seven DkPME and eight DkPMEI genes exhibited constitutive expression (FPKM > 1 in all tested developmental stages), showing an indispensable role of DkPME and DkPMEI genes during fruit development. Six DkPME genes from clade IV were highly expressed at 70 DAF, suggesting that these genes might be associated with fruit growth. Three DkPMEs and two DkPMEIs were highly expressed in 160 DAF, including DkPME8, DkPME9, DkPME20, DkPMEI22, and DkPMEI24. Therefore, these genes might have essential roles in fruit maturation. Moreover, some DkPMEs and DkPMEIs from the same clade exhibited diverse expression patterns. For example, the expression of 7 DkPMEI genes from clade I peaked at 140 DAF, whereas 8 DkPMEI genes, also from clade I, expressed highly at 70 DAF, indicating that these PMEI genes might have diverse functions.
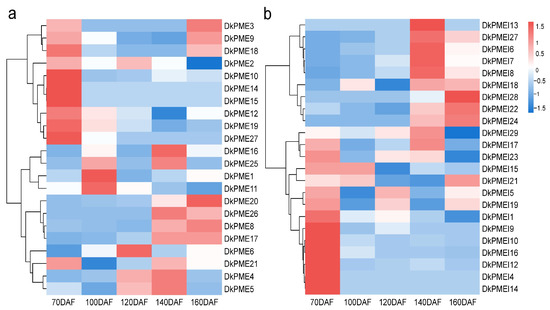
Figure 8.
Gene expression of (a) DkPME and (b) DkPMEI genes during fruit development.
3.7. Protein Interaction Analysis of DkPMEs and DkPMEIs
To analyze the function of DkPMEs and DkPMEIs, the protein–protein interaction network was constructed using the STRING database according to A. thaliana homologous proteins (Figure 9a). As a result, DkPMEI21 and DkPMEI15 were prominent core nodes in this interaction network. The protein sequence of DkPMEI21 was highly similar to that of AtPMEI10, a negative transcriptional regulator related to carbohydrate metabolism, stress responses, and sugar signaling. Furthermore, the sequence of DkPMEI15 was homology to that of AtPMEI78, a crucial transcriptional activator related to carbohydrate metabolism, stress responses, and sugar signaling with a positive regulatory function. DkPMEI21 and DkPMEI15 were closely related to DkPMEI1 and DkPMEI8, which may form a solid interaction network (Figure 9b).
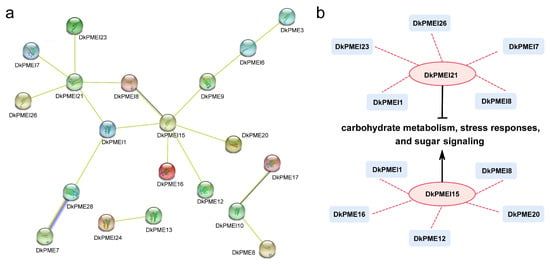
Figure 9.
Functional network of DkPMEs and DkPMEIs: (a) Network of DkPMEs and DkPMEIs based on the orthologous genes in A. thaliana. (b) A schematic representation of a regulatory network among DkPMEs and DkPMEIs.
4. Discussion
The PME and PMEI gene families are involved in cell wall modifications, and thus play a crucial role in organ development, fruit maturation and softening, and responses to various biotic and abiotic stresses in plants [14,17,34]. Due to their essential functions, the PME and PMEI gene families were identified in multiple plants [27,29]. However, systematic analysis of PME and PMEI genes is lacking in persimmon. This study identified 28 PME and 29 PMEI genes in D. kaki via genome-wide analysis. The number of DkPME genes (28) was comparable with that of Selaginella moellendorffii (23) [52] and O. sativa (30) [52] but was smaller than that of Gossypium arboretum (80) [53] and A. thaliana (66) [10]. The number of DkPMEI genes (29) was comparable to that observed in O. sativa (35) [52] and Carica papaya (23) [52] but smaller than that observed in A. thaliana (71) [34] and C. sinensis (45) [54]. The number of PMEs and PMEIs in persimmon was lower than that in A. thaliana, which might be associated with the genome of the persimmon (D. oleifera), only undergoing an ancient γ whole-genome duplication event [55], but A. thaliana genome underwent at least three duplication events [56].
Gene duplication events generate new gene copies that encode proteins with novel or improved functions, and it can increase genome evolution, diversity, and complexity [57]. Our study found two pairs of DkPME and two pairs of DkPMEI genes with tandem duplication events, two pairs of DkPME genes, and 1 DkPMEI gene pair with segmental duplication events. These results are similar to those found in A. thaliana, V. vinifera, and O. sativa [52], indicating that tandem and segmental duplication events are crucial in expanding PME and PMEI gene families. By measuring the Ka/Ks ratios between duplicated DkPME or DkPMEI gene copies, we observed that all duplicated DkPME or DkPMEI genes underwent purifying selection. The differentiation time of Ebenaceae with Actinidia chinensis and C. sinensis was approximately 73.6–95.6 MYA, and that of Diospyros species (D. oleifera and D. lotus) was nearly 2.9–18.3 [58]. However, the differentiation time of the PME and PMEI gene pairs in persimmon was primarily at 26.599–49.717 MYA and 39.777 MYA, respectively [58]. These results showed that the time of gene differentiation was later than that of Ebenaceae with the other plants (like A. thaliana) differentiation but earlier than the differentiation time between Diospyros species. Besides, synteny analysis showed that the PME and PMEI genes of persimmon had more homologous gene pairs in Diospyros species (D. oleifera and D. lotus) than A. thaliana. Thus, we inferred that the PME and PMEI genes were conserved between persimmon species but might show high inter-species polymorphism.
Analysis of gene structure and conserved motif provided potential feature information for resolving phylogenetic relationships [59]. DkPMEs and DkPMEIs could divide into 4 and 5 clades based on the phylogenetic analysis. DkPME and DkPMEI members of the same clade contained similar exon–intron and motifs compositions. For example, nearly all DkPMEI genes lacked introns and UTRs except genes in Clade I, which is in accordance with the results for C. sinensis [54], Zea mays [60], and G. max [61]. These intronless genes could reduce post-transcriptional processing and respond immediately to abiotic stresses [62]. PME-related motifs and PMEI-related motifs were distributed in most DkPME and DkPMEI proteins. Moreover, a unique motif was conserved across subgroups, like motif 2 of PMEs is unique in Clade IV, and motif 9 of DkPMEIs was mainly present in Clade V. The same phenomenon has been observed in other plants like C. sinensis [54]. These results showed that these conserved and clade clade-specific motifs keep the fundamental functions and improve the gene diversities during evolution [53].
Cis-elements are essential regulatory units as they regulate various biological processes, including developmental, hormone, and abiotic or biotic stress response [63]. In this study, the promoter of four DkPMEI genes (DkPMEI12, DkPMEI17, DkPMEI22, and DkPMEI29) contained a flavonoid biosynthetic MBSI cis-element. MYB family members could recognize the MBSI region [64]; in persimmon, tannin insolubilization, the MBSI, and the other motif participated in the trans-activating of the DkERF19 promoter by DkMYB6, DkERF18, and DkERF19 [65]. Thus, these four DkPMEI genes contained an MBSI cis-element, which might have potential functions in tannin biosynthesis under MYB transcription factor regulation and need further confirmation. Six cis-regulatory elements (LTR, ARE, and MBS, etc.) were essential to stress response in the promoter of DkPME and DkPMEI genes, in accord with the vital role of these genes in the abiotic stress response [20,21,32]. Gene family members usually formed protein complexes during a long evolutionary process, and diverse PPI networks could represent protein physical relationships [66]. The interaction network found two key nodes, DkPMEI21 and DkPMEI15. They could be used as candidate genes for studying carbohydrate metabolism, sugar signaling, and stress responses of DkPMEs and DkPMEIs. These results further proved the function of PME and PMEI genes for regulating growth, development, and stress response.
Fruit development is complicated, directly affecting fruit qualitative traits such as appearance, flavor, and texture [67]. The development, ripening, and softening of fruits is regulated by environmental factors and transcription regulatory factors, such as PME, PMEI, etc. [68]. VvPMEI1 is expressed during fruit development and could control PME activity at the early developmental stage in grape berry [68]. The Tomato SlPMEI gene forms an inactive complex with the significant fruit-specific PME-1 subtype and inhibits pectin degradation during fruit ripening [69]. The temporal expression patterns provide essential information for gene function investigation. A large proportion of DkPME family members (around 79%, 22 out of 28) was expressed in persimmon than that in navel orange (about 64%, 34 out of 53) [11] and tomato (about 47%, 27 out of 57) [70] during fruit development. The same phenomenon was also found in DkPMEI genes. These results firmly indicated that DkPME and DkPMEI genes might play indispensable roles in persimmon fruit development.
In summary, 28 DkPME and 29 DkPMEI genes were identified in the D. kaki genome. Subsequently, comprehensive bioinformatics analyses of DkPME and DkPMEI genes/proteins on phylogeny, gene structure, conserved motif, chromosomal location, gene duplication, and collinearity, cis-elements, and temporal expression patterns in persimmon fruit were performed. This study provides a systematic and comprehensive analysis that may help elucidate the DkPME and DkPMEI genes for further structural and functional characterization.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8121159/s1, Table S1: List of all PME and PMEI genes identified in the Diospyros oleifera; Table S2: Ka, Ks and Ka/Ks values calculated for homologous PME and PMEI gene pairs; Table S3: List of light responsive elements in the PME and PMEI genes.
Author Contributions
Q.Z. and T.P. performed the majority of this study; Y.W. helped to finish the experiments and analyze partial data; Q.Z. and T.P. wrote this manuscript; Y.B., Y.S. and J.F. designed the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the by National Key R&D Program of China (2019YFD1001200).
Data Availability Statement
Not applicable.
Acknowledgments
The processing and analysis of transcriptome data were completed with the help of Novogene.
Conflicts of Interest
There were no conflict of interest in the submission of this manuscript.
References
- Zhao, Y.; Man, Y.; Wen, J.; Guo, Y.; Lin, J. Advances in imaging plant cell walls. Trends Plant Sci. 2019, 24, 867–878. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef]
- Anderson, C.T. We be jammin’: An update on pectin biosynthesis, trafficking, and dynamics. J. Exp. Bot. 2015, 67, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef]
- Di, M.A.; Giovane, A.; Raiola, A.; Camardella, L.; Bonivento, D.; De, L.G.; Cervone, F.; Bellincampi, D.; Tsernoglou, D. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 2005, 17, 849–858. [Google Scholar] [CrossRef]
- Balestrieri, C.; Castaldo, D.; Giovane, A.; Quagliuolo, L.; Servillo, L. Aglycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur. J. Med. Chem. 1990, 193, 183–187. [Google Scholar] [CrossRef]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in Plant Physiol. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef]
- Louvet, R.; Cavel, E.; Gutierrez, L.; Guénin, S.; Roge, D.; Gillet, F.; Guerineau, F.; Pelloux, J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 2006, 224, 782–791. [Google Scholar] [CrossRef]
- Li, Z.X.; Wu, L.M.; Wang, C.; Wang, Y.; He, L.G.; Wang, Z.J.; Ma, X.F.; Bai, F.X.; Feng, G.Z.; Liu, J.H.; et al. Characterization of pectin methylesterase gene family and its possible role in juice sac granulation in navel orange (Citrus sinensis Osbeck). BMC Genom. 2022, 23, 185. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, Y.; Wang, S.; Zhang, Q.; Yang, S. Genome-wide identification of PME genes, evolution and expression analyses in soybean (Glycine max L.). BMC Plant Biol. 2021, 21, 578. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, H.; He, L.F. Genome-wide analysis of the pectin methylesterase gene family in potato. Potato Res. 2021, 64, 1–19. [Google Scholar] [CrossRef]
- Kagan, Z.V.; Tieman, D.M.; Marlow, S.J.; Handa, A.K. Differential regulation of polygalacturonase and pectin methylesterase gene expression during and after heat stress in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Mol. Biol. 1995, 29, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Ström, A.; Tasker, A.; West, G.; Tucker, G.A. Effect of silencing the two major tomato fruit pectin methylesterase isoforms on cell wall pectin metabolism. Plant Biol. 2013, 15, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Guénin, S.; Hardouin, J.; Paynel, F.; Müller, K.; Mongelard, G.; Driouich, A.; Lerouge, P.; Kermode, A.; Lehner, A.; Mollet, J.C.; et al. AtPME3, a ubiquitous cell wall pectin methylesterase of Arabidopsis thaliana, alters the metabolism of cruciferin seed storage proteins during post-germinative growth of seedlings. J. Exp. Bot. 2017, 68, 1083–1095. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005, 17, 3219–3226. [Google Scholar] [CrossRef]
- Jiang, L.X.; Yang, S.L.; Xie, L.F.; Li, F.; Puah, C.S.; Zhang, X.Q.; Yang, W.C.; Sundaresan, V.; Ye, D. Vanguard1 encodes a pectin methylesterase that enhances pollen tube growth in the arabidopsis style and transmitting tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef]
- Pilling, J.; Willmitzer, L.; Fisahn, J. Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 2000, 210, 391–399. [Google Scholar] [CrossRef]
- Yan, J.; He, H.; Fang, L.; Zhang, A. Pectin methylesterase31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 496, 497–501. [Google Scholar] [CrossRef]
- Angelica, G.; Vincenzo, L.; Stefania, L.G.; Daniela, Z.; Eleonora, F.; Nathan, R.; Olga, A.Z.; Elisabetta, D.A.; Linda, M.; Daniela, B.; et al. Cell wall features transferred from common into durum wheat to improve Fusarium Head Blight resistance. Plant Sci. 2018, 274, 121–128. [Google Scholar] [CrossRef]
- Kohli, P.; Kalia, M.; Gupta, R. Pectin Methylesterases: A Review. J. Bioprocess. Biotech. 2015, 5, 1. [Google Scholar] [CrossRef]
- Giovane, A.; Balestrieri, C.; Quagliuolo, L.; Castaldo, D.; Servillo, L.A. Glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Eur. J. Med. Chem. 1995, 233, 926–929. [Google Scholar] [CrossRef]
- Müller, K.; Levesque, T.G.; Fernandes, A.; Wormit, A.; Bartels, S.; Usadel, B.; Kermode, A. Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal. Behav. 2013, 8, e26464. [Google Scholar] [CrossRef]
- Zhu, X.; Tang, C.; Li, Q.H.; Qiao, X.; Li, X.; Cai, Y.L.; Wang, P.; Sun, Y.Y.; Zhang, H.; Zhang, S.L.; et al. Characterization of the pectin methylesterase inhibitor gene family in Rosaceae and role of PbrPMEI23/39/41 in methylesterified pectin distribution in pear pollen tube. Planta 2021, 253, 118. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Jeong, H.Y.; Kim, H.; Kim, Y.C.; Lee, C. Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol. Biochem. 2016, 101, 105–112. [Google Scholar] [CrossRef]
- Li, B.; Wang, H.; He, S.; Ding, Z.T.; Wang, Y.; Li, N.N.; Hao, X.Y.; Wang, L.; Yang, Y.J.; Qian, W.J. Genome-Wide Identification of the PMEI Gene Family in Tea Plant and Functional Analysis of CsPMEI2 and CsPMEI4 Through Ectopic Overexpression. Front. Plant Sci. 2022, 12, 807514. [Google Scholar] [CrossRef]
- Ren, A.; Ahmed, R.I.; Chen, H.; Han, L.H.; Sun, J.H.; Ding, A.M.; Guo, Y.F.; Kong, Y.Z. Genome-wide identification, characterization and expression patterns of the pectin methylesterase inhibitor genes in sorghum bicolor. Genes 2019, 10, 755. [Google Scholar] [CrossRef]
- Pinzon-Latorre, D.; Deyholos, M.K. Pectinmethylesterases (PME) and Pectinmethylesterase Inhibitors (PMEI) enriched during phloem fiber development in Flax (Linum usitatissimum). PLoS ONE 2014, 9, e105386. [Google Scholar] [CrossRef]
- Liu, T.T.; Yu, H.; Xiong, X.P.; Yu, Y.J.; Yue, X.Y.; Liu, J.L.; Cao, J.S. Genome-wide identification and characterization of pectin methylesterase inhibitor genes in Brassica oleracea. Int. J. Mol. Sci. 2018, 19, 3338. [Google Scholar] [CrossRef]
- Liu, T.T.; Yu, H.; Xiong, X.P.; Yu, Y.J.; Yue, X.Y.; Liu, J.L.; Cao, J.S. Genome-wide identification, molecular evolution, and expression profiling analysis of pectin methylesterase inhibitor genes in Brassica campestris ssp. chinensis. Int. J. Mol. Sci. 2018, 19, 1338. [Google Scholar] [CrossRef]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef]
- Sénéchal, F.; Mareck, A.; Marcelo, P.; Lerouge, P.; Pelloux, J. Arabidopsis PME17 activity can be controlled by pectin methylesterase Inhibitor4. Plant Signal. Behav. 2015, 10, e983351. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Levesque, T.G.; Bartels, S.; Weitbrecht, K.; Wormit, A.; Usadel, B.; Haughn, G.; Kermode, A.R. Demethylesterification of cell wall pectins in arabidopsis plays a role in seed germination. Plant Physiol. 2012, 161, 305–316. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Jeong, H.Y.; Jeon, S.H.; Kim, D.; Lee, C. Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels. J. Plant Physiol. 2017, 208, 17–25. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, R. Persimmon in China: Domestication and traditional utilizations of genetic resources. JSTOR 2008, 22, 239–243. Available online: http://www.jstor.org/stable/42883463 (accessed on 6 December 2022).
- Giordani, E.; Doumett, S.; Nin, S.; Del Bubba, M. Selected primary and secondary metabolites in fresh persimmon (Diospyros kaki Thunb.): A review of analytical methods and current knowledge of fruit composition and health benefits. Food Res. Int. 2011, 44, 1752–1767. [Google Scholar] [CrossRef]
- Taira, S.; Ono, M.; Matsumoto, N. Reduction of persimmon astringency by complex formation between pectin and tannins. Postharvest Biol. Technol. 1997, 12, 265–271. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.K.; Li, C.M. Effects of interaction between pectin and tannin on the deastringency of different varieties of persimmons during maturing. Xiandai Shipin Keji 2019, 35, 87–94. [Google Scholar] [CrossRef]
- Awad, M. Persimmon Pectinmethylesterase: Extraction and variation during ripening. J. Food Sci. 1985, 50, 1643–1645. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Julio, C. 2-D Proteome Analysis Protocols (ed Andrew J. Link). Protein Sci. 1999, 8, 531–552. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.J.; Hao, C.; Yi, Z.; Hannah, R.T.; Margaret, H.F.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Li, J.F.; Miao, B.B.; Wang, S.X.; Dong, W.X.; Hou, S.; Si, C.C.; Wang, M.J. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.J.; Yuan, D.J.; Gao, W.H.; Li, Y.; Tan, J.F.; Zhang, X.L. A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE 2013, 8, e72082. [Google Scholar] [CrossRef] [PubMed]
- Li, W.J.; Shang, H.H.; Ge, Q.; Zou, C.S.; Cai, J.; Wang, D.J.; Fan, S.M.; Zhang, Z.; Deng, X.Y.; Tan, Y.N.; et al. Genome-wide identification, phylogeny, and expression analysis of pectin methylesterases reveal their major role in cotton fiber development. BMC Genom. 2016, 17, 1000. [Google Scholar] [CrossRef]
- Li, Z.X.; Wang, C.; Long, D.; Jiang, Y.C.; He, L.G.; Wang, Z.J.; Ma, X.F.; Bai, F.X.; Liu, J.H.; Wu, L.M.; et al. Genome-wide identification, bioinformatics characterization and functional analysis of pectin methylesterase inhibitors related to low temperature-induced juice sac granulation in navel orange (Citrus sinensis Osbeck). Sci. Hortic. 2022, 298, 110983. [Google Scholar] [CrossRef]
- Zhu, Q.G.; Xu, Y.; Yang, Y.; Guan, C.F.; Zhang, Q.Y.; Huang, J.W.; Grierson, D.; Chen, K.S.; Gong, B.C.; Yin, X.R. The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic. Res. 2019, 6, 15. [Google Scholar] [CrossRef]
- De, S.R.; Sabaghian, E.; Li, Z.; Saeys, Y.; Van-de, P.Y. Coordinated functional divergence of genes after genome duplication in Arabidopsis thaliana. Plant Cell 2017, 29, 2786–2800. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.J.; Sun, P.; Cheng, H.H.; Han, W.J.; Diao, S.F.; Li, H.W.; Mai, Y.N.; Zhao, X.; Li, F.D.; Fu, J.M. A high-quality chromosomal genome assembly of Diospyros oleifera Cheng. GigaScience 2020, 1–10. [Google Scholar] [CrossRef]
- Sun, P.W.; Gao, Z.H.; Lv, F.F.; Yu, C.C.; Jin, Y.; Xu, Y.H.; Wei, J.H. Genome-wide analysis of basic helix–loop–helix (bHLH) transcription factors in Aquilaria sinensis. Sci. Rep. 2022, 12, 7194. [Google Scholar] [CrossRef]
- Zhang, P.P.; Wang, H.; Qin, X.; Chen, K.; Zhao, J.R.; Zhao, Y.X.; Yue, B. Genome-wide identification, phylogeny and expression analysis of the PME and PMEI gene families in maize. Sci. Rep. 2019, 9, 19918. [Google Scholar] [CrossRef]
- Wang, J.; Ling, L.; Cai, H.; Guo, C. Gene-wide identification and expression analysis of the PMEI family genes in soybean (Glycine max). 3 Biotech 2020, 10, 335. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lyu, H.M.; Zhu, K.K.; Van-de, P.Y.; Cheng, Z.M.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Omodele, I.; Christiaan, E.J.B.; Graeme, B. In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 2010, 34, 268–283. [Google Scholar] [CrossRef]
- Xu, W.J.; Grain, D.; Bobet, S.; Le, G.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Zhu, Q.G.; Gong, Z.Y.; Wang, M.M.; Li, X.; Grierson, D.; Yin, X.R.; Chen, K.S. A transcription factor network responsive to high CO2/hypoxia is involved in deastringency in persimmon fruit. J. Exp. Bot. 2018, 69, 2061–2070. [Google Scholar] [CrossRef]
- Damian, S.; Gable, A.L.; David, L.; Alexander, J.; Stefan, W.; Jaime, H.C.; Milan, S.; Nadezhda, T.D.; John, H.M.; Peer, B.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.F.; Tang, Y.; Li, J.G.; Zhao, M.L. RNA-Seq provides new insights into the molecular events involved in “ball-skin versus bladder effect” on fruit cracking in litchi. Int. J. Mol. Sci. 2021, 22, 454. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Mattei, B.; Bellincampi, D. The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS ONE 2015, 10, e0133810. [Google Scholar] [CrossRef]
- Reca, I.B.; Lionetti, V.; Camardella, L.; D’Avino, R.; Giardina, T.; Cervone, F.; Bellincampi, D. A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol. Biol. 2012, 79, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhang, F.; Wu, X.; Li, H. Characterization of the tomato (Solanum lycopersicum) pectin methylesterases: Evolution, activity of isoforms and expression during fruit ripening. Front. Plant Sci. 2020, 11, 238. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).