Abstract
Basic helix–loop–helix (bHLH) proteins belong to one of the largest families involved in plant growth, development, signal transduction, and secondary metabolism. Although bHLH genes have been previously identified in persimmon (Diospyros kaki), systematic studies have not been reported. A total of 59 bHLH family members have been identified from the “Xiaoguotianshi” persimmon transcriptome. These proteins were clustered into 12 groups from I to XII based on their phylogenetic relationships with Arabidopsis thaliana. Combined with the phylogenetic analysis, in silico expression patterns of five developmental stages, the protein–protein interaction analysis between DkbHLH and DkMYB proteins showed that the bHLH_Cluster-15548.1 protein sequence was identified to be highly similar to the AtGL3 (AT5G41315.1) protein, which is associated with flavonoid and proanthocyanidin (PA) biosynthesis. This study presents the systematic analysis of bHLH genes from D. kaki and provides valuable information for further research on the involvement of bHLH protein in anthocyanin biosynthesis.
1. Introduction
Persimmon (Diospyros kaki) is an important fruit tree. The high soluble proanthocyanin content leads to the astringency taste of persimmon fruits, the key characteristic of persimmon. Transcription factors (TFs) involved in proanthocyanin biosynthesis have been identified previously; for example, the R2R3MYB, bHLH, and WD40 (MBW) complex regulates the structural genes of the flavonoid/phenylpropanoid pathway. Basic helix–loop–helix (bHLH) proteins belong to one of the largest families involved in plant growth, development, signal transduction, and secondary metabolism [1,2,3]. bHLH proteins contain two conserved domains: (1) the HLH domain consisting of two functional segments (the basic and helix–loop–helix (HLH) regions), which possesses approximately 50–60 amino acids, and (2) the bHLH-MYC_N domain consisting of approximately 15–17 amino acids. The bHLH gene family has been recently identified in different plants including Arabidopsis [4], potato [5], tomato [6], apple [7], and rice [8]. Furthermore, 162 Arabidopsis AtbHLH proteins phylogenetically cluster into 26 subfamilies [4].
The bHLH family members, particularly the bHLH III (d and e) subfamily genes, play important roles in regulating anthocyanin biosynthesis via the jasmonic acid signaling pathway. In addition, the III (f) subfamily is involved in anthocyanin synthesis [9,10]. The first anthocyanin accumulation-regulating bHLH genes R and B were identified and characterised in maize [11]. The GLABRA3 (AtbHLH1) gene in group III (f), encoding an R homolog, is essential for anthocyanin biosynthesis and trichome formation in A. thaliana [12]. Overexpression of the III (f) subfamily member VdbHLH037 in grapes increases the accumulation of anthocyanins [13].
Multiple studies have indicated that bHLH gene family members regulate anthocyanin and proanthocyanin biosynthesis via interaction with MYB members [14]. For example, bHLH members (i.e., R1 and B1) are involved in purple anthocyanin synthesis by interacting with R2R3-MYB members C1 and PL1 [15,16]. MYB10.1 and MYB10.3 interact with bHLH3 to activate anthocyanin production in peaches by regulating NtCHS, NtDFR, and NtUFGT [17]. VvMYC1 interacts with MYB family members to mediate the PA biosynthesis in grapes by inducing gene promoters in the flavonoid pathway [18]. The overexpression of Arabidopsis CPC and GL3 in tomatoes enhances anthocyanin accumulation [19].
This study identified 59 bHLH transcription factors from the D. kaki transcriptome data. Their phylogenetic relationships, motifs, and expression patterns at different developmental stages were analysed to identify the bHLH gene family members that may be associated with PA biosynthesis. This study provides a basic understanding of the bHLH gene association with natural astringency loss in Chinese pollination-constant non-astringent (C-PCNA) persimmon.
2. Materials and Methods
2.1. Plant Materials
Diospyros kaki Thunb. ‘Xiaoguotianshi’ cultivar plants were grown in Yuanyang County, Henan Province, China (34°55′18″~34°56′27″ N, 113°46′14″~113°47′35″ E). Five different stages (T1 = 70, T2 = 100, T3 = 120, T4 = 140, T5 = 160 days after flowering (DAF)) of persimmon fruit flesh were flash frozen in liquid nitrogen and stored at −80 °C for further analysis.
2.2. RNA Sequencing
‘Xiaoguotianshi’ transcriptome data were collected from a previously published study [20]. A total of 15 cDNA libraries (three independent biological replicates) of fruit samples (T1–T5) were constructed for RNA-Seq. Transcriptome sequencing was performed by the Beijing Novogene Bioinformatics Technology Company (Beijing, China).
2.3. Identification of bHLH Transcription Factors (TFs) in the D. kaki Transcriptome
All putative bHLH sequences were queried from the previously published D. kaki transcriptome using “bHLH”. Pfam rechecked and filtered the predicted sequences (https://pfam.xfam.org/ (accessed on 12 June 2022)) [21] and the NCBI-CDD online software. The molecular weight (MW), isoelectric point (pI), and instability index of the proteins were calculated using the ExPASy server (https://www.expasy.org/ (accessed on 14 June 2022)) [22].
2.4. Multiple Sequence Alignment, Conserved Motif Identification, and Phylogenetic Analysis
To investigate the phylogenetic relationship of the bHLH gene families between D. kaki and A. thaliana, AtbHLH proteins were downloaded from the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/index.php (accessed on 18 June 2022)), with redundant sequences removed [23,24,25]. bHLH proteins were aligned using the ClustalX 2 program [26]. A phylogenetic tree was constructed for these proteins using the neighbour-joining (NJ) method with 1,000 bootstrap reiterations using MEGA5.0 software [27]. DkbHLH proteins were classified according to the distance between the homologous sequences of A. thaliana [28]. MEME v5.0.0 online software (https://meme-suite.org/tools/meme (accessed on 22 June 2022)) was used to identify motifs with default parameters, with the maximum number of motifs set to nine [29].
2.5. Gene Ontology Annotation
Gene Ontology (GO) enrichment analysis of the identified DkbHLH proteins was analysed and visualised using agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php (accessed on 2 July 2022)). The results of the GO analysis were grouped into three categories: biological process, cellular component, and molecular function.
2.6. Protein Interaction Network Analysis
All putative bHLH sequences were queried from the previously published D. kaki transcriptome using “MYB”. The interaction network between DkbHLH and DkMYB proteins using A. thaliana on the String protein interaction database (http://string-db.org/ (accessed on 14 July 2022)) [30].
2.7. Calculating Ka and Ks of the Homologous bHLH Gene Pairs
Non-synonymous substitutions per non-synonymous site (Ka) and synonymous substitutions per synonymous site (Ks) were used to assess the selection pressure and divergence time of the bHLH gene family [31]. The number of Ks and Ka of the orthologous bHLH gene pairs between A. thaliana and D. kaki and the paralogous DkbHLH gene pairs were calculated using the TBtools v1.0971 software [32]. The divergence time (T) of the duplication events was calculated using the formula T = Ks/(2 × 6.1 × 10−9) × 10−6 million years ago (MYA) [33].
2.8. In Silico Analysis of bHLH Genes in Different Tissues
To analyse the expression levels of DkbHLH genes during fruit development, the expression profiles of these genes at five different fruit developmental stages were represented using the Fragments Per Kilobase per Million (FPKM) value of the high-throughput sequencing data. The heatmap was generated using the log10-transformed FPKM values. The expression heatmaps and k-means clustering of DkbHLH genes were performed and generated using HEATMAP tools in Hiplot (https://hiplot.com.cn (accessed on 28 July 2022)) [34].
3. Results
3.1. Identification and Phylogenetic Analysis of bHLH Genes in A. thaliana and D. kaki
Following the removal of the redundant sequences without complete conserved domains, a total of 59 DkbHLH proteins were identified from the D. kaki transcriptome. The molecular weights of these DkbHLH proteins ranged from 8641.81 (Cluster-6987.43499) to 79,061.33 Da (Cluster-6987.28853), and the theoretical isoelectric points (pI) ranged from 4.52 (Cluster-6987.10953) to 10.79 (Cluster-17857.0). The length of these proteins varied from 76 (Cluster-6987.43499) to 720 amino acids (Cluster-6987.28853) with an average length of 334 aa (Table 1). The instability index ranged from 34.57 (Cluster-6987.29876) to 93.67 (Cluster-6987.40617), and only two DkbHLH proteins (Cluster-6987.29876 and Cluster-6987.44311) were stable in vitro. The DkbHLH protein GRAVY values ranged from −0.016 (Cluster-6987.29876) to −0.841 (Cluster-6987.19844). All of the DkbHLH proteins with a lower GRAVY index (GRAVY < 0) were considered to be more water-soluble (Table 1).

Table 1.
A list of all bHLH genes identified in the D. kaki from the transcriptome unigenes.
3.2. Phylogenetic Analysis of the bHLH Proteins
To investigate the evolutionary relationships of the bHLH proteins, the NJ phylogenetic tree was constructed between D. kaki and A. thaliana (Figure 1). The DkbHLH proteins were classified under the previous classification of AtbHLH [28]. The DkbHLH proteins were divided into 12 groups, from I to XII. Group III contained 12, the largest number of DkbHLH proteins, while groups II, VI, and X did not have any DkbHLH proteins. Combining the phylogenetic analysis with conserved domain analysis, DkbHLH proteins in groups III (d, e, and f) contained the bHLH-MYC and R2R3-MYB transcription factor N-terminal domains.
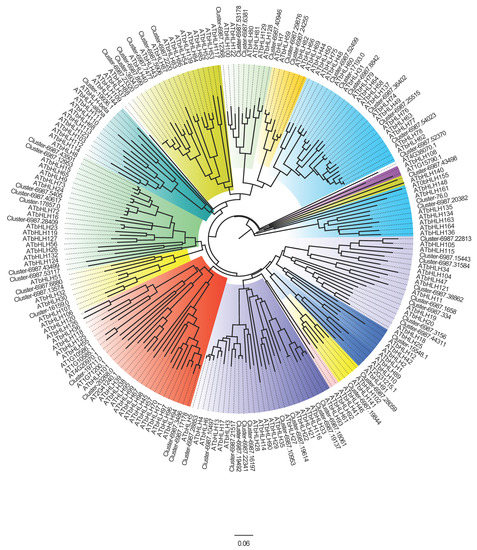
Figure 1.
Phylogenetic analysis of the D. kaki bHLH proteins compared to A. thaliana. Different colours represent different family groups, and the detail groups are shown in Table 1.
3.3. Conserved Motif Analysis
The predicted DkbHLH amino acid sequence conserved motif characteristics were analysed using the MEME tool (Figure 2). A total of nine motifs were predicted in the 59 bHLH members and labelled as Motifs 1–9. The lengths of the motifs ranged from 9 to 35 amino acids. Motifs 1 and 2 were present in nearly all DkbHLH proteins. Proteins in Groups VII and Ⅸ only contained these two motifs. Furthermore, Motifs 6 and 7 were identified in Groups I, III (d and e), and V; Motifs 8 and 9 were only present in Group XII; Motif 4 was identified in Group VIII; and Motif 5 was only observed in Group III (f). Most of the DkbHLH proteins in the same group possessed the same conserved motifs as in the phylogenetic analysis.
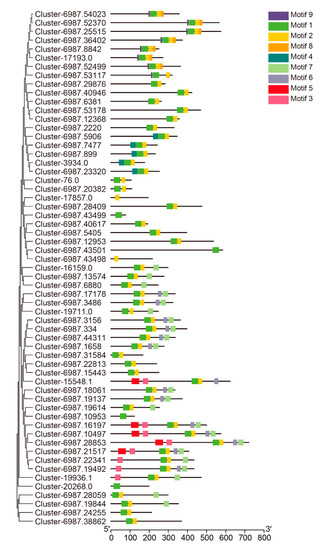
Figure 2.
Phylogenetic tree (left) and the conserved motifs (right) of the D. kaki bHLH proteins. Different colours represent different motifs.
3.4. Gene Ontology Annotation
The functional associations were identified using GO term enrichment analysis among the DkbHLH proteins with agriGO (Figure 3). Sixty annotations were assigned to GO terms and summarised in three primary functional categories, including the cellular component, molecular function, and biological process. Three groups, including binding (GO:0005488), protein binding (GO:0005515), and protein dimerisation activity (GO:0046983) were the significant classifications for all of the DkbHLH genes.
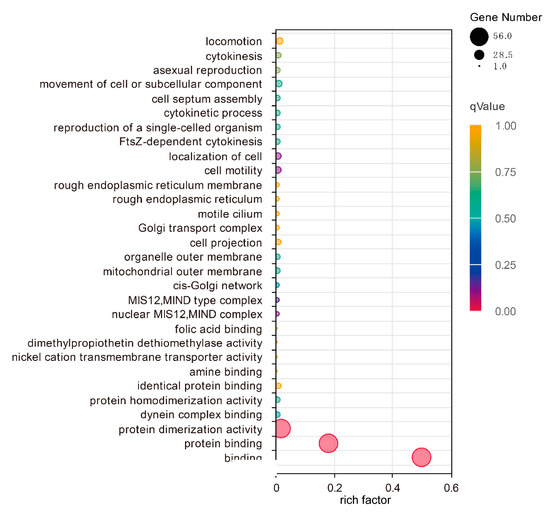
Figure 3.
GO annotation of 59 bHLH transcription factors. The dot size represents gene number, and different colours represent the values of the qValue.
3.5. Selection Pressure and Differentiation Time of the Homologous bHLH Genes
To further analyse the selection pressure and differentiation time of paralogous DkbHLH gene pairs and orthologous bHLH gene pairs between D. kaki and A. thaliana, the Ka:Ks ratio was calculated (Table 2). Surprisingly, the Ka:Ks ratio of all paralogous and orthologous gene pairs was less than one, demonstrating that these bHLH genes are under purifying selection. The differentiation time of paralogous bHLH gene pairs in D. kaki was between 77 and 136 MYA, and the differentiation time of orthologous bHLH genes between D. kaki and A. thaliana was between 137 and 266 MYA.

Table 2.
Ka, Ks, and Ka:Ks values between homologous bHLH gene pairs.
3.6. bHLH and MYB Protein–Protein Interaction Network Analysis
The protein–protein interactions between DkbHLH and DkMYB proteins were analysed using the STRING database (Figure 4). The D. kaki protein sequence of bHLH_Cluster-15548.1 showed similarity to the AtGL3 (AT5G41315.1) protein with a 538.5-bit score and 48.7 identities. bHLH_Cluster-15548.1 interacted with other bHLH and MYB proteins with combined scores from 0.411 to 0.99. The predicted functional partners of Cluster-15548.1, including MYB_Cluster-6987.24679, MYB_Cluster-6987.15450, MYB_Cluster-6987.38210, bHLH_Cluster-6987.21517, MYB_Cluster-6987.43699, MYB_Cluster-6987.3798, MYB_Cluster-6987.38499, and MYB_Cluster-6987.4705, were associated with a flavonoid biosynthetic process and RNA polymerase II transcription regulator recruiting activity. bHLH_Cluster-6987.21517 is an AtbHLH3 (AT4G16430) homolog with a 339-bit score and 46.1 identities and could interact with other bHLH and MYB proteins with combined scores from 0.525 to 0.567. The predicted functional partners of bHLH_Cluster-6987.21517, including MYB_Cluster-6987.24679, MYB_Cluster-6987.15450, bHLH_Cluster-6987.21517, and MYB_Cluster-6987.38499, were associated with flavonoid and proanthocyanidin biosynthetic processes.
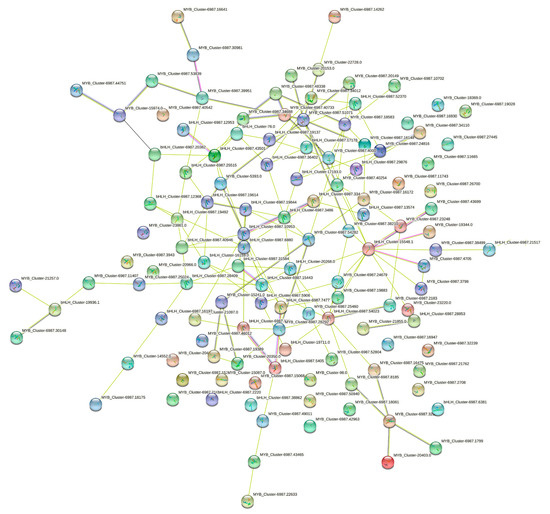
Figure 4.
The protein–protein interaction network between bHLH and MYB proteins. Different coluors have no additional meaning.
3.7. In Silico Analysis of bHLH Genes in D. kaki Fruit at Five Different Stages
The expression patterns of DkbHLH genes in the fruit at five different developmental stages were analysed using RNA-seq. Distinct bHLH gene clusters exhibited time-specific expression profiles in persimmon fruit development (Figure 5a,b). Genes in Cluster 3 were highly expressed in T1 and showed a downregulated trend; genes in Cluster 2 reached the highest expression in T2; genes in Cluster 1, whose expression peaked at T5, were differentially upregulated between T4 and T5; genes in Clusters 4, 5, and 6 peaked at T4, T3, and T1, respectively.
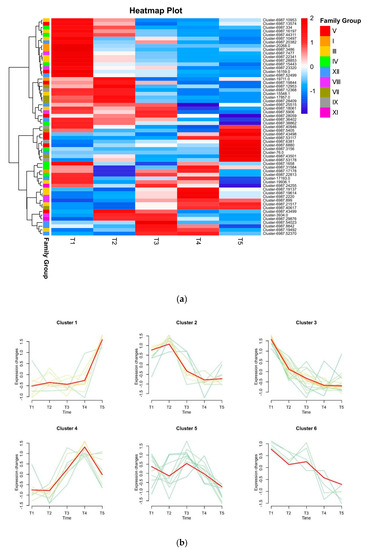
Figure 5.
Gene expression analysis of bHLH genes in D. kaki fruit at five stages. (a) Heat map clustering of DkbHLH gene expression levels at five developmental stages; heatmaps depict the normalised gene expression values, which represent the mean value of three biological replicates. (b) Clustering analysis of DkbHLH genes.
4. Discussion
Persimmon is an important fruit crop in east Asia. The high PA content leads to fruit astringency, the critical characteristic of persimmon. The natural de-astringency of C-PCNA persimmon is associated with both the “coagulation effect” and “dilution effect”. Previous studies have shown that bHLH and MYB may associate with PA biosynthesis in Lotus species [35], Freesia hybrida [36], and Anthurium andraeanum [37]. However, only a few MYB and bHLH genes, such as DkMYB4 and DkMYB14, are the key transcription factors for the natural astringency of Japanese PCNA (J-PCNA) and C-PCNA persimmon, respectively [38,39]. Su et al. [40] isolated a novel bHLH gene DkMYC1 from persimmon that may be associated with PA biosynthesis via the regulation of structural genes such as DkDHD, DkF3′5′H, and DkANR. In addition, DkbHLH1 was strongly induced by an artificially high CO2 atmosphere in three astringent-type persimmons [41]. The mechanisms of the bHLH TFs’ involvement in PA biosynthesis are currently unknown. In this study, 59 bHLH TFs were identified, and their phylogenetic relationships with A. thaliana were analysed. The expression patterns of five developmental stages of the C-PCNA persimmon were analysed, providing a theoretical basis for further study of the natural de-astringency of PCNA persimmon.
Phylogenetic, motif, and conserved domain analyses of proteins were used to infer biological functions. Phylogenetic analysis demonstrated that five DkbHLH proteins were clustered into groups III (d, e, and f) containing bHLH-MYC and R2R3-MYB TF N-terminal domains. bHLH members of III (f) play essential roles in anthocyanin synthesis [9,10]. Genes clustered in the same group commonly exhibited similar structures and functions. The AtbHLH001 (GL3), AtbHLH002 (EGL3), and AtbHLH042 (TT8) genes are the critical regulators of anthocyanin and PA biosynthesis [41,42,43]. Only one bHLH member (Cluster-15548.1) clustered into Group III (f) contained the bHLH-MYC and R2R3-MYB TF N-terminal domains, possibly involved in PA biosynthesis. Motif 5 was only detected in group III (f) and may be associated with its specific function in PA biosynthesis.
Furthermore, the protein–protein interaction analysis between DkbHLH and DkMYB demonstrated that the D. kaki bHLH_Cluster-15548.1 protein sequence exhibited high similarity to the AtGL3 (AT5G41315.1) protein, which is associated with flavonoid and PA biosynthesis. Previous studies have shown that 120 (T3) to 140 (T4) DAF may be the critical phase for the “dilution effect”, and 140 (T4) to 160 (T5) DAF may be the crucial phase for the “coagulation effect” [20]. Therefore, the expression patterns of the bHLH gene family members were analysed in the D. kaki ‘Xiaoguotianshi’ cultivar at five developmental stages. Surprisingly, Cluster-15548.1 was differentially downregulated at T4 compared to T3, which may be associated with the “dilution effect”. However, the function of Cluster-15548.1 still requires further investigation.
5. Conclusions
A total of 59 bHLH family members have been identified from the “Xiaoguotianshi” persimmon transcriptome data. These proteins were clustered into 12 groups from I to XII based on their phylogenetic relationships with Arabidopsis thaliana. Combined with the phylogenetic analysis, in silico expression patterns of five developmental stages, and the protein–protein interaction analysis between DkbHLH and DkMYB proteins, the bHLH_Cluster-15548.1 protein sequence was identified to be highly similar to the AtGL3 (AT5G41315.1) protein, which is associated with flavonoid and PA biosynthesis. This study presented the systematic analysis of bHLH genes from D. kaki and provided valuable information for further research on the involvement of bHLH protein in PA and anthocyanin biosynthesis.
Author Contributions
L.H. and J.F. designed the experiments. Q.Z., Y.S., H.L., S.D. and P.S. offered meaningful feedback and assisted form the research. W.H. conducted the analysis and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32071801) and the National Key R&D Program of China (2019YFD1000600).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Castillon, A.; Shen, H.; Huq, E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Duek, P.D.; Fankhauser, C. HFR1, a putative bHLH transcription factor, mediates both phytochrome a and cryptochrome signaling. Plant J. 2003, 34, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Kondou, Y.; Nakazawa, M.; Kawashima, M.; Ichikawa, T.; Yoshizumi, T.; Suzuki, K.; Ishikawa, A.; Koshi, T.; Matsui, R.; Muto, S. RETARDED GROWTH OF EMBRYO1, a new basic helix-loop helix protein, in endosperm to control EMBRYO growth. Plant Physiol. 2008, 147, 1924–1935. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Peng, Z.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H. Genome-wide identification and characterization of the Potato bHLH transcription factor family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 2017, 8, 480. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.; Guo, H.; Xie, D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef]
- Liu, X.; An, X.; Liu, X.; Hu, D.; Wang, X.; You, C.; Hao, Y. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017, 68, 2977–2990. [Google Scholar] [CrossRef]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [PubMed]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Li, M.; Sun, L.; Gu, H.; Cheng, D.; Guo, X.; Chen, R.C.; Wu, Z.; Jiang, J.; Fan, X.; Chen, J. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 2021, 11, 6863. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed]
- Neuffer, M.G.; Coe, E.H.; Wessler, S.R. Mutants of Maize, 1st ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 54–59. [Google Scholar]
- Petroni, K.; Cominelli, E.; Consonni, G.; Gusmaroli, G.; Gavazzi, G.; Tonelli, C. The developmental expression of the maize regulatory gene Hopi determines germination-dependent anthocyanin accumulation. Genetics 2000, 155, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Rahim, M.A.; Busatto, N.; Trainotti, L. Regulation of anthocyanin biosynthesis in peach fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Leon, C.; Czemmel, S.; Delrot, S.; Virginie, L.; Bogs, J. The basic helix-loop-helix transcription factor MYC1 is involved in the gegulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Wada, T.; Kunihiro, A.; Tominaga-Wada, R.; Takaya, M. Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycopersicum) anthocyanin biosynthesis. PLoS ONE 2014, 9, e109093. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.; Pu, T.; Suo, Y.; Han, W.; Diao, S.; Li, H.; Sun, P.; Fu, J. Transcriptomic profiling analysis to identify genes associated with PA biosynthesis and insolubilization in the late stage of fruit development in C-PCNA persimmon. Sci. Rep. 2022, 12, 19140. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J. The Pfam protein families database. Nucleic Acids Res. 2004, 28, 263–266. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.C.; Gao, G. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 21, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Li, W.H.; Gojobori, T.; Nei, M. Pseudogenes as a paradigm of neutral evolution. Nature 1981, 292, 237–239. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yu, J.; Ma, X.; Sun, S.; Yang, N.; Xia, R.; Lei, T. Hierarchically aligning 10 legume genomes establishes a family-level genomics platform. Plant Physiol. 2017, 174, 1981–2016. [Google Scholar] [CrossRef]
- Li, J.F.; Miao, B.B.; Wang, S.X.; Dong, W.X.; Hou, S.; Si, C.C.; Wang, M.J. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef] [PubMed]
- Escaray, F.J.; Passeri, V.; Perea-García, A.; Antonelli, C.J.; Damiani, F.; Ruiz, O.A.; Paolocci, F. The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of Lotus species. Planta 2017, 246, 243–261. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Zhou, L.; Gao, R.; Yang, S.; Wang, S.; Wang, L.; Gao, X. The R2R3-MYB factor FhMYB5 from Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis. Front. Plant Sci. 2019, 9, 1935. [Google Scholar] [CrossRef]
- Li, C.; Qiu, J.; Huang, S.; Yin, J.; Yang, G. AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)—Another strategy to modulate pigmentation. Hortic. Res. 2019, 6, 14. [Google Scholar] [CrossRef]
- Akagi, T.; Ikegami, A.; Tsujimoto, T.; Kobayashi, S.; Sato, A.; Kono, A.; Yonemori, K. DkMyb4 is a myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009, 151, 2028–2045. [Google Scholar] [CrossRef]
- Chen, W.; Xiong, Y.; Xu, L.; Zhang, Q.; Luo, Z. An integrated analysis based on transcriptome and proteome reveals deastringency-related genes in CPCNA persimmon. Sci. Rep. 2017, 7, 44671. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Jia, H.; Zhang, Q.; Luo, Z. Isolation and characterization of a basic Helix-Loop-Helix transcription factor gene potentially involved in proanthocyanidin biosynthesis regulation in persimmon (Diospyros kaki Thunb.). Sci. Hortic. 2012, 136, 115–121. [Google Scholar] [CrossRef]
- Feyissa, D.N.; Lovdal, T.; Olsen, K.M.; Slimestad, R.; Lillo, C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation asinduced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 2009, 230, 747–754. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Le Gourrierec, J.; Harscoët, E.; Berger, A.; Jauvion, V.; Dubos, C. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 2013, 198, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).